Abstract
The circadian clock is an autonomous oscillator that produces endogenous biological rhythms with a period of about 24 h. This clock allows organisms to coordinate their metabolism and development with predicted daily and seasonal changes of the environment. In plants, circadian rhythms contribute to both evolutionary fitness and agricultural productivity. Nevertheless, we show that commercial barley varieties bred for short growing seasons by use of early maturity 8 (eam8) mutations, also termed mat-a, are severely compromised in clock gene expression and clock outputs. We identified EAM8 as a barley ortholog of the Arabidopsis thaliana circadian clock regulator EARLY FLOWERING3 (ELF3) and demonstrate that eam8 accelerates the transition from vegetative to reproductive growth and inflorescence development. We propose that eam8 was selected as barley cultivation moved to high-latitude short-season environments in Europe because it allowed rapid flowering in genetic backgrounds that contained a previously selected late-flowering mutation of the photoperiod response gene Ppd-H1. We show that eam8 mutants have increased expression of the floral activator HvFT1, which is independent of allelic variation at Ppd-H1. The selection of independent eam8 mutations shows that this strategy facilitates short growth-season adaptation and expansion of the geographic range of barley, despite the pronounced clock defect.
The timing of flowering during the year is an important adaptive trait that strongly influences reproductive fitness. Many plants use the environmental cue of day length (photoperiod) to regulate flowering and this response can vary within species along a latitudinal cline (1–3). Photoperiod response has been extensively studied in Arabidopsis, where daily light oscillations entrain the circadian clock (4), which is the internal timepiece by which photoperiod is measured. In Arabidopsis, the circadian clock is composed of several feedback loops that interlock to provide robustness (5, 6). The related Myb transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) have expression peaks in the morning and act antagonistically to the pseudoresponse regulator (PRR) TIMING OF CAB EXPRESSION1 (TOC1), which peaks in the evening. CCA1 and LHY promote the expression of PRR7 and PRR9, which themselves repress CCA1 and LHY, forming a morning feedback loop (5–7). This loop also involves EARLY FLOWERING3, EARLY FLOWERING4, and LUX ARHYTHMO (ELF3, ELF4, LUX), which repress PRR7 and PRR9 (8–10). An additional evening feedback loop involves GIGANTEA (GI), which promotes TOC1, which in turn represses GI (5). ELF3 also affects the evening loop by regulating GI protein turnover (11). Clock circuitry thus functions continuously during the day and this provides capacity for timed physiological outputs in response to the time of day.
The clock is important for providing diurnal and seasonal control of flowering. In Arabidopsis, transcriptional outputs from the clock regulate CONSTANS (CO) expression so that it peaks late in the day (12). When CO protein is produced in the light period, which only occurs in long days, it activates the floral inductive target FLOWERING LOCUS T (FT) (13, 14). Circadian clock and core photoperiod genes are well conserved in barley (15), suggesting that they retain conserved functions and that information from Arabidopsis will be useful for identifying and interpreting barley mutations affecting the circadian clock and photoperiod response.
Wild-type barley is early flowering under long days (LDs) and later flowering under short days (SDs). The main locus affecting LD photoperiod response is Ppd-H1 (16), a PRR gene, the closest ortholog of which in Arabidopsis is the morning clock gene PRR7 (15, 16). The ppd-H1 mutation has no effect under SDs, but delays flowering under LDs, and this is advantageous in long growth season environments (3, 16). Delayed flowering is associated with reduced expression of HvFT1, the barley counterpart of Arabidopsis FT (16, 17). In contrast, the barley early maturity8 (eam8) mutation, also termed mat-a, causes an early-flowering day-neutral phenotype with rapid flowering under either SDs or LDs (18, 19). The eam8 mutations are recessive, indicating loss of gene function. Spontaneous and induced eam8 mutations have been used commercially in varieties bred for environments with short growth seasons, such as those of Scandinavia (20).
To understand how eam8 affects flowering and adaptation, we used transcript-based cloning to identify the EAM8 gene, which we showed to be the ortholog of Arabidopsis ELF3. We then showed that eam8 mutations affect the expression of genes that are components of the circadian clock, severely attenuating clock function, and affect the downstream photoperiod pathway. We analyzed flowering time and expression of clock and clock-output genes in two contrasting genetic backgrounds: the spring barley Bowman(ppd-H1) and the winter barley Igri(Ppd-H1). We show that eam8 produces an early flowering phenotype in a background with a wild-type or mutated Ppd-H1 allele, and in this context we discuss reasons why eam8 mutations provided an effective adaptive solution despite their effects on the circadian clock.
Results
eam8 Mutants Are Early Flowering in SD or LD Conditions.
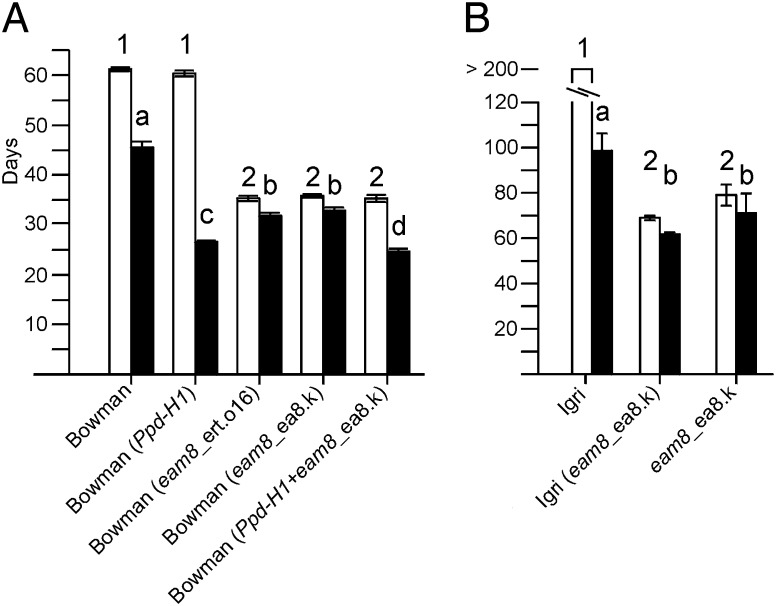
We recorded days to flowering of Bowman (ppd-H1) and derived introgression lines carrying Ppd-H1, eam8_ea8.k (naturally occurring allele), eam8_ert.016 (induced allele), or Ppd-H1+eam8_ea8.k (Fig. 1A). Under SDs (10-h light), Ppd-H1 had no effect, but the eam8 and Ppd-H1+eam8 lines were equivalent and flowered 25 d earlier than Bowman. Under LDs (18-h light), Ppd-H1 plants were ∼20 d earlier flowering than the Bowman (ppd-H1) parent, consistent with previous findings (16). The eam8 lines flowered earlier than Bowman, but were significantly later than lines carrying a functional Ppd-H1 allele. Ppd-H1 and Ppd-H1+eam8 lines flowered at the same time. A separate controlled-environment experiment compared vernalized plants of Igri (Ppd-H1), eam8_ea8.k and Igri(eam8_ea8.k) under SD and LD conditions (Fig. 1B). The Igri (eam8) line accelerated flowering compared with Igri and behaved similarly to Bowman(eam8). Under SDs, therefore, the early-flowering phenotype of eam8 mutants was not dependent on, or enhanced by, a functional Ppd-H1 allele. Under LD, variation at Ppd-H1 had a small effect on flowering time of eam8 plants in the Bowman background.
Fig. 1.
Flowering time of wild-type and eam8 mutant plants. (A) Bowman (ppd-H1) and introgression lines carrying Ppd-H1, eam8 (two alleles), or Ppd-H1+eam8 grown under SD (10-h light, white bars) or LD (18-h light, black bars). (B) Igri(Ppd-H1), Igri(eam8), and the eam8 parent grown under SDs (9-h light, white bars) or LDs (16-h light, black bars). Flowering time is shown as mean days to awn emergence on the main stem. Error bars (standard deviation) are for 10 (A) or 8 (B) plants of each genotype. Genotypes with the same number (SDs) or letter (LDs) were not significantly different by paired t tests.
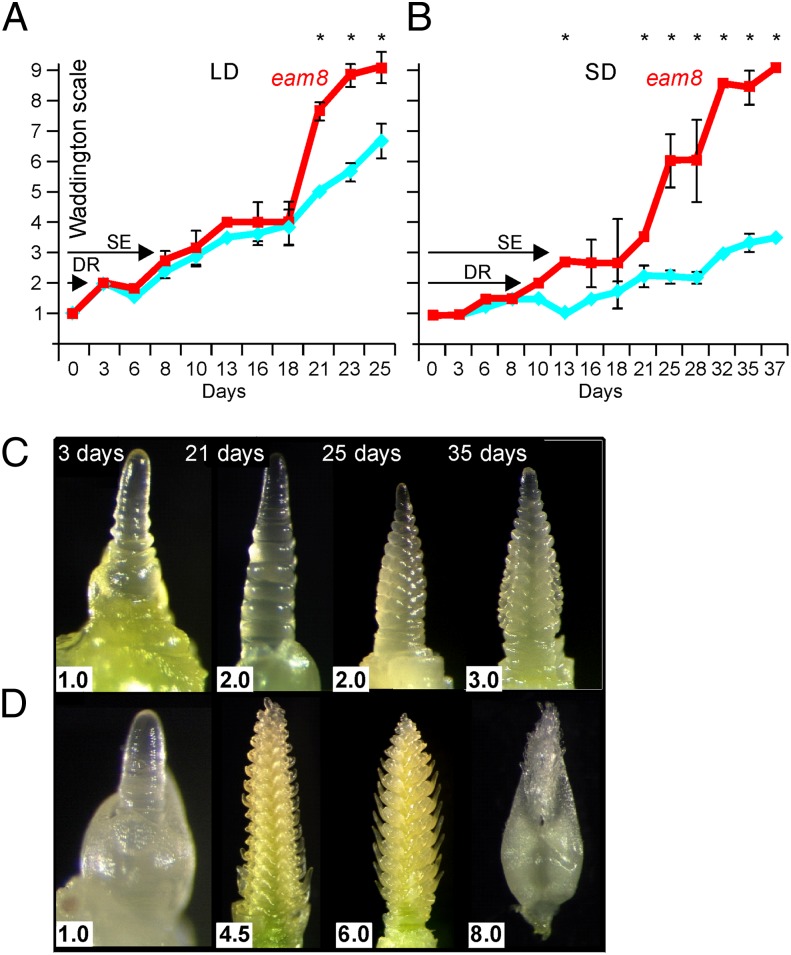
We then assessed the effect of eam8 on meristem development. Under LDs, Bowman and Bowman(eam8) apices reached the double-ridge stage (the first visible sign of floral identity) and started stem elongation at the same times. However, inflorescence development was significantly faster in Bowman(eam8) (Fig. 2A), which flowered after 23 d when Bowman was still in the stem elongation phase. Under SDs, Bowman(eam8) reached the double-ridge and stem elongation stages earlier than Bowman (Fig. 2 B–D). Thus, early flowering in Bowman(eam8) was caused by faster inflorescence development under LDs, but under SDs the transition to a reproductive meristem and inflorescence development were accelerated.
Fig. 2.
Meristem development in wild-type and eam8 mutant plants. Apex development assessed by the Waddington scale (WS) (21) for plants of Bowman (blue diamonds and lines) and Bowman(eam8.w) (red squares and lines). Mean values and standard deviations are shown for plants (A) under LD (16-h light) or (B) under SDs (8-h light). Between three and four plants per genotype and treatment were sampled at each time point. Timings of double-ridge formation (DR; WS2) and the initiation of stem elongation (SE) are arrowed. Significant differences in WS score by ANOVA are shown (*P < 0.05). Example meristems from four time points are shown for Bowman (C) and Bowman(eam8.w) (D) grown under SD conditions. Figures in white boxes are WS scores.
EAM8 Encodes a Barley Ortholog of the Arabidopsis ELF3 Gene.
We isolated the EAM8 gene by transcript-based cloning. Barley1 Affymetrix microarrays compared gene expression at the end of the light period in vernalized plants of Igri, Igri(eam8), and Igri(eam7) (phenotypically similar to eam8 but genetically distinct) (Fig. S1 and Table S1) after 10 d growth at 16 °C under SDs or LDs. Misexpressed genes in eam7 and eam8 suggested a defect in circadian-clock function and an analysis of transcripts specifically missing from Igri(eam8) identified one as a homolog of the Arabidopsis circadian-clock gene ELF3 (Table S1). The gene was fully sequenced from a Morex BAC clone and then amplified in segments by PCR and sequenced from Igri, Bowman, and Golden Promise (all with a genetically wild-type allele at EAM8). The predicted barley protein was aligned with Arabidopsis ELF3 and other previously identified ELF3-like proteins using the ClustalW method, and the alignment was used to construct a phylogenetic tree that placed the barley protein with wheat and Brachypodium sequences previously identified as ELF3 homologs (8, 15) (Fig. S2).
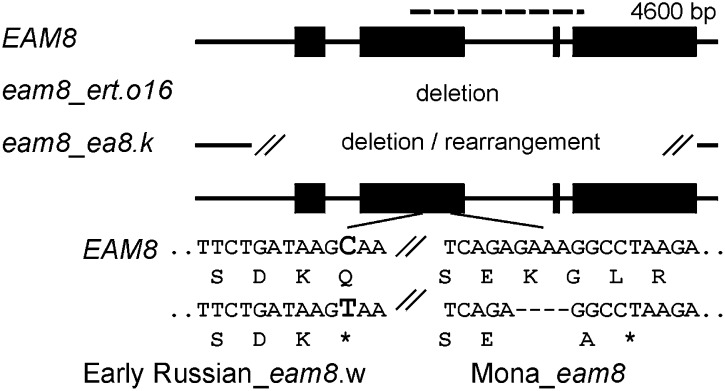
The ELF3-like gene was then sequenced from four independent eam8 mutant alleles. The ert-016 (X-ray induced) allele had a complete deletion of the gene and the ea8.k allele had a partial deletion. The variety Mona had a 4-bp deletion in exon 2 and Early Russian (eam8.w) had a C-to-T point mutation in exon 2, both of which would encode a truncated protein (Fig. 3). Polymorphism identified by sequencing allowed the barley ELF3-like gene to be mapped in the Igri × Igri(eam8) mapping population, where it cosegregated with the eam8 mutation (Fig. S1). We concluded from these data that EAM8 was the ELF3 ortholog and that loss-of-function mutations produced the eam8 phenotype.
Fig. 3.
Structure of the barley EAM8 gene in wild-type (Bowman or Igri) and four eam8 mutant lines. Exons are shown as black rectangles and introns, 5′ and 3′ flanking sequences as lines. Dotted line above EAM8 shows a region used for haplotype analysis. The eam8_ert.016 allele lacks the entire sequence. Early Russian and Mona have a point mutation (C to T) or a 4-bp deletion, respectively, in exon 2 that result in premature stop codons.
EAM8 Is Required for Normal Circadian Function.
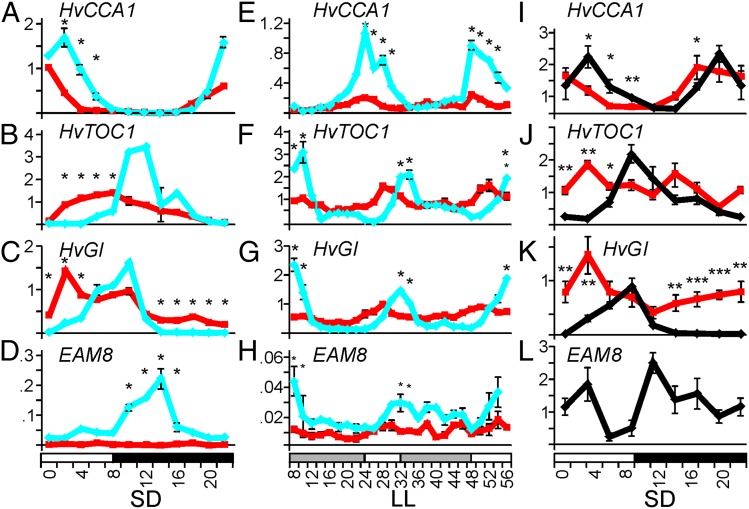
Because Arabidopsis elf3 is early flowering and defective in circadian clock function (8), we tested clock-gene expression. Bowman and Bowman(eam8.w) were grown for 18 d under SDs (8-h light/16-h dark), at which point the shoot apical meristem was vegetative in Bowman and had transitioned to a floral meristem in Bowman(eam8.w). Plants were sampled at 2-h intervals over 24 h in a light-dark cycle and for 48 h under constant light (LL) conditions. We measured expression of barley orthologs of Arabidopsis core clock genes (HvCCA1 and HvTOC1) and the evening loop gene HvGI and EAM8 (HvELF3) itself. Expression in Bowman oscillated under SDs and the timing of expression peaks was consistent with studies in rice (22) (Fig. 4 A–D). Expression peaks were maintained under LL conditions, which showed that these genes have circadian regulation (Fig. 4 E–H). In Bowman(eam8.w), EAM8 exhibited lower expression than in Bowman (Fig. 4D). HvCCA1, HvTOC1, and HvGI showed a strongly dampened amplitude, especially under LL, and the evening-expressed genes HvTOC1 and HvGI peaked earlier (6 h in SDs and 4 h under LL) (Table S2). Expression patterns of HvCCA1, HvTOC1, HvGI, and EAM8 in Igri and Igri(eam8) were essentially identical to those in the Bowman background (Fig. 4 I–L), showing that clock effects were equivalent in Ppd-H1 and ppd-H1 backgrounds. Igri and Igri(eam8) plants were harvested 10 d after sowing and Bowman plants were harvested 18 d after sowing. Microscopic dissection and analysis of apex development showed that in both cases the wild-type was vegetative and the mutant was at the transition to flowering. Hence, the developmental stages were comparable.
Fig. 4.
Expression patterns of core circadian clock genes (HvCCA1 and HvTOC1), the evening-loop gene HvGI and EAM8 (HvELF3) in wild-type, and eam8 mutant plants. (A–H) Expression patterns for Bowman (blue diamonds and lines) and Bowman(eam8.w) plants (red squares and lines). (A–D) Plants were grown under SDs (8-h light, white bar) for 14 d and sampled at 2-h intervals over 24 h. (E–H) Plants sampled every 2 h during a further 48 h after transfer from SD to LL at the end of the light period. Gray bars show subjective nights. Error bars are standard deviation of two technical replicates derived from a pool of three plants. Significant differences (P < 0.05) were calculated by ANOVA using expression data of three consecutive time points in a sliding-window approach across 24 h. (I–L) Expression patterns for Igri (black diamonds and lines) and Igri(eam8_ea8.k allele) plants (red squares and lines). Plants were grown under SDs (9-h light) for 10 d and sampled at 3-h intervals over 24 h. EAM8 was undetectable in the Igri(eam8_ea8.k) line as the gene is deleted in this mutant (Fig. 3). Error bars are standard deviations of three biological replicates. Significant expression differences by ANOVA are shown: *P < 0.05, **P < 0.01, ***P < 0.001.
Additional clock and clock output genes, including homologs of PRR59, FKF1, and chlorophyll A-B binding protein genes, displayed altered expression in the eam8 mutant compared with wild type in the analysis of the Affymetrix arrays (Table S1). Misexpression over a 24-h time course was confirmed for a chlorophyll a/b binding protein gene by quantitative RT-PCR (qRT-PCR) (Fig. 5). As the wild-type and eam8 mutant plants reached specific developmental stages at different rates (Fig. 2), the plants were at different developmental stages when sampled for gene expression (Figs. 4 and 5). To assess whether developmental state affected gene expression, we measured the diurnal expression of circadian clock orthologs in Bowman under SDs before (at 8 d) and after (at 21 d) the transition to a reproductive meristem. The expression profiles showed no qualitative differences (Fig. S3). We therefore concluded that differences in gene expression and phenotype were because of the eam8 mutation and that eam8, like Arabidopsis elf3 (8), consistently displayed significant disruption in the expression of core clock genes and clock output genes.
Fig. 5.
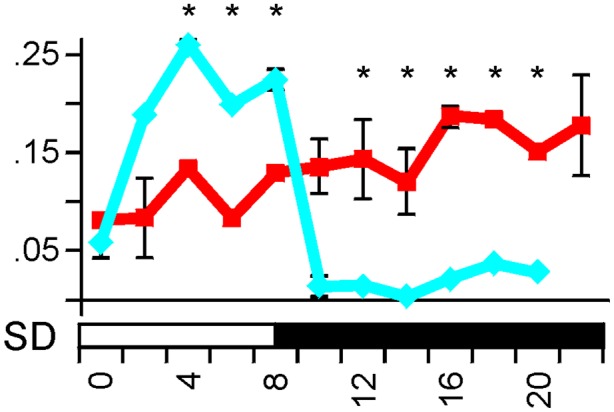
Expression patterns of a chlorophyll a/b binding protein gene in wild-type and eam8 mutant plants. The expression pattern for Bowman (blue diamonds and line) and Bowman(eam8.w) plants (red squares and line) are shown. Plants were grown under SDs (8-h light, white bar) for 14 d and sampled at 2-h intervals over 24-h. Error bars are standard deviations of two technical replicates derived from a pool of three plants. Significant differences (*P < 0.05) were calculated by ANOVA using expression data of three consecutive time points in a sliding-window approach across 24 h.
eam8 Affects the Expression of Floral Integrator Genes.
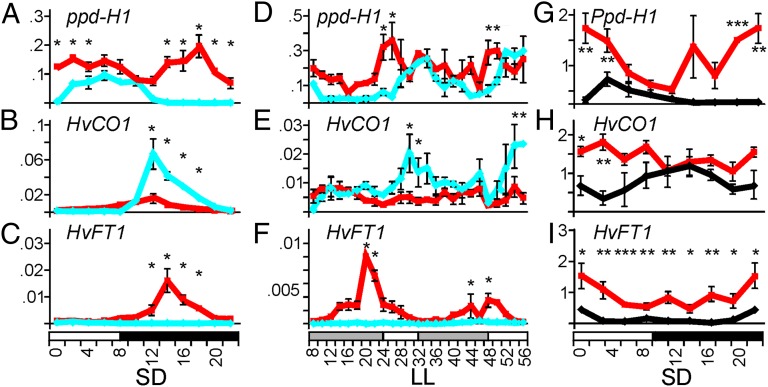
To understand how eam8 affected flowering time, we measured expression of photoperiod pathway genes Ppd-H1, HvCO1, and HvFT1 under SD and LL conditions. SD is the condition where the difference in flowering behavior between mutant and wild-type plants was greatest. The mutant ppd-H1 allele had a morning peak of expression in Bowman under SD and LL, consistent with previous studies (16, 23). Expression was higher in Bowman(eam8) at all time points, and particularly during the dark or subjective dark during LL when expression was lowest in Bowman. eam8 therefore de-repressed transcript accumulation of ppd-H1. Increased expression of Ppd-H1 (Fig. 6 A, D, and G) might be the result of a conserved ELF3 function because its primary targets in Arabidopsis are related members of the PRR family (8).
Fig. 6.
Expression patterns of photoperiod pathway genes (Ppd-H1, HvCO1, and HvFT1) in wild-type and eam8 mutant plants. Expression levels (A–C) are for Bowman (blue diamonds and lines) and Bowman(eam8.w) plants (red squares and lines) grown under SDs (8-h light) for 14 d and sampled at 2-h intervals over 24 h. (D–F) Bowman and Bowman(eam8.w) plants sampled during a further 48 h after transfer from SD to LL at the end of the light period. Error bars are standard deviation of two technical replicates derived from a pool of three plants. Significant differences (*P < 0.05) were calculated by ANOVA using expression data of three consecutive time points in a sliding-window approach across 24 h. (G–I) Igri (black diamonds and lines) and Igri(eam8_ea8.k) (red squares and lines) plants grown under SDs (9-h light) for 10 d and sampled at 3-h intervals over 24 h. Bowman plants (A–F) have the mutant ppd-H1 allele, Igri plants (G–I) have the functional Ppd-H1 allele. Error bars are standard deviations of three biological replicates. Significant expression differences by ANOVA are shown: *P < 0.05, **P < 0.01, ***P < 0.001.
In Bowman, HvCO1 peaked 4 h after the end of the light period under LD, consistent with previous studies (16, 23), and continued to cycle under LL with peaks at the beginning of the subjective night. HvCO1 expression was lower in Bowman(eam8) under SD and LL. HvFT1 expression was nearly undetectable in Bowman, but was strongly increased in Bowman(eam8), consistent with the respective flowering phenotypes (Fig. 1). This result shows that activation of HvFT1 did not require increased HvCO1 expression. Parallel experiments comparing Igri and Igri(eam8) showed increased expression of Ppd-H1, HvCO1, and HvFT1, suggesting the Ppd-H1 allele had an effect on HvCO1 (Fig. 6). In summary, eam8 mutations significantly affected expression of circadian clock and clock output genes and induced flowering by up-regulating HvFT1 in a way that is not dependent on wild-type Ppd-H1.
eam8 Mutations Are Found in Different Clades of Accessions Containing the ppd-H1 Mutation.
Because the eam8 mutation caused early flowering independently of Ppd-H1, we investigated the distribution of eam8 relative to the widespread functional polymorphism in Ppd-H1, which is of long standing and might even have arisen before the domestication of barley (3). A 1.6-kb fragment of EAM8 covering part of exon 2, exon 3, and part of exon 4 (Fig. 3) was sequenced from 80 diverse barley lines comprising the genotypes described in above, plus a collection of 76 barley accessions originating from Europe, East Asia, West Asia, North Africa, and America (Table S3). These lines are a subset of a core collection (24) capturing the widest range of diversity for European areas, with a focus on genotypes from short growing seasons. Sequences were aligned by the ClustalW method using MEGA5 (25) and phylogenetic analysis was performed using the neighbor-joining method (26) within the MEGA5 package with bootstrap values from 10,000 trials (Fig. S4). Evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site; the differences in the composition bias among sequences were considered in evolutionary comparisons.
The phylogenetic analysis showed that functional mutations in EAM8 are rare compared with the widely distributed ppd-H1 mutation (Fig. S4), suggesting that eam8 mutations arose within germplasm containing ppd-H1. eam8 mutations are thus relatively recent, consistent with the observation that there were only widely introduced into breeding programs during the second half of the 20th century (20).
Discussion
In Arabidopsis, ELF3 and LUX physically associate with the promoters of PRR7 and PRR9 to repress transcription. PRR7 and PRR9, in turn, repress CCA1 and LHY (8, 10, 27). Gene-expression results in barley were consistent with a conserved role for EAM8. In comparison with wild-type plants, the eam8 mutants showed increased expression of Ppd-H1 (a PRR gene), particularly during the night or subjective night, and expression of core clock and clock-output genes was also altered (Figs. 4–6). Expression patterns of HvCCA1, HvGI, and HvTOC1 showed significantly dampened amplitude in the eam8 mutant lines compared with wild-type, both under SD and LL. Furthermore, expression of the evening-expressed genes HvGI and HvTOC1 peaked earlier under SD and LL in the eam8 mutants. These effects of eam8 are reminiscent of the effect of an Arabidopsis elf3-12 allele, which encodes an amino acid replacement in a conserved domain and resulted in a short circadian period under ambient light (8). The circadian clock can be entrained by temperature fluctuations as well as light/dark cycles (4). In the barley experiments, the use of controlled environment rooms with limited temperature variation or glasshouse environments, where temperatures fluctuated between day and night, gave consistent results in terms of eam8 phenotype. Therefore, neither diurnal changes in temperature nor light could rescue the clock defects and restore normal flowering behavior in the eam8 mutants. Thus, the barley eam8 genotype is severally attenuated in core clock function and we propose that this is the cause of the early-flowering phenotype.
The eam8 mutants had increased expression of HvFT1 compared with wild-type plants, consistent with their early-flowering phenotype. In the Igri background this may result from elevated levels of Ppd-H1, because this is similar to wheat plants with dominant Ppd-1 mutations that show elevated expression of the Ppd-1 gene itself, elevated FT1 expression, and an early-flowering phenotype (28). However, in eam8 mutants of barley the early-flowering phenotype and the effects on clock gene expression were also observed in the Bowman background with the recessive ppd-H1 mutation. This finding suggests that EAM8 affects the circadian clock by regulating PRR genes in addition to Ppd-H1, and that there is a pathway independent of Ppd-H1 that leads to increased HvFT1 expression in the eam8 mutants. An alternative explanation is that the ppd-H1 mutation is hypomorphic and has sufficient function to affect clock gene expression and flowering when expressed at the increased level observed in eam8 mutants. Differences at the Ppd-H1 locus may therefore explain the difference in HvFT1 expression pattern in Igri and Bowman plants (Fig. 6).
The eam8 mutants had inconsistent effects on the expression pattern of HvCO1, with increased expression in the Igri background and decreased expression in the Bowman background (Fig. 6). The reason for this finding is unclear but it demonstrates that increased expression of HvCO1 was not required for the high HvFT1 expression observed in eam8 mutants. This finding is consistent with other results in barley showing that variation at Ppd-H1 affects HvFT1 expression and flowering independently of HvCO1 expression levels (23) and in wheat, where dominant Ppd-1 mutations induce increased TaFT1 expression without increased TaCO1 expression (28). These results suggest that CO homologs in temperate cereals are less important for photoperiod response than CO in Arabidopsis.
The ubiquity of clocks in the natural world, including crop plant ancestors, strongly points to an adaptive advantage, and a functional circadian clock has been shown experimentally to benefit fitness (29). It could therefore be seen as an enigma that modern breeding has selected a clock-attenuated mutant in eam8. We suggest that this enigma has a historical explanation. The adaptation of crops to specific eco-geographical regions—in this case, the expansion of cultivated barley in Europe—is likely to have occurred in discrete steps. Movement of barley to areas with cold winters and warm, wet summers favored genotypes with no vernalization requirement and reduced photoperiod response (the ppd-H1 mutation) suitable for spring sowing (3, 16). Further expansion into shorter growing seasons in Scandinavia required the selection of early flowering in the context of a ppd-H1 genetic background. Adaptation was provided by mutations in EAM8 (20). Alternative routes to earliness that do not disrupt the circadian clock can be envisaged for a ppd-H1 genetic background; for example, this could be through a gain-of-function mutation at HvFT1. We suggest that loss of EAM8 function was selected because it provided a more effective solution for northern environments with short growing seasons, but long day lengths, which can exceed 20-h light in 1 d. The eam8 mutations might be effective in such environments because they could combine early flowering for the short growing season with reduced gating of physiological processes, as illustrated by the expression of a chlorophyll a/b binding protein gene (Fig. 5). This process could potentially increase light harvesting and photosynthesis and increase biomass accumulation and seed production under very long days.
The analysis of eam8 in relation to plant adaptation to northern environments shows that in some circumstances there are benefits from the loss of circadian clock function. This idea has an intriguing conceptual parallel in work on vertebrate animals from Scandinavia, as it has been shown that reindeer (Rangifer tarandus) do not show circadian organization of behavior under continuous light or dark conditions (30). This finding is not because of disconnection of the clock from behavior but because of reduced function of the clock itself, as two key genes (Bma1 and Per2) assayed in fibroblasts failed to show circadian rhythms of expression (31). Thus, plants and animals from environments that experience extreme variation in light period during the year have adapted by reducing their dependency on the circadian clock.
Because of their wide eco-geographical distribution, crop species like barley provide a rich source of variation for studying adaptation. By combining this finding with knowledge of plant development and genetic pathways, it is possible to understand the mechanisms underlying adaptation and their benefits for specific environments. In turn, this provides the potential for further improvement of crop performance.
Materials and Methods
Plant Material and Growth Conditions.
The spring barley cultivar Bowman and an introgression line Bowman(eam8.w) (kindly provided by R. Waugh, James Hutton Institute, Dundee, United Kingdom) were used to test diurnal and circadian expression of core clock and flowering-time genes. Replicate plants were grown in soil in a controlled environment growth chamber (8-h light, 20 °C/16-h dark, 18 °C, SD treatments). After 14 d replicate plants per genotype were harvested every 2 h for a total of 24 h from the start of the light period (T0). Night samples were collected in the dark. After SD sampling, Bowman and Bowman(eam8.w) plants were released into continuous light (20 °C; LL treatment). Sampling started at T8 and continued every 2 h for 48 h. A minimum of three biological replicates were taken per time point, each comprising the second youngest leaves of three independent plants. Samples were pooled and immediately frozen in liquid nitrogen and stored at −80 °C until processed.
Igri(eam8) lines were developed by two backcrosses with marker selection for Igri alleles at Vrn-H1 (32) and Vrn-H2 (33) (determining vernalization requirement), and Ppd-H1 (16), followed by self-pollination and selection of early flowering in vernalized plants under SDs. BC2F2 populations confirmed the map position of the eam8 locus in the distal region of chromosome 1HL (Fig. S1). Plants carrying the phenotypically similar eam7 mutation were developed in the same way. The eam8 donor was provided by R. Ellis, University of Reading, Reading, United Kingdom, and the eam7 donor was provided by A. Börner, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany. For gene expression, replicate plants were grown in soil in a controlled environment growth chamber (9-h light, 16 °C/15-h dark, 16 °C, SD treatments). Three biological replicates of three plants were taken every 3 h over a 24-h period.
Flowering times (days to awn emergence of the main stem) were recorded for Bowman, Bowman introgression lines carrying Ppd-H1, eam8, or Ppd-H1+eam8, and Igri and Igri(eam8) plants (10 per genotype per experiment). Plants were grown in soil in a glass house providing 10-h natural light or 18-h natural plus supplementary light. Bowman introgression lines (developed by J. D. Franckowiak, North Dakota State University, Fargo, ND) were obtained from the US Department of Agriculture National Germplasm Resources Laboratory, Aberdeen, Idaho. The results bring together work from two different laboratories, which worked with different SD and LD conditions, but tests showed that these differences made minimal difference to plant behavior (Fig. S5). The day lengths used clearly separate SD and LD effects and make it valid to combine results from 8-h/10-h SD or 16-h/18-h LD experiments.
Identification of EAM8.
Sequence of Affymetrix array feature Barley_13595 (Table S1) (http://www.plexdb.org/plex.php?database=Barley) was used for tblastn searches of EST databases at the National Center for Biotechnology Information Web site (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene sequence was used to develop primers for PCR and a partial sequence was used to screen a barley BAC library (cv. Morex) (34). The gene was fully sequenced from BAC clone 537I19. PCR was then used to amplify and sequence alleles from Igri (GenBank accession no. HQ850272), Bowman (GenBank accession no. HQ850273), Bowman(eam8_ea8.k allele), Mona (GenBank accession no. HQ850275), and Early Russian.
Gene Expression Analysis.
HvCCA1 and HvTOC1 sequences, PCR primers for all of the genes analyzed, and methods used to measure gene expression are described in Table S4.
Statistical Analysis.
Significant differences in flowering time and meristem development between eam8, eam8+Ppd-H1, and wild-type genotypes grown under SD or LD were calculated with paired t tests using a 95% confidence level (P < 0.05). Significant differences (P < 0.05) between expression profiles of Igri and Igri(eam8) were calculated for each time point based on three biological replicates with a one-factorial ANOVA. Significant differences in gene expression between eam8 mutant and wild-type Bowman measured under LL for 2 consecutive days were calculated using a general linear model in the SAS software, v9.1 (35) with the factor’s genotype, time point, and first-order interaction effects. For calculating significant differences (P < 0.05) between least-squares means of the genotype by time interactions, expression data over 2 d were considered as replicates of a 24-h day and a Tukey-Kramer adjustment for multiple comparisons was used. Significant differences in gene expression measured during SD were calculated using a general linear model where three consecutive time point measurements were considered as replicates in a sliding window approach across the 24 h. Significant differences are indicated in Figs. 4–6, if gene expression for three consecutive time points were significantly different between genotypes.
Note: As this manuscript was under review and decision appeal, EAM8 (Mat-a) was also identified as an ELF3 ortholog by fine-scale mapping and allele sequencing by Zakhrabekova et al. (36).
Supplementary Material
Acknowledgments
We thank Felix Schneider and Filiz Guerel for support, and Kerstin Luxa and Amanda Davis for excellent technical assistance. This study was supported in part by United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) Grant 208/D19952 (to S.F., A.S.T., and D.A.L.); Grant-in-Aid (to S.F., A.S.T., D.G., and D.A.L., and the John Innes Centre); a European Union Erasmus Student placement (D.G.); a BBSRC studentship (V.C.); the Max Planck Society (M.v.K. and S.J.D.); and by grants from the Deutsche Forschungsgesellschaft (to M.v.K. and S.J.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ850272, HQ850273, HQ850275, HQ850269, and HQ850270).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120496109/-/DCSupplemental.
References
- 1.Worland AJ, et al. The influence of photoperiod genes to the adaptability of European winter wheats. Euphytica. 1998;100:385–394. [Google Scholar]
- 2.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 3.Jones H, et al. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol. 2008;25:2211–2219. doi: 10.1093/molbev/msn167. [DOI] [PubMed] [Google Scholar]
- 4.Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 5.Locke JC, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ., 3rd A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolmos E, et al. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell. 2011;23:3230–3246. doi: 10.1105/tpc.111.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrero E, et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 13.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 14.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE. 2010;5:e10065. doi: 10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 17.Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Börner A, Buck-Sorlin GH, Hayes PM, Malyshev S, Korzun V. Molecular mapping of major genes and quantitative trait loci determining flowering time in response to photoperiod in barley. Plant Breed. 2002;121:129–132. [Google Scholar]
- 19.Franckowiak JD, Lundqvist U, Konishi T, Gallagher LW. Description of Stock number: BGS 214. Barley Genet Newsl. 1997;26:213–215. [Google Scholar]
- 20.Lundqvist U. Eighty years of Scandinavian barley mutation genetics and breeding. In: Shu QY, editor. Induced Mutations in the Genomics Era. Rome: FAO; 2009. pp. 39–43. [Google Scholar]
- 21.Waddington SR, Cartwright PM, Wall PC. A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot (Lond) 1983;51:119–130. [Google Scholar]
- 22.Murakami M, Tago Y, Yamashino T, Mizuno T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:110–121. doi: 10.1093/pcp/pcl043. [DOI] [PubMed] [Google Scholar]
- 23.Campoli C, Drosse B, Searle I, Coupland G, von Korff M. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 2012;69:868–880. doi: 10.1111/j.1365-313X.2011.04839.x. [DOI] [PubMed] [Google Scholar]
- 24.Stracke S, et al. Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor Appl Genet. 2009;118:259–273. doi: 10.1007/s00122-008-0896-y. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw LM, Turner AS, Laurie DA. Plant J. Rome: 2012. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum) [DOI] [PubMed] [Google Scholar]
- 29.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 30.van Oort BEH, et al. Circadian organization in reindeer. Nature. 2005;438:1095–1096. doi: 10.1038/4381095a. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Meng QJ, Tyler NJ, Stokkan KA, Loudon AS. A circadian clock is not required in an arctic mammal. Curr Biol. 2010;20:533–537. doi: 10.1016/j.cub.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 32.Fu D, et al. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- 33.Dubcovsky J, Chen C, Yan L. Molecular characterization of the allelic variation at the Vrn-H2 vernalization locus in barley. Mol Breed. 2005;15:395–407. [Google Scholar]
- 34.Yu Y, et al. A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet. 2000;101:1093–1099. [Google Scholar]
- 35.SAS Institute . The SAS system for Windows, release 9.1. Cary, N.C.: SAS Institute; 2003. [Google Scholar]
- 36.Zakhrabekova S, et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA. 2012;109:4326–4331. doi: 10.1073/pnas.1113009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.