Abstract
The LSD1–CoREST histone demethylase complex is required to repress neuronal genes in nonneuronal tissues. Here we show that sumoylation of Braf35, one of the subunits of the complex, is required to maintain full repression of neuron-specific genes and for occupancy of the LSD1–CoREST complex at its gene targets. Interestingly, expression of Braf35 was sufficient to prevent neuronal differentiation induced by bHLH neurogenic transcription factors in P19 cells and in neuronal progenitors of the chicken embryo neural tube. Sumoylation of Braf35 is required for this antineurogenic activity. We also show that iBraf, a paralogue of Braf35, forms heterodimers with Braf35. Braf35–iBraf heterodimerization impairs Braf35 interaction with the LSD1–CoREST complex and inhibits Braf35 sumoylation. Consistent with these results, iBraf prevents the antineurogenic activity of Braf35 in vivo. Our data uncover a mechanism of regulation of the LSD1–CoREST complex and provide a molecular explanation for the antagonism between Braf35 and iBraf in neuronal differentiation.
Keywords: chromatin, neurogenesis, KDM1, HMG20A, HMG20B
Cell differentiation involves large modifications of gene expression that require extensive changes of chromatin epigenetic marks (1). Epigenetic marks are DNA or histone posttranslational modifications that are inherited through cell division and that inform about the transcriptional state of loci. Among histone modifications, histone lysine methylation is of particular interest in development for the broad range of processes in which it is involved, including maintenance of stem cell pluripotency, germ-line determination, cell differentiation, control of HOX genes expression, and so forth (2, 3). Histone lysine methylation was considered a stable posttranslational modification until the discovery of histone demethylases. LSD1/KDM1 (lysine-specific demethylase 1) was the first demethylase identified and catalyzes demethylation of both di- and monomethylated lysine 4 (K4) or lysine 9 (K9) of histone H3 (H3K4me2/1 or H3K9me2/1) (4, 5). The lysine specificity of LSD1 seems to depend on its molecular partners. Thus, when LSD1 is associated with CoREST in the LSD1–CoREST corepressor complex (also called BHC, BRAF–histone deacetylase complex), the preferred substrate is H3K4me2/1, consistent with the fact that methylation of H3K4 is a mark of transcriptionaly active genes. In addition to LSD1 and CoREST, the LSD1–CoREST complex also contains HDAC1-2, BHC80, and BRAF35 (also called HMG20B) (6–9). BRAF35 contains a high-mobility group (HMG) domain and a coiled-coil domain, but its function within the complex is not well-understood (7, 10). Several functions of the LSD1–CoREST complex in differentiation and development have been reported (2). One of the best-characterized functions of the complex is its role in repression of neuronal genes in nonneuronal tissues and neuronal progenitors through its interaction with repressor factor REST (RE1 silencing transcription factor) (11, 12).
iBRAF (inhibitor of BRAF35, also called HMG20A) is a close paralogue of BRAF35 (13). As BRAF35, iBRAF contains an HMG domain in its amino terminus and a coiled-coil domain in the carboxyl-terminal half of the protein. In the mouse developing brain, Braf35 is predominantly expressed in immature neurons at the edges of the ventricles whereas iBraf is expressed in mature neurons, with the highest level being present in the outer cortex (13). Consistent with this expression pattern it has been shown that iBRAF improves neuronal differentiation of P19 cells. Furthermore, iBRAF activates expression of neuronal specific genes, whereas BRAF35 represses neuronal specific genes. Wynder et al. showed that iBRAF promotes recruitment of the histone methyltransferase MLL to neuronal specific genes (13). However, the molecular mechanism by which iBRAF antagonizes BRAF35 activity was unclear.
Posttranslational modification by small ubiquitin-related modifier (SUMO) regulates many cellular processes such as proliferation, intracellular trafficking, and transcription (reviewed in ref. 14). SUMO conjugation often occurs at the consensus sequence ΨKxE/D and creates new protein–protein interaction surfaces that modulate localization and/or activity of the target substrate. In transcription, sumoylation is normally associated with repression (reviewed in refs. 15 and 16). However, very little is known about how sumoylation is regulated.
We have investigated the role of sumoylation in controlling the function of LSD1–CoREST during neuronal differentiation. We found that Braf35 is modified by SUMO and that this modification is relevant for transcriptional repression of neuronal specific genes and to inhibit neuronal differentiation. Furthermore, we found that iBraf specifically inhibits sumoylation of Braf35 and that this inhibition is dependent on the formation of a Braf35–iBraf heterodimer. Heterodimer formation also impairs Braf35 incorporation into the LSD1–CoREST complex. Our data indicate that the interplay between Braf35 and iBraf controls the activity of the LSD1–CoREST complex during neuronal differentiation.
Results
Sumoylation of Braf35 Is Involved in LSD1–CoREST-Dependent Repression.
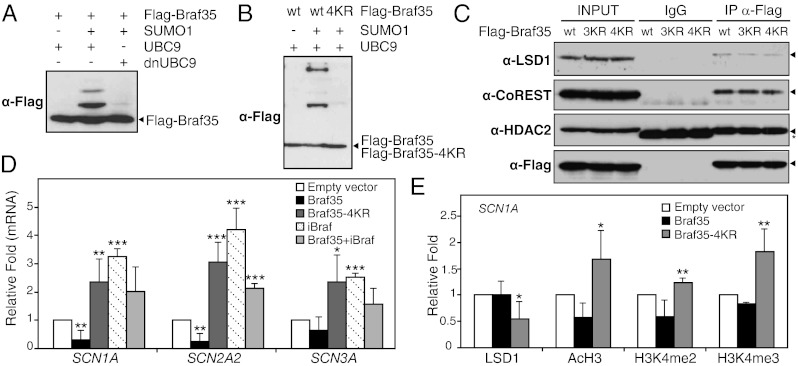
Analysis of the mouse Braf35 sequence with the SUMOsp 2.0 program (17) revealed the presence of four putative sumoylation sites: two sites that matched the consensus ΨKxE/D (30-VKQE-33 and 155-IKKE-158, putative sumoylated lysines are in boldface) and two nonconsensus sites (124-EKEK-127 and 156-KKED-159) (Fig. S1). Three of these sites are extensively conserved in vertebrates (Fig. S1). To verify whether these sites were substrate for SUMO conjugation in vivo, SUMO1 and UBC9 (E2 SUMO-conjugating enzyme) were coexpressed with Braf35 in 293T cells. Two Braf35 slow-migrating bands were observed only when SUMO1 was expressed (Fig. 1A). Similar results were observed when SUMO2 was coexpressed with Braf35 (Fig. S2A). Expression of a dominant-negative UBC9 protein strongly impaired sumoylation of Braf35 (Fig. 1A). Next, a lysine-to-arginine mutational analysis was performed to identify sumoylated lysines. First, we decided to mutate lysines of conserved sites. Because two of the putative target lysines were contiguous (K156 and K157), both lysines were mutated at the same time. Mutants of one (Braf35-K31R), two (Braf35-K156,157R), or three conserved putative sumoylation sites (Braf35-K31,156,157R, called Braf35-3KR) still exhibited some residual sumoylation. Then, a quadruple mutant was constructed where all putative SUMO target lysines were mutated (Braf35-K31,125,156,157R, called Braf35-4KR), and it was confirmed that sumoylation was completely abolished in this mutant (Fig. 1B).
Fig. 1.
Braf35 sumoylation is required for LSD1–CoREST repression. (A and B) Braf35 is sumoylated. 293T cells were transfected with expression vectors for the indicated proteins. wt, wild type Flag-Braf35; 4KR, Flag-Braf35-4KR mutant; dnUBC9, dominant-negative UBC9 (C93S). Flag-tagged proteins were detected by Western blot using anti-Flag antibodies. (C) Sumoylation of Braf35 is not required for its assembly in the LSD1–CoREST complex. 293T cells were transfected with expression vectors encoding Flag-Braf35 (wt), Flag-Braf35-3KR (3KR), or Flag-Braf35-4KR (4KR) sumoylation-defective mutants. Whole-cell extracts were subjected to immunoprecipitation with anti-Flag antibody (IP α-Flag) or mouse purified IgG as a control and analyzed by Western blot using the indicated antibodies. The asterisk denotes bands corresponding to immunoglobulins. (D) Derepression of SCN1A, SCN2A2, and SCN3A genes in HeLa cells. SCN1A, SCN2A2, and SCN3A mRNA levels were determined by RT-qPCR from HeLa cells transfected with expression vectors for the indicated proteins. (E) Overexpression of Braf35-4KR is coupled with dissociation of LSD1 and elevated levels of AcH3, H3K4me2, and H3K4me3 at the SCN1A promoter. HeLa cells transfected with Braf35, Braf35-4KR, or empty vector were subjected to ChIP assays with the indicated antibodies. Relative fold values indicate occupancies relative to empty vector set at 1. (D and E) Values are the average of multiple experiments (n ≥ 3) ±SD. *P < 0.1, **P < 0.05, ***P < 0.01 by ANOVA analysis, compared with cells transfected with empty vector.
Mutation of the sumoylation sites of Braf35 did not affect either nuclear localization (Fig. S3) or assembly of the mutant protein into the LSD1–CoREST complex (Fig. 1C). Then, we analyzed the functional consequences of sumoylation of Braf35 in LSD1–CoREST-mediated transcriptional repression. It has been shown that the LSD1–CoREST complex represses the neuronal specific voltage-gated sodium-channel α-subunit genes SCN1A, SCN2A2, and SCN3A in HeLa cells (4, 9, 18). As shown in Fig. 1D, expression of Braf35 provoked a slight down-regulation of the three genes. In contrast, expression of Braf35-4KR significantly derepressed the three genes (2.4-, 3.0-, and 2.4-fold induction for SCN1A, SCN2A2, and SCN3A genes, respectively), suggesting that the assembly of a nonsumoylatable Braf35 protein in the LSD1–CoREST complex impairs its repressive function. Then, we analyzed how Braf35 sumoylation affected LSD1–CoREST occupancy at the SCN1A gene promoter by chromatin immunoprecipitation. Expression of Braf35 had no effect on LSD1 occupancy. However, expression of the Braf35-4KR mutant protein led to a reduced binding of LSD1 to the SCN1A promoter (Fig. 1E). Consistently, expression of Braf35-4KR provoked an increase in both acetylated histone H3 (AcH3) and methylated histone H3 lysine 4 (H3K4me2/3). Therefore, our data suggest that sumoylation of Braf35 is required for the targeting or the stabilization of the LSD1–CoREST complex in chromatin.
Sumoylation-Dependent Inhibition of Neurogenesis by Braf35.
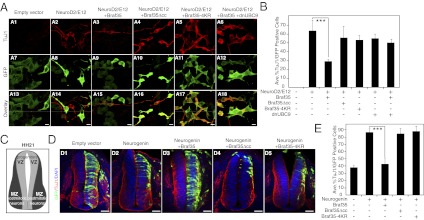
To analyze the biological consequences of Braf35 sumoylation, we used the P19 embryonal carcinoma stem cell line, a multipotent progenitor cell line that can be differentiated into neurons (19). It has been reported that expression of the neurogenic transcription factor NeuroD2 with its dimerization partner E12 efficiently promotes differentiation of P19 cells into fully functional neurons (20, 21). Fig. 2A shows that 3 d after expression of NeuroD2 and E12, a high percentage (64 ± 12%) of P19 transfected cells induced expression of the neuron-specific βIII-tubulin (TUBB3 gene) detected by the antibody TuJ1 (see Fig. 2B for quantification) or by RT-quantitative (q)PCR (Fig. S4A). Coexpression of Braf35 together with NeuroD2 and E12 strongly impairs differentiation (27 ± 3% TuJ1-positive cells) (Fig. 2A3), consistent with the role of Braf35 in repressing neuronal genes. This antineurogenic activity of Braf35 requires interaction of Braf35 with the LSD1–CoREST complex, because coexpression of a Braf35 mutant in the coiled-coil domain (Braf35Δcc, deletion of amino acids 195–210), which is not assembled into the complex (Fig. S5), was unable to inhibit neurogenesis (Fig. 2 A and B and Fig. S4A). More importantly, coexpression of the Braf35-4KR mutant failed to inhibit NeuroD2/E12-promoted differentiation (Fig. 2 A5 and B and Fig. S4A), indicating that sumoylation of Braf35 is essential for inhibition of neuronal differentiation. Consistently, inhibition of sumoylation by coexpression of the dominant-negative mutant of UBC9 (dnUBC9) together with Braf35 strongly impaired Braf35 antidifferentiation activity (Fig. 2 A6 and B and Fig. S4A). As a control, we verified that expression of dnUBC9 together with NeuroD2 and E12 did not affect differentiation (Fig. 2B and Fig. S4A). Interestingly, all sumoylation sites were not equally important for inhibition of neurogenesis. Thus, mutation of the conserved lysines K31 (Braf35-K31R), K156 and K157 (Braf35-K156,157R), or the three conserved lysines (Braf35-3KR) impaired inhibition of differentiation similar to the Braf35-4KR mutant (Fig. S4B). However, mutation of the nonconserved K125 had little effect on the antidifferentiation activity of Braf35, highlighting the importance of evolutionarily conserved sumoylation sites.
Fig. 2.
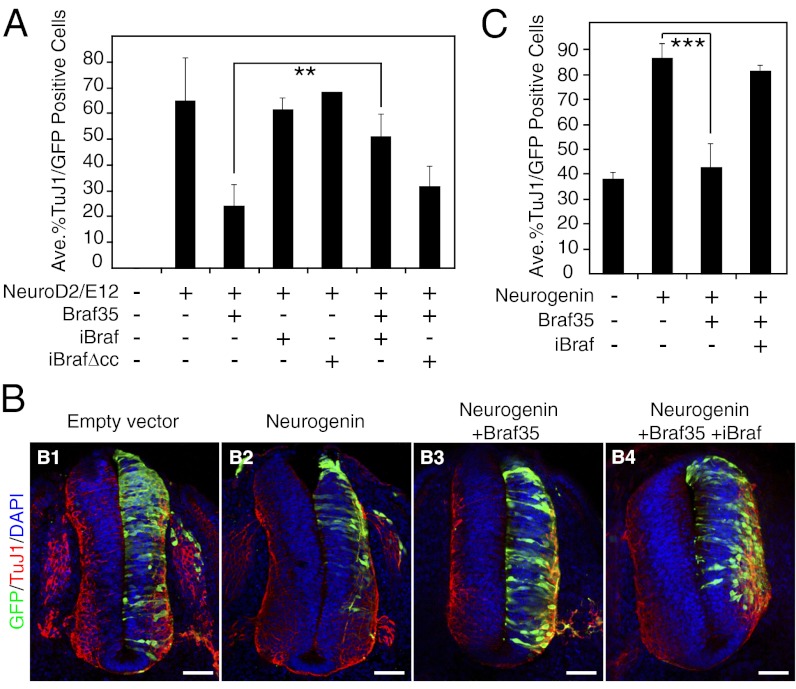
Expression of Braf35 inhibits neuronal differentiation. (A) P19 cells were transfected with a GFP expression vector and with expression vectors encoding the indicated proteins or empty vector. Three days after transfection, cells were analyzed by TuJ1 immunostaining (A1–A6) and GFP expression (A7–A12). A13–A18 are overlay images. (Scale bars, 10 μm.) (B) TuJ1-positive cells were scored as a percentage of GFP-positive transfected cells. (C) Diagram showing regions occupied by proliferating progenitors (ventricular zone; VZ) and postmitotic neurons (mantle zone; MZ) in HH21 chicken embryo spinal cord. (D) The neural tube of chicken embryos was electroporated with constructs expressing the proteins indicated at the top of each panel or empty vector. GFP was used to monitor electroporation. Thirty hours postelectroporation, embryos were immunostained for TuJ1 and DAPI. (Scale bars, 50 μm.) (E) Quantification of data presented in D as a percentage of TuJ1-positive cells per total number of GFP-positive cells. (B and E) Data are the average of three independent experiments ±SD. ***P < 0.01 by ANOVA analysis.
To investigate whether Braf35 ability to inhibit neurogenesis is conserved in other vertebrates, we performed in ovo electroporation experiments of chicken embryo neural tube. Analysis of chicken genome databases demonstrated the conservation of the consensus sumoylation sites in the chicken Braf35 protein (Fig. S1). Two zones are distinguished in the chicken embryo neural tube: the ventricular zone, formed by proliferating neuroblasts (neuronal progenitors), and the mantle zone, where postmitotic neuroblasts migrate and differentiate into neurons (Fig. 2C). Progression of differentiation was followed by monitoring migration of neuroblasts to the mantle layer. Mantle neurons were stained with the TuJ1 antibody. Expression of the neurogenic factor Neurogenin2 strongly promoted migration of the electroporated cells to the mantle zone (Fig. 2D2). However, coexpression of Braf35 strongly inhibited Neurogenin2-mediated migration of the electroporated cells (Fig. 2D3; quantification in Fig. 2E), indicating that Braf35 inhibits neuronal differentiation in the chick neural tube. Braf35Δcc was not able to inhibit differentiation (Fig. 2 D4 and E), suggesting that Braf35 interaction with the LSD1–CoREST complex is also required for the antineurogenic activity in the chick neural tube. Inhibition of differentiation by Braf35 was sumoylation-dependent, because expression of Braf35-4KR had no effect on neuroblast migration to the mantle layer (Fig. 2 D5 and E). Therefore, our data demonstrate, in two different systems, not only the ability of Braf35 to repress neuronal differentiation but also that this activity depends on Braf35 sumoylation.
iBraf Forms Homodimers and Heterodimers with Braf35.
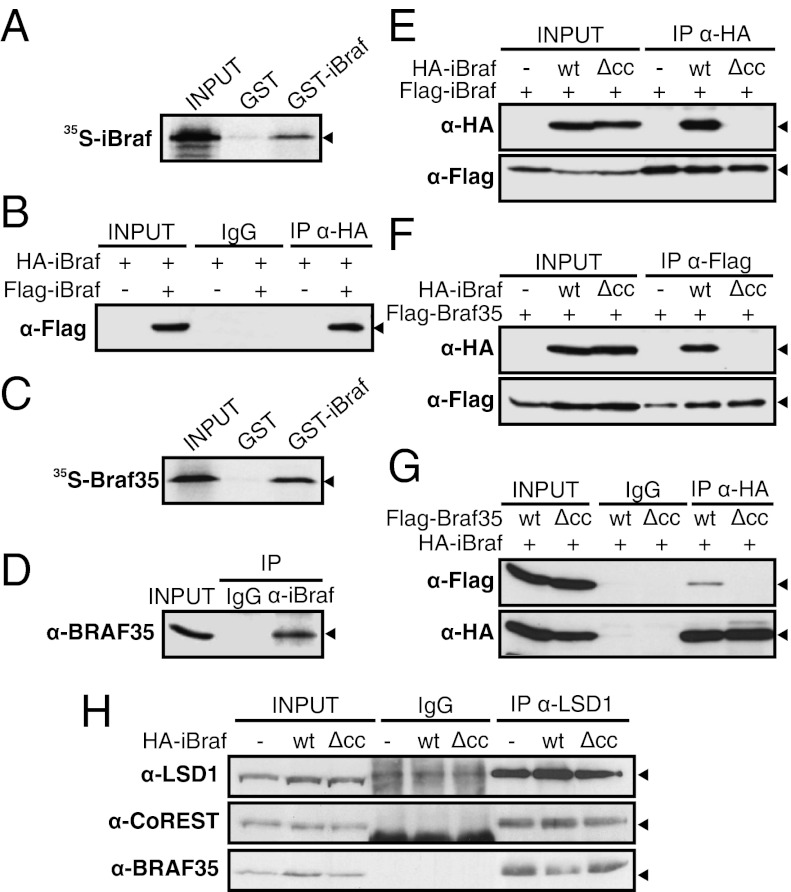
To investigate the role of iBraf in transcription and its possible relationship with the LSD1–CoREST complex, a yeast two-hybrid screening of a library of 8.5 days post coitum mouse embryo cDNAs was performed, using full-length iBraf protein as bait. A total of 14 yeast clones able to grow in the restrictive medium was selected, and their respective cDNAs were subjected to nucleotide sequence analysis. Two of them corresponded to α-Dystrobrevin, a cytoplasmic component of the dystrophin-associated protein complex (Fig. S6A). Interestingly, an interaction between iBraf and β-Dystrobrevin has recently been reported (22), indicating that iBraf can interact with both homologous proteins. Two other selected clones contained full-length and truncated (encoding amino acids 201–346) iBraf cDNAs, suggesting that iBraf is able to form homodimers (Fig. S6B). GST pull-down and immunoprecipitation experiments also indicated that iBraf forms homodimers in vitro and in vivo, respectively (Fig. 3 A and B). Analysis of the domain responsible for the interaction by the yeast two-hybrid system demonstrated that an intact coiled-coil region was required for the homodimerization (Fig. S6B).
Fig. 3.
iBraf forms homodimers and heterodimers with Braf35. (A and C) iBraf homodimerizes and heterodimerizes with Braf35 in vitro. One microgram of GST-iBraf or GST proteins bound to glutathione-Sepharose beads was incubated with in vitro translated 35S-labeled iBraf (A) or 35S-labeled Braf35 (C). Bound proteins and 20% of the input were subjected to SDS/PAGE. Gels were dried and autoradiographed. (B) iBraf homodimerizes in vivo. 293T cells were transfected with expression vectors encoding HA-iBraf or Flag-iBraf. Whole-cell extracts were subjected to immunoprecipitation with anti-HA antibodies (IP α-HA) or mouse purified IgG as a control and analyzed by Western blot using α-Flag antibodies. (D) Endogenous iBRAF and BRAF35 interact in vivo. Whole-cell extracts from 293T cells were subjected to immunoprecipitation with anti-iBraf antibody or rabbit purified IgG as a control. Immunoprecipitated proteins and 3% of the input were analyzed by Western blot with anti-Braf35 antibodies. (E–G) The coiled-coil domains of iBraf and Braf35 are required for iBraf–iBraf and iBraf–Braf35 interactions. Plasmids expressing the indicated proteins were transfected in 293T cells. Flag-iBraf (E), Flag-Braf35 (F), or HA-iBraf (G) were immunoprecipitated and analyzed by Western blot using the indicated antibodies. (H) Composition of LSD1–CoREST complexes upon overexpression of iBraf. 293T cells were transfected with expression vectors encoding HA (-), HA-iBraf (wt), or HA-iBrafΔcc (Δcc). Cell extracts were subjected to immunoprecipitation with anti-LSD1 antibodies or purified IgG as a control. Immunoprecipitated proteins and 3% of the input were analyzed by Western blot with the indicated antibodies.
Another positive clone isolated in the yeast two-hybrid screening contained a truncated cDNA of Braf35 (encoding amino acids 176–317), suggesting that iBraf can form heterodimers with Braf35 (Fig. S6B). GST pull-down experiments confirmed that iBraf interacts directly with Braf35 in vitro (Fig. 3C). Furthermore, BRAF35 was coimmunoprecipitated with iBRAF from 293T cell extracts by using anti-iBraf antibodies (Fig. 3D), demonstrating that both endogenous proteins interact in vivo. Mapping the interaction domains by the two-hybrid system indicated that the coiled-coil domain of iBraf is again essential for the heterodimerization (Fig. S6B). To confirm the role of this domain in dimerization, an iBraf coiled-coil mutant was generated by deleting 15 amino acids (residues 252–266) of the coiled-coil region (iBrafΔcc). Immunoprecipitation experiments demonstrated that iBrafΔcc was not able to form homodimers or heterodimers with Braf35 (Fig. 3 E and F). Equally, Braf35Δcc was also unable to form heterodimers with iBraf (Fig. 3G). All these experiments demonstrate that the coiled-coil domains of Braf35 and iBraf are essential for the formation of iBraf–iBraf and Braf35–iBraf dimers. Taking into account the homology between iBraf and Braf35, we tested, both by two-hybrid assay and immunoprecipitation, whether Braf35 was able to form homodimers. None of these experiments confirmed the homodimerization of Braf35 (Fig. S6 C and D). Then, we investigated whether heterodimer formation affects assembly of Braf35 into the LSD1–CoREST complex. Fig. 3H shows that overexpression of iBraf, but not of iBrafΔcc, provoked a decrease in the level of Braf35 coimmunoprecipitated with LSD1, suggesting Braf35–iBraf heterodimer formation interferes with the interaction of Braf35 with the complex.
iBraf Inhibits Sumoylation of Braf35.
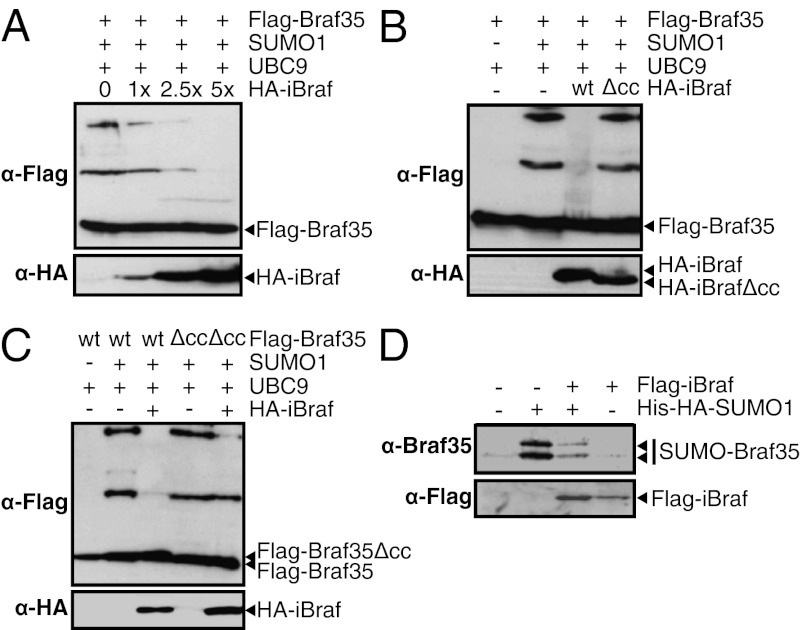
Despite the homology between Braf35 and iBraf, analysis of the iBraf amino acid sequence indicated that this protein lacked sumoylation consensus sites. Consistently, iBraf was not sumoylated in SUMO1 or SUMO2 transfected 293T cells (Fig. S7A). Given the fact that Braf35 forms heterodimers with iBraf, we checked whether heterodimerization affects sumoylation of Braf35. Interestingly, we observed that coexpression of Braf35 and iBraf significantly inhibited sumoylation of Braf35 (Fig. 4A). Furthermore, increasing quantities of iBraf were able to completely inhibit Braf35 sumoylation. Then, we checked whether iBraf was a general inhibitor of sumoylation. For that, sumoylation of the carboxyl-terminal part of RanGAP1 (RanGAP1-Cter), a well-known sumoylation substrate (23), was assayed. Upon coexpression with SUMO1, RanGAP1-Cter was sumoylated regardless of the presence of Braf35, iBraf, or both, indicating that iBraf is not a general inhibitor of the sumoylation enzymatic machinery (Fig. S7B).
Fig. 4.
Inhibition of Braf35 sumoylation by iBraf. (A) iBraf inhibits Braf35 sumoylation. Numbers are the amount of transfected HA-iBraf–expressing vector with respect to the amount of Flag-Braf35–expressing vector. (B) The coiled-coil domain of iBraf is required for inhibition of Braf35 sumoylation. (C) The coiled-coil domain of Braf35 is required for iBraf-dependent inhibition of Braf35 sumoylation. (A–C) 293T cells were transfected with expression vectors for the indicated proteins. Flag-tagged, or HA-tagged proteins were detected by Western blot using anti-Flag or anti-HA antibodies. (Lower) Inputs of the indicated proteins. (D) Sumoylation of endogenous BRAF35 depends on the level of iBraf. 293T cells were transfected with expression vectors for His-HA-SUMO1 and Flag-iBraf as indicated. Sumoylated proteins were purified using nickel-affinity chromatography and subjected to Western blot analysis using anti-Braf35 antibodies. (Lower) The presence of Flag-iBraf in the extracts was verified by Western blot using anti-Flag antibodies.
Then, we investigated whether iBraf-dependent inhibition of Braf35 sumoylation required iBraf–Braf35 heterodimerization. We have shown above that Braf35Δcc and iBrafΔcc are unable to form heterodimers (Fig. 3). Fig. 4B shows that iBrafΔcc was unable to inhibit sumoylation of Braf35. Furthermore, sumoylation of the Braf35Δcc mutant was not correctly inhibited by iBraf (Fig. 4C). All these experiments indicate that iBraf is a specific inhibitor of Braf35 sumoylation and that inhibition of sumoylation requires heterodimerization of both proteins.
Then, we analyzed whether the level of sumoylation of endogenous Braf35 was controlled by iBraf. For that, His-SUMO1 was expressed in 293T cells and all endogenous sumoylated proteins were purified by nickel-affinity chromatography. As shown in Fig. 4D, two bands of sumoylated BRAF35 were clearly observed, demonstrating sumoylation of endogenous BRAF35. Most importantly, the level of sumoylated BRAF35 was strongly decreased upon expression of iBraf. Therefore, our data demonstrate that iBraf controls the degree of sumoylation of endogenous BRAF35.
Functional Consequences of iBraf–Braf35 Interaction.
Next, the effect of iBraf in Braf35-dependent repression of SCN genes was investigated. Expression of iBraf led to derepression of SCN1A, SCN2A2, and SCN3A genes in HeLa cells (Fig. 1D). Furthermore, expression of iBraf was able to overcome the repressive effect of Braf35 in this assay. Then, we investigated the effect of Braf35–iBraf interaction in P19 neuronal differentiation. Coexpression of iBraf with NeuroD2 and E12 did not increase neuronal differentiation with respect to that obtained by only expressing NeuroD2 and E12 (Fig. 5A). We have previously shown that sumoylation of Braf35 is essential for inhibition of neurogenesis and that iBraf inhibits Braf35 sumoylation. Furthermore, iBraf–Braf35 heterodimer formation impairs association of Braf35 with the LSD1–CoREST complex. Therefore, we hypothesized that iBraf should impair Braf35-dependent inhibition of neurogenesis. Indeed, we observed that coexpression of iBraf together with Braf35 in P19 cells abolished Braf35-dependent inhibition of differentiation. This effect was dependent on Braf35–iBraf heterodimerization, because iBrafΔcc was unable to suppress the effect of Braf35 (Fig. 5A).
Fig. 5.
iBraf antagonizes Braf35-dependent inhibition of differentiation. (A) P19 cells were transfected with expression vectors for the indicated proteins together with a GFP expression vector. Three days after transfection, cells were analyzed by TuJ1 immunostaining and GFP expression. TuJ1-positive cells were scored as a percentage of GFP-positive transfected cells. (B) Sections from HH21 chicken embryos stained with TuJ1 and DAPI 30 h postelectroporation of plasmids encoding Neurogenin2 (B2), Neurogenin2 and Braf35 (B3), and Neurogenin2, Braf35, and iBraf (B4), or an empty vector (B1) together with GFP expression vector. (Scale bars, 50 μm.) (C) Quantification of data presented in B as a percentage of TuJ1-positive cells per total number of GFP-positive cells. Data are the average of three independent experiments ±SD. **P < 0.05, ***P < 0.01, by ANOVA analysis.
The antagonism between iBraf and Braf35 in neuronal differentiation was also investigated in the chick neural tube. Down-regulation of endogenous chicken iBRAF (ciBRAF), but not of cBRAF35, impaired neuronal differentiation (Fig. S8), indicating that ciBRAF is required for differentiation. As described above, expression of Braf35 inhibited differentiation. However, coexpression of iBraf together with Braf35 impaired the antidifferentiation effect of Braf35 (Fig. 5B; quantification in Fig. 5C), confirming the opposite effect of iBraf and Braf35 also in chick neural tube development.
Discussion
We have demonstrated that overexpression of Braf35, one of the subunits of the LSD1–CoREST complex, is sufficient to inhibit neurogenesis. Furthermore, we show that sumoylation of Braf35 is important for inhibition of neurogenesis and for repression of neuronal genes. Our study also reveals that Braf35 is able to form heterodimers with its homologous protein iBraf. This interaction controls Braf35 association with the LSD1–CoREST complex and impairs its sumoylation.
How the LSD1–CoREST histone demethylase complex is recruited to its gene targets is being actively investigated. Interactions of several LSD1–CoREST subunits with some transcription factors (11, 24–26) or with long noncoding RNAs (27) have been reported. Furthermore, Ouyang et al. recently showed that CoREST1 contains a SUMO interaction domain (SIM) responsible for noncovalent interaction with SUMO-2/3 (28). They suggested that interaction of CoREST with sumoylated transcription factors through its SIM mediates recruiting of the complex. The LSD1–CoREST complex contains two DNA-binding domains: the SANT2 domain of CoREST and the HMG domain of Braf35. The SANT2 domain of CoREST is required for the efficient demethylation of nucleosomes by the LSD1–CoREST dimer and seems to be essential for a correct presentation of the substrate to the LSD1 catalytic site (29). The role of the HMG domain of Braf35 is less clear. Braf35 HMG domain binds structured DNA, such as four-way junction DNA, without sequence specificity (10). The Shiekhattar group reported that a point mutation in the HMG domain of Braf35 abrogates binding of this protein to its target genes, suggesting that the DNA-binding activity of Braf35 is important for its function (7). We show that sumoylation of Braf35 is relevant for LSD1 occupancy, H3 acetylation, and H3K4 methylation at the SCN1A promoter, suggesting a role of Braf35 in recruiting the LSD1–CoREST complex or in stabilization of the complex at the chromatin. Nonsumoylatable Braf35 retains its ability to interact with the LSD1–CoREST complex. Therefore, our data suggest that sumoylated Braf35 increases interaction with DNA or with other components of the chromatin, promoting recruitment or a higher stability of the LSD1–CoREST complex at its genomic targets.
Recent reports have highlighted the role of LSD1–CoREST components in stem cell and neural stem cell maintenance (30–32). Interestingly, our data demonstrate that overexpression of Braf35 is enough to inhibit neuronal differentiation of the embryonal carcinoma stem cell line P19 and of the neuronal progenitors of the chicken embryo neural tube, suggesting an essential role of Braf35 in maintaining the undifferentiated state, despite the expression of proneurogenic transcription factors. A Braf35Δcc mutant that cannot interact with the complex was not able to inhibit differentiation, suggesting that the LSD1–CoREST complex is required for the antineurogenic activity of Braf35. Sumoylation of Braf35 is also essential for this antineurogenic activity. Altogether, these data suggest that high levels of sumoylatable Braf35 promote stabilization of the LSD1–CoREST complex in its targets. To our knowledge, it has not been reported that overexpression of other subunits of the LSD1–CoREST complex inhibit differentiation, which highlights the important role that Braf35 plays in the complex.
iBraf was described as an antagonist of Braf35 during neuronal differentiation, based on the fact that iBraf recruits the H3K4 methyltransferase MLL to neuronal specific genes (13). Now we show that iBraf can form homodimers and heterodimers with Braf35. Importantly, heterodimerization of Braf35 with iBraf displaces Braf35 out from the LSD1–CoREST complex and inhibits sumoylation of Braf35. How is sumoylation inhibited? One possibility is that Braf35 cannot be sumoylated out of the LSD1–CoREST complex. However, we ruled out this possibility because the Braf35Δcc mutant protein, which does not interact with the complex, was normally sumoylated. Another obvious possibility is that heterodimerization sterically impairs access of the sumoylation machinery. It has been previously shown that in the developing central nervous system, Braf35 and iBraf display complementary expression patterns. Thus, Braf35 is mostly expressed in immature neurons whereas iBraf is widely expressed, with the highest level being present in the mature neurons of the outer cortex (13). Therefore, we propose a model where heterodimerization between Braf35 and iBraf controls LSD1–CoREST activity. In our model, there is a dynamic equilibrium in which Braf35 could be in three different forms: free, or associated with the LSD1–CoREST complex or with iBraf. However, upon an increase in the level of iBraf, the equilibrium will be displaced toward the formation of more iBraf–Braf35 heterodimers. This has two consequences. Part of the Braf35 pool is sequestered out of the LSD1–CoREST complexes and cannot be sumoylated, which we have shown to be essential for its activity. Therefore, the model predicts that an increase of the iBraf:Braf35 ratio is important for neuronal differentiation. Consistently, we show that overexpression of Braf35 during the process of neuronal differentiation, which decreases the iBraf:Braf35 ratio, impairs neurogenesis. In contrast, if both proteins are expressed at the same time, neurogenesis is not altered. Furthermore, silencing of ciBRAF impairs neuronal differentiation in the developing chick neural tube. Also in agreement with our model, Wynder et al. reported that overexpression of iBraf stimulates neuronal differentiation (13). We cannot rule out the possibility that other factors, in addition to just the concentration of the proteins such as posttranslational modifications or the presence of additional interacting factors, also affect the Braf35–iBraf equilibrium.
Vertebrates contain genes encoding Braf35 and iBraf; however, simple chordata such as Branchiostoma and all invertebrates including Drosophila melanogaster and the worm Caenorhabditis elegans have only one representative from the Braf35/iBraf subfamily. It is tempting to speculate that development of a complex nervous system might require the fine regulation of the activity of the LSD1–CoREST complex, favoring the duplication of the ancestral Braf35/iBraf gene, the diversification of functions of both proteins, and the implementation of the complex mechanism of regulation analyzed in our work.
Materials and Methods
All mammalian expression constructs were derived from vector pAdRSV-Sp (RSV) (33). Mouse cDNAs of Braf35 and iBraf were kindly provided by L. Sumoy (Center for Genomic Regulation, Barcelona, Spain) (34). Gene truncations or point mutations were generated by standard PCR techniques. 293T and HeLa cells were cultured in DMEM supplemented with 10% FBS. P19 cells were cultured in α-modified Eagle’s medium supplemented with 7.5% (vol/vol) calf and 2.5% (vol/vol) fetal bovine sera. Sumoylation assays in cells and purification of endogenous sumoylated proteins from 293T cells were performed as described (35). In ovo electroporation and embryo immunofluorescence were carried out as described (33). Total RNA was prepared by using the RNeasy Kit (Qiagen) as described in the manufacturer’s instructions. cDNA was generated by using the SuperScript First Strand Synthesis System (Invitrogen). Quantification of gene products was performed by qPCR with the Applied Biosystems 7500 FAST Real-Time PCR System. Values were normalized to the expression of the GAPDH housekeeping gene. Each experiment was performed at least in triplicate. ChIP assays were performed as previously described (36). Further details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank L. Sumoy, K. Kroll, and T. Baba for generously providing different plasmids; R. Losson for P19 cells; and R. Shiekhattar for anti-iBraf antibody. We thank G. Arribas for construction of the iBrafΔcc expression plasmid and P. Domínguez for microscopy technical support. We also thank F. Prado for critical reading of the manuscript and discussion and D. Haun for style supervision. This work was supported by Grants BFU2008-00238 and CSD2006-00049 from the Spanish Ministerio de Ciencia e Innovacion, Grant P06-CVI-4844 from Junta de Andalucía and Fundación Ramón Areces (to J.C.R.), and Grant P09-CTS-04967 from Junta de Andalucía (to S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121522109/-/DCSupplemental.
References
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Nottke A, Colaiácovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 6.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakimi MA, et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 9.Shi YJ, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Marmorstein LY, et al. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 11.Andrés ME, et al. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballas N, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 13.Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat Cell Biol. 2005;7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 14.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18(1):1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Ren J, et al. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 18.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossant J, McBurney MW. The developmental potential of a euploid male teratocarcinoma cell line after blastocyst injection. J Embryol Exp Morphol. 1982;70:99–112. [PubMed] [Google Scholar]
- 20.Farah MH, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 21.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132(1):105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 22.Artegiani B, et al. The interaction with HMG20a/b proteins suggests a potential role for β-dystrobrevin in neuronal differentiation. J Biol Chem. 2010;285:24740–24750. doi: 10.1074/jbc.M109.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137(1):47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 27.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34(2):145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M, et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Sun G, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamo A, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 32.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giudicelli F, Gilardi-Hebenstreit P, Mechta-Grigoriou F, Poquet C, Charnay P. Novel activities of Mafb underlie its dual role in hindbrain segmentation and regional specification. Dev Biol. 2003;253(1):150–162. doi: 10.1006/dbio.2002.0864. [DOI] [PubMed] [Google Scholar]
- 34.Sumoy L, et al. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet Cell Genet. 2000;88(1-2):62–67. doi: 10.1159/000015486. [DOI] [PubMed] [Google Scholar]
- 35.García-Gutiérrez P, Juárez-Vicente F, Gallardo-Chamizo F, Charnay P, García-Domínguez M. The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators. EMBO Rep. 2011;12:1018–1023. doi: 10.1038/embor.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subtil-Rodríguez A, Reyes JC. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 2010;11:751–757. doi: 10.1038/embor.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.