Abstract
We recently reported a deletion of exon 2 of the trimethyllysine hydroxylase epsilon (TMLHE) gene in a proband with autism. TMLHE maps to the X chromosome and encodes the first enzyme in carnitine biosynthesis, 6-N-trimethyllysine dioxygenase. Deletion of exon 2 of TMLHE causes enzyme deficiency, resulting in increased substrate concentration (6-N-trimethyllysine) and decreased product levels (3-hydroxy-6-N-trimethyllysine and γ-butyrobetaine) in plasma and urine. TMLHE deficiency is common in control males (24 in 8,787 or 1 in 366) and was not significantly increased in frequency in probands from simplex autism families (9 in 2,904 or 1 in 323). However, it was 2.82-fold more frequent in probands from male-male multiplex autism families compared with controls (7 in 909 or 1 in 130; P = 0.023). Additionally, six of seven autistic male siblings of probands in male-male multiplex families had the deletion, suggesting that TMLHE deficiency is a risk factor for autism (metaanalysis Z-score = 2.90 and P = 0.0037), although with low penetrance (2–4%). These data suggest that dysregulation of carnitine metabolism may be important in nondysmorphic autism; that abnormalities of carnitine intake, loss, transport, or synthesis may be important in a larger fraction of nondysmorphic autism cases; and that the carnitine pathway may provide a novel target for therapy or prevention of autism.
The role of carnitine in biology and disease has been studied for decades (1, 2). Carnitine has been proposed to be a conditionally essential nutrient, and even termed vitamin BT. Carnitine content of foods varies widely, being very low in fruits, vegetables, and grains; intermediate in milk products, eggs, chicken, and fish; and very high in red meats. The proportion of carnitine derived from the diet varies widely in humans, being quite low in vegetarians and especially low with a vegan diet that excludes dairy products and eggs. In contrast, about 75% of carnitine is derived from the diet in meat eaters. Carnitine homeostasis in humans (Fig. 1) is maintained by a modest rate of endogenous synthesis, absorption from dietary sources, and efficient tubular reabsorption by the kidney. Apart from the dietary intake, carnitine is synthesized in humans in kidney, liver, and brain from protein-derived 6-N-trimethyllysine (TML) via 3-hydroxy-6-N-trimethyllysine (HTML), 4-N-trimethylaminobutyraldehyde (TMABA), and 4-N-trimethylaminobutyric acid [γ-butyrobetaine (γBB)] (3). Renal resorption plays an important role in carnitine metabolism, with considerable excretion if carnitine intake is abundant, but there is extremely efficient resorption if body stores of carnitine are low. Carnitine is present as free carnitine and as acylcarnitines, of which the latter reflect the cellular acyl-CoA ester pool. Up to 99% of carnitine is intracellular and is essential for mitochondrial function, where its role is to enable transport of fatty acids into mitochondria, where β-oxidation takes place (3); it is also involved in the transport of peroxisomal oxidation products to the mitochondria.
Fig. 1.
Carnitine biosynthesis and homeostasis in humans. Carnitine is synthesized in four enzymatic steps. After release of TML by lysosomal protein degradation, this compound is hydroxylated by TMLD, producing HTML. HTML is cleaved by HTML aldolase (HTMLA) into TMABA and glycine. Subsequently, TMABA is oxidized by TMABA-dehydroxygenase (TMABA-DH) to form 4-N-trimethylaminobutyrate, also named γ-butyrobetaine (γBB). Finally, γBB is hydroxylated by γBBD, yielding l-carnitine. Because TMLD is located in mitochondria, TML needs to be transported out of the lysosome and across the inner mitochondrial membrane into the mitochondrial matrix by means of transporters, which are unknown at this time. Depending on the subcellular localization of the HTLMA (also uncertain, likely the cytosol), HTML or γBB needs to be transported back to the cytosol (where γBBD is located). In cells that do not contain γBBD, γBB is exported from the cell and imported into tissues (liver, kidney, and brain in humans) that do express γBBD by means of at least one specific transporter, presumably SLC6A13. Carnitine is transported by OCTN2 and other lower affinity transporters (not shown).
Carnitine deficiency can develop secondary to dietary inadequacy or as an adverse effect of medical treatments. Although humans can synthesize carnitine, nutritional deficiency can occur, as when infants were fed early preparations of soy formulas that were deficient in carnitine (4). Similarly, deficiency can arise with parenteral alimentation in neonates (5). Carnitine deficiency can also occur secondary to administration of pivalate-conjugated antibiotics or valproic acid (2). Various disease processes and medical interventions, such as renal tubular disorders and chronic hemodialysis, respectively, can also be associated with carnitine deficiency.
There are primary and secondary genetic forms of carnitine deficiency (6). Secondary deficiency is caused by various fatty oxidation defects and organic acidemias that lead to carnitine deficiency through urinary loss of acylcarnitines that accumulate related to the enzyme deficiency. Primary systemic carnitine deficiency is caused by biallelic loss-of-function mutations in the SLC22A5 gene that encodes the plasma membrane organic cation transporter-2 (OCTN2). OCTN2 deficiency is characterized by excessive urinary loss of carnitine, leading to systemic deficiency with associated skeletal myopathy, cardiomyopathy, fatty liver, and hypoglycemia. Although the possibility of a primary systemic carnitine deficiency caused by a defect in carnitine biosynthesis was postulated long ago, no primary disorders of carnitine biosynthesis have been described until now (7).
Administration of carnitine is the centerpiece of therapy for systemic carnitine deficiency, and it is beneficial in some genetic forms of secondary carnitine deficiency. Administration of carnitine and acetylcarnitine has been explored as an antioxidant and for treatment of many disorders, including diabetic peripheral neuropathy (8), heart failure (9), and mitochondrial disorders.
Recently, we discovered a deletion of the 6-N-trimethyllysine dioxygenase (TMLD) gene [also known as trimethyllysine hydroxylase, epsilon (TMLHE)] while studying probands with autism, raising the question of whether there might be an association of autism with TMLHE mutations (10). TMLHE maps to the long arm of the X chromosome near the boundary of the pseudoautosomal region and encodes TMLD. TMLD is the first enzyme of the carnitine biosynthesis pathway (3) and is localized in mitochondria (11).
The etiology of severe, dysmorphic autism with a male/female ratio of 3.2:1 (12) has become increasingly well defined as often attributable to de novo mutations or recent mutations transmitted for a few generations. These mutations include large copy number variants (CNVs), which are detectable by chromosomal microarray analysis in up to 25% of the most severe cases with phenotypes that include major intellectual disability, which restricts reproduction (13). De novo point mutations are also being discovered as causes of autism using next-generation sequencing of genomic DNA (14). Disease-causing CNVs are found in ∼10% of patients with intermediate phenotypes, often with less severe intellectual disability (15, 16). In these cases, penetrance may be incomplete and the phenotype can be highly variable, with diagnoses of intellectual disability, autism, schizophrenia, and idiopathic epilepsy seen with the same CNV (17–19). These examples typify single-locus conditions, perhaps with genetic and nongenetic modifier effects. Autism spectrum disorders and related neurocognitive phenotypes blend into even more complex genotype-phenotype relationships with evidence for two-hit or two-locus pathogenesis (20). At the milder end of the autism spectrum are patients who often have speech, have an intelligence quotient (IQ) ranging from low to the normal range, and are nondysmorphic. This milder population, some of whom meet diagnostic criteria for Asperger syndrome, can display up to an 8:1 male/female ratio (21, 22), and will be referred to herein as having nondysmorphic autism (NDA). This includes the milder portion of the autism spectrum, but patients who have NDA can have severe cognitive and behavioral phenotypes. The etiology of NDA remains almost completely unknown, but the extreme sex ratio may provide a clue as to its etiology.
In this paper, we show that TMLHE deficiency is a very common inborn error of metabolism in males and suggest that it may be significantly more frequent in autistic male-male sib pairs than in controls.
Results
Deletions of Exon 2 Are Heterogeneous and Common in Autistic and Healthy Males.
Given the discovery of a deletion of exon 2 in TMLHE in a male simplex proband with autism (10), we examined the frequency of TMLHE mutations in autism and control populations. We studied simplex families primarily from the Simons Simplex Collection (SSC) and multiplex families primarily from the Autism Genetic Resource Exchange (AGRE) collection, and we recruited multiple collaborators to study additional families (Table 1). We have now identified a total of 16 male autism probands, six affected male siblings of probands, and 24 healthy adult males with deletions of exon 2, indicating that this is a relatively common CNV (Table 1). For 28 of the 29 deletions characterized more thoroughly, size ranged from 5.7 to 15.9 kb and only exon 2 was deleted; one additional deletion of 59.6 kb removed exons 2–6 (Fig. 2 A and B and Table S1). Based on position, size, and sequence, there was a minimum of 14 different deletion junctions among 29 unrelated families. Sequencing of the breakpoints of many deletions showed that almost all junctions occurred in long interspersed elements and short interspersed elements in the introns flanking exon 2 (Fig. 2B and Table S2), as has been seen in other loci (23). For all SSC samples (probands, heterozygous mothers, and healthy fathers), the deletions were present in both DNA extracted directly from blood and DNA extracted from lymphoblastoid cell lines (LCLs).
Table 1.
Sources of male autism and control samples and methods of testing
| No. | Deletion | |
| Simplex autism | ||
| SSC | 1,887 | 6* |
| SCAP | 80 | 0 |
| Houston | 24 | 0 |
| Toronto | 328 | 3 |
| Paris | 333 | 0 |
| New York | 252 | 0 |
| Totals | 2,904 | 9 |
| Male probands from male-male sibling pairs† | ||
| AGRE + NIMH | 752 | 7 |
| Toronto | 93 | 0 |
| Paris | 53 | 0 |
| New York | 11 | 0 |
| Totals | 909 | 7 |
| Male probands from male-female sibling pairs† | ||
| Paris | 38 | 0 |
| New York | 5 | 0 |
| Controls | ||
| SSC, NIMH, and AGRE autism fathers | 2,197 | 7 |
| NIMH controls | 897 | 3 |
| BPR | 49 | 1 |
| Houston fathers | 36 | 0 |
| Multiplex fathers | 615 | 0 |
| WTCCC | 3,018 | 9 |
| Toronto | 1,975 | 4 |
| Totals | 8,787 | 24 |
SCAP, South Carolina Autism Project; WTCCC, Wellcome Trust Case–Control Consortium.
*Screening for the deletion and confirmation were performed as described in Materials and Methods.
†Excludes affected sibling.
Fig. 2.
Exon 2-containing deletions found in TMLHE and structure of TMLHE. (A) Array CGH showing eight exonic deletions in TMLHE, with the relative location of TMLHE exons 2–6 aligned above the array CGH plots (GRCh37/hg19 assembly; http://genome.ucsc.edu). The horizontal axis shows chromosome position, and the vertical axis shows the log2 ratio of array signal. Semitransparent filled boxes on CGH plots highlight the regions of aberration; all samples have deletions involving exon 2, and most have a separate deletion in intron 1 (red circle). All samples are autism probands with identifiers found in Tables S1, S4, and S5. (B) Twenty-four unrelated individuals with deletions involving exon 2 of TMLHE are mapped. Deletion coordinates were determined by array CGH unless specified by PCR assay. White arrowheads indicate the continuation of the deletion for SSC 13928.p1. NA 12003 is an unaffected individual whose deletion is published (38) and was better characterized in this study. BPR 664 is an unaffected individual. (C) Diagram of gene structure. Large open arrows represent near-identical inverted repeats. Fa, father; P1, proband; HI, AGRE individuals; #, individual first described by Celestino-Soper et al. (10).
In addition, we identified an extremely common intronic deletion (Fig. 2A) that appeared on the basis of comparative genomic hybridization (CGH) array to be indistinguishable in all cases. This is equivalent to Database of Genomic Variants numbers 115349 and 104572, which appear to be identical, and/or to number 97130, which is very similar. The intronic deletion was present in 74% of 93 autism male probands and 71% of 48 control males examined, and it should not be misinterpreted as causing enzyme deficiency. This intronic deletion was present on 24 of 29 chromosomes from unrelated families with exon 2 deletion of TMLHE (Table S1).
Genomic sequencing of exons for TMLHE is complicated by the presence of two pseudoexons (7aP and 8aP) that are highly homologous to exons 7a and 8a, and are imbedded in a large inverted repeat downstream of TMLHE (24) (Fig. 2C). In addition, there are two alternative exons 7b and 8b, which are located between the two inverted repeats and have sequences unrelated to 7a and 8a. Sequencing of exons 1–8 of TMLHE from genomic DNA in 536 SSC autism male probands, 98 AGRE probands, and 443 National Institute of Mental Health (NIMH) controls identified very few point mutations (Table S3). In addition to exon 2 deletion, sequencing in a multiplex AGRE family (AU 0177) identified an arginine-to-glutamine change in codon 241 (R241Q) in the mother and unaffected half-brother of the two autistic males (Fig. S1). More recently, we have been able to study plasma from 156 male SSC probands for carnitine biosynthesis metabolites. We identified one male (Table S3) with biochemical abnormalities similar to those described below, and sequencing identified an R70H mutation likely causing TMLHE deficiency.
Exon 2 Deletion Results in Loss of TMLD Activity and Absence of TMLD Protein.
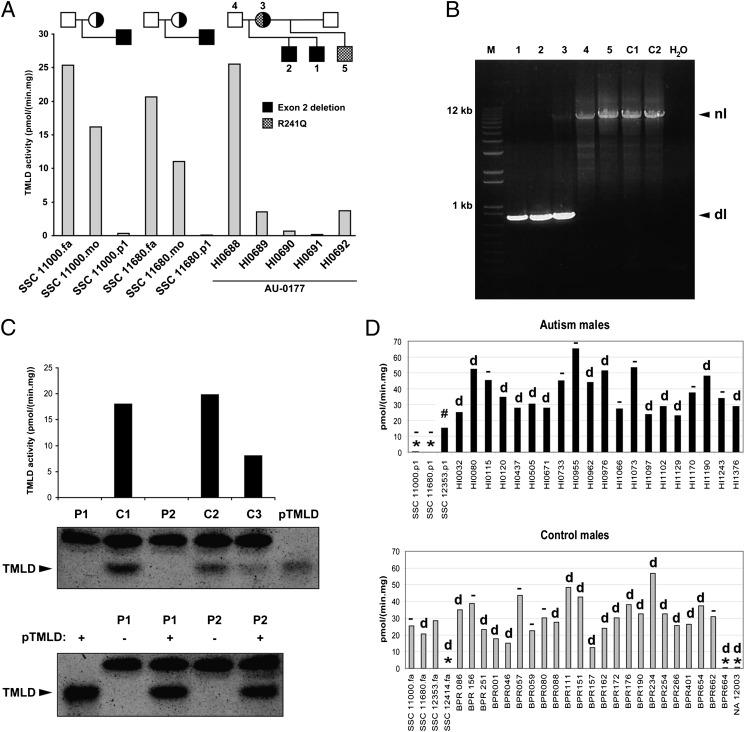
The functional effect of deletion of exon 2 of TMLHE was examined as TMLD enzyme activity based on its role in carnitine biosynthesis. Cultured LCLs from males with deletion of exon 2 had low or undetectable TMLD enzyme activity, and heterozygous mothers had reduced activity compared with healthy males (Fig. 3A). Results from family AU 0177 were complex, with the two affected brothers with deletion of exon 2 having very low or undetectable enzyme activity but the unaffected mother and half-brother showing low activity but higher than that of the affected brothers (Fig. 3 A and B). The mother is a compound heterozygote and transmitted the R241Q mutation to the unaffected half-brother of the siblings with autism. Cells with exon 2 deletion also lacked immunodetectable protein by Western blot analysis (Fig. 3C). Many of the control and autism cell lines in Fig. 3D have the intron 1 deletion, and many do not.
Fig. 3.
Genetic and enzymatic characterization of hemizygous deletion of exon 2. (A) TMLD activity measured in lymphoblast homogenates of three families with exon 2 deletion. (B) PCR assay results for the AU 0177 family showing the deletion in the two affected brothers (1, 2) and in the mother (3) but not in the father (4), unaffected maternal half-brother (5), or unaffected controls (C1 and C2). There is bias of amplification in the mother, such that the normal band is faint. dl, deletion; nl, normal. (C) (Upper) TMLD activity and Western blot analysis of 2 individuals with exon 2 deletion (P1, HI0690; P2, BPR664) and three controls (C1–C3). Purified TMLD (pTMLD) is used as a positive control. (Lower) Western blot analysis of 2 individuals (P1 and P2) with (+) or without (−) addition of pTMLD, showing the complete absence of protein in cases of exon 2 deletion and confirmation of the identity of the immunoreactive material as TMLD. The upper band in the Western blot is an irrelevant protein. (D) (Upper) TMLD activity measured in lymphoblast homogenates from several autism males. *,TMLHE exon 2 deletion; #, E287K; d, deletion in intron 1 in 13 individuals; −, no deletion in intron 1 in 9 individuals. SSC 12353.p1 was not tested for the presence of intron 1 deletion. (Lower) TMLD activity measured in lymphoblast homogenates from male controls. BPR indicates local unaffected controls, and NA 12003 is an unaffected individual. SSC 12353.fa was not tested for presence of intron 1 deletion. There was no apparent correlation of the level of enzyme activity with the presence or absence of the intronic deletion. For A, C, and D, assays were run in duplicate and the average is plotted without error bars. fa, father; mo, mother; p1, proband.
Analysis of RNA from LCLs using RT-PCR revealed low levels of skipping of exon 2 in most samples and a stable transcript with complete absence of exon 2 in cells from males with deletion of this exon (Fig. S2A). Thus, nonsense-mediated decay is not prominent for the exon 2 deletion transcript. This was confirmed using a quantitative RT-PCR assay, which showed normal levels of transcript for the exon 5/6 junction in all samples but complete absence of the exon 1/2 junction in deletion samples (Fig. S2B).
Diagnostic Metabolite Abnormalities in Plasma and Urine.
The two affected brothers from AGRE family AU 0177 had a normal facial appearance in childhood and were otherwise nondysmorphic. They both had normal plasma free carnitine levels (33 and 34 μmol/L, normal = 22–65 μmol/L) at the recent ages of 15 and 17 y. In urine of the affected brothers, HTML and γBB were undetectable and the excretion of TML was threefold that of controls (Fig. 4A). Plasma from the two brothers and from 5 SSC probands showed a significant increase in TML, complete absence of HTML, and severely reduced levels of γBB, except for one case (Fig. 4B). The (HTML + γBB)/TML ratio was very low in patients compared with control plasma and may be an excellent index of TMLD activity (Fig. 4C). These data indicate that TMLHE deficiency represents a unique inborn error of carnitine biosynthesis. To search for other evidence of a common TMLHE deficiency and for other defects in carnitine biosynthesis, urine from 29 SSC probands who did not have any known TMLHE mutations was studied and did not reveal any abnormal carnitine metabolites.
Fig. 4.
Increased TML and decreased HTML and γBB in patients with TMLHE exon 2 deletions. (A) Bar diagram of concentrations (mean ± SD) of carnitine biosynthesis intermediates in urine of two patients with exon 2 deletion (1, HI0690; 2, HI0691) compared with controls. (B) Box and whisker plot of carnitine biosynthesis intermediates in plasma of seven patients with exon 2 deletion (HI0690, HI0691, SSC 13928.p1, SSC 13489.p1, SSC 11000.p1, SSC 11229.p1, and SSC 11680.p1) compared with controls. (C) Box and whisker plot showing the diagnostic potential of the (HTML + γBB)/HTML ratio as an indicator of TMLHE deficiency. All seven patients have a very low ratio. The black square represents the mean of the controls, and whiskers show the minimum and maximum values of the control group.
Sex Ratio in NDA Is Not Caused by a Common Inherited Mutation in TMLHE.
It was important to determine whether there was a common mutation or epigenetic mechanism causing TMLD enzyme deficiency, and perhaps explaining the male predominance in some forms of autism. To address the possibility of a common but difficult to detect inherited mutation, we analyzed SNP data from Illumina arrays on 411 AGRE families; this revealed no evidence of linkage for TMLHE, which is the most telomeric gene on Xq that is not on Y, or for VAMP7, which is nearby but across the pseudoautosomal boundary and present on both the X and Y chromosomes. For the TMLHE region, a maximum nonparametric linkage score of 1.25 and logarithm of odds (LOD) score of 0.34 were observed for markers located within and flanking the TMLHE gene. For the VAMP7 gene region, a maximum nonparametric linkage score of 0.76 and LOD score of 0.15 were observed. Thus, there was no evidence for linkage at either locus. This is not surprising, given the extensive genetic heterogeneity in autism.
TMLHE Deficiency Likely Is a Risk Factor for Autism.
Because deletion of exon 2 was more common than any other mutations detectable by genomic sequencing, and because it was associated with loss of enzyme activity, it was expedient to analyze a large series of autism cases and controls for exon 2 deletion. A PCR assay was designed with primers slightly outside the boundaries of exon 2 to give a product of 538 bp in normal males but no product for males with deletion of exon 2. An example of the PCR assay with an internal control product is shown in Fig. S3. If a sample failed to give a PCR product for exon 2, the presence or absence of the deletion was then confirmed using CGH custom array with densely spaced oligonucleotides interrogating the TMLHE region, as shown in Fig. 2A. For PCR analysis, we focused entirely on males because the assay did not reliably detect the deletion in heterozygous females.
Using the PCR assay, we tested simplex male probands primarily from the SSC with lesser numbers of probands from the South Carolina Autism Project and from probands from Houston, TX. We also tested male controls, including SSC fathers of autism probands, NIMH controls, and Baylor Polymorphism Resource (BPR) controls. With the collaboration of the laboratories of two of the authors (J.S.S. and D.H.G.), we tested multiplex probands (here, multiplex refers to male-male sibling pairs both affected with autism) from the AGRE, NIMH, and Nashville collections. We subsequently developed collaborations with the laboratories of four of the authors (S.W.S., C.B., J.D.B., and M.E.H.) to expand the data for exon 2 deletion in autism male probands and control males. Some collaborating laboratories used quantitative PCR (qPCR) or existing Affymetrix 6.0 array data as the primary test for deletion of exon 2, as specified in Table 1. All deletion probands were validated, and approximate coordinates were determined using the custom TMLHE array. Deletions in control males from the laboratories of S.W.S. and M.E.H. were not validated.
Comparison of the data for male probands from simplex families (9 in 2,904 or 1 in 323 deleted) with all controls (24 in 8,787 or 1 in 366 deleted) did not provide evidence for an association (P = 0.44) (Table 1). One SSC proband (11680.p1) also had deletion of chromosome 16p11.2 and was eliminated from these calculations and from the phenotypic data. We hypothesized for numerous reasons reviewed in the discussion that the frequency of exon 2 deletion might be substantially higher in probands from male-male affected sib-pairs. The frequency of exon 2 deletion was 2.85-fold higher in these multiplex probands (7 in 909 or 1 in 130) compared with all male controls, with a P value of 0.023 (Table 1). For each multiplex family, only one affected male (identified as the proband) was tested initially. All genotypes were consistent with the X-linked inheritance; all 17 mothers of probands with deletion of exon 2 were tested and were heterozygous for the deletion, confirming that the deletions were not cell culture artifacts.
We next examined the affected male siblings of the multiplex male probands and found that six of the seven had the same deletion as the proband. Based on analysis using the Transmission Disequilibrium Test (TDT), the probability of obtaining this result, if there were no association, is 0.012. Using metaanalysis, we calculated a Stouffer’s z-statistic to combine the data for multiplex probands compared with control males and data from the TDT. We obtained a Z-score of 2.90 and a P value of 0.0037 using Stouffer’s method, suggesting that TMLHE deficiency is a risk factor for autism. If the data from the simplex families are included in the metaanalysis, the Z-score is −2.81 and the P value is 0.0051, which is only slightly higher than the P value of 0.0037.
Cognitive Function of TMLHE-Deficient Males with Autism Varies Widely.
Significant phenotypic information was available for seven SSC probands, two of three Canadian Genetic probands, seven SSC unaffected fathers, all seven multiplex probands and six affected brothers, and 3 NIMH control males with deletion of exon 2 of TMLHE (Tables S4 and S5). The levels of cognitive and language functioning varied considerably across patients. The full-scale IQ of autistic males with deletion of exon 2 ranged from 38 to 143; 5 of 21 with available data were in the range of intellectual disability, and 3 of 21 were reported as untestable. One proband had seizures. For six of six cases in which information was available, patients were described as nondysmorphic. With respect to the controls, two of the seven SSC fathers had at least one domain with an elevated broader autism phenotype score based on the self-report Broad Autism Phenotype Questionaire, but the Social Responsiveness Scale rating by significant other and Family History Interview-Interviewee Impression scores were not consistent with the broader autism phenotype.
Assuming a True Association, the Penetrance for Autism in TMLHE Deficiency Would Be Very Low.
The majority of males with an exon 2 deletion in the US and UK populations are expected to be phenotypically “normal” as adults. TMLHE deficiency was present in slightly less than 1% of probands from male-male affected sibling pairs; thus, it would be present in substantially less than 1% of all cases of autism. If we assume an overall frequency of 1 in 100 for autism, with a 4:1 male/female ratio, a frequency of 1 in 350 for TMLHE deficiency in normal males, and a frequency of TMLHE deficiency of 1 in 250 or 1 in 150 in males with autism, the penetrance would calculate at 2.2% or 3.6%, respectively (SI Materials and Methods and Table S6).
Discussion
TMLHE deficiency is a previously undescribed inborn error of metabolism discovered about 100 y after Garrod described such conditions in his 1908 Croonian Lectures to the Royal College of Surgeons. The frequency of TMLHE deficiency is startling, at ∼1 in 350 control males of European descent, making it at least 20-fold more frequent than phenylketonuria in males. The enzyme deficiency and metabolite changes in plasma and urine are typical for an inborn error of metabolism. TMLHE appears to be a gene in which deletions are much more common than point mutations. There is precedent for this at the DMD, PMP22, UBE3A, and other loci causing Duchenne muscular dystrophy, hereditary neuropathy with liability to pressure palsies, Angelman syndrome, and other phenotypes, respectively. These biases are usually explained, in part, by genome architecture, as is likely the case for TMLHE.
It might be of some concern that we did not detect any indication that TMLHE deficiency is a risk factor for simplex autism. However, simplex and multiplex groups of autism families are significantly different, with an expectation of higher rates of de novo mutations in simplex compared with multiplex families and a higher rate of inherited mutations in multiplex vs. simplex families. In addition, shared genetic modifiers and shared environment are potential factors in multiplex families. Given these differences between simplex and multiplex families, the apparent low penetrance of TMLHE deficiency for autism, and the modest sample size, the negative result for simplex families is not surprising. Assuming that the association with multiplex autism is replicated, we would expect that there would be a significant association with simplex autism males with a much larger sample size, because all multiplex families are initially simplex before the birth of a second affected sibling. We would argue that it is not appropriate in this circumstance of low penetrance to combine the probabilities from simplex and multiplex families for an association of TMLHE deficiency with autism, because they are significantly different samples. However, if one were to do so, the failure to detect an association in a sample of simplex families of this size, given the necessarily low penetrance, has relatively weak statistical significance and does not detract substantially from the P value of 0.0037 observed with male-male multiplex families. We conclude that TMLHE deficiency is likely to be a weak risk factor for autism, but replication studies are needed, particularly those focusing on male-male multiplex families. The data make it clear that TMLHE deficiency is neither necessary nor sufficient to cause autism. With roughly 4 million births per year in the United States, this would equate to about 5,600 deficient males born per year, which, in turn, would equate to 168 males with TMLHE deficiency and autism assuming a 3% penetrance.
One might ask whether carnitine metabolism plays a broader role in the etiology of NDA. One possibility is that TMLHE deficiency is entirely benign, as is generally believed to be the case for pentosuria and histidinemia. Alternatively, TMLHE deficiency could mediate harmful effects either through toxic accumulation of TML or through deficiency of downstream metabolites, including HTML, TMABA, γBB, or carnitine. All these are possible, but we believe that the most attractive hypothesis at this time is that there is an increased risk for autism, and that this risk is modified by dietary intake of carnitine from birth through the first few years of life. Carnitine intake of the pregnant or nursing mother could also be important. There are extensive reports of mitochondrial abnormalities in autism, as reviewed recently (25), and some of the mitochondrial dysfunction could be secondary to carnitine deficiency. There are reports of low plasma carnitine in autism (26–29), but these reports have not prompted intensive investigations into a possible role of carnitine deficiency in autism and further studies are needed.
Another hypothesis could be that other genetic abnormalities involving the carnitine pathway might confer a risk for autism. Features of autism generally are not reported in children with systemic carnitine deficiency, although cases of autism with carnitine deficiency have been reported (26). Given the neurological basis of autism and the prominent expression of TMLHE in hippocampal neurons and Purkinje cells, one possibility would be that symptoms of autism might be secondary to carnitine deficiency in the brain. If that were the case, the pathophysiology of systemic carnitine deficiency would be very different from TMLHE deficiency. The former has low plasma carnitine, but ability to synthesize carnitine in the brain and elsewhere is intact. In the latter, plasma carnitine may be normal or low-normal based on dietary intake, but neurons are unable to synthesize carnitine and become completely dependent on transfer across the blood–brain barrier. If carnitine deficiency in the brain was deleterious, dietary deficiency, excess renal losses, disorders of transport (especially across the blood–brain barrier), and defects in synthesis might be risk factors for autism. Relatively little is known about transport across the blood–brain barrier, but this transport may be a limiting factor, because the concentration of carnitine in cerebrospinal fluid is 10- to 15-fold lower than in plasma (30, 31). As shown in Fig. 1, not all tissues are capable of complete carnitine biosynthesis because of the differential expression of the last enzyme, γBB dioxygenase (γBBD), which is only expressed in kidney, liver, and brain in humans. After degradation of proteins that contain TML residues, TML is converted to γBB, which is then transported to the tissues that express γBBD and converted into carnitine. The plasma membrane γBB transporter likely is encoded by the SLC6A13 gene, which is known as a betaine/GABA transporter and has recently been suggested to function in carnitine biosynthesis as the liver γBB transporter (32). Transport of either or both carnitine and γBB across the blood–brain barrier could be important.
One important question is whether the association with autism is valid and can be replicated in future studies. Although TMLHE deficiency was discovered by a genome-wide molecular analysis, a P value of genome-wide significance is not needed here, because this is a simple test of the hypothesis that a newly discovered inborn error of metabolism is a risk factor for autism or not. The metaanalysis P value of 0.0037, indicating an association with multiplex autism, suggests that the data indicating an association are unlikely to have occurred by chance. If penetrance for autism is influenced by carnitine intake during infancy, the risk for autism associated with TMLHE deficiency may be greater in countries with a high frequency of vegetarian diets and lower meat or beef intake. China, India, and South Korea are all countries where some studies of the incidence of autism are available (22, 33, 34) and there is a more vegetarian diet and/or much lower beef intake.
Two clinical investigations are of immediate interest and are being initiated. One is studying carnitine metabolites in cerebrospinal fluid of infants with autism with and without TMLHE deficiency near the age of onset, and the other is treating very young infants with autism with and without TMLHE deficiency with carnitine or γBB supplementation. Whether increased carnitine intake before onset of autism might prevent the development of symptoms would require a more complex study. There is a recent report of a trial of carnitine supplementation in autism suggesting clinical improvement (35), but the study included some patients up to 10 y of age who might be unlikely to respond; it would be desirable to have data from very young patients, preferably nondysmorphic, with and without TMLHE deficiency. The data reported here suggest that TMLHE deficiency is a risk factor for NDA and that carnitine metabolism could be a target for therapeutic intervention in this and related disorders.
Materials and Methods
Human Subjects and DNA.
All work with direct involvement of human subjects was approved by the relevant institutional review boards or equivalents, and informed consent was obtained from all subjects. For SSC, AGRE, and NIMH samples, DNA derived from LCLs was obtained from the Rutgers University Cell and DNA Repository. Additional information is provided in SI Materials and Methods. The numbers of simplex probands, multiplex male-male sib pairs, and controls from various sources are specified in Table 1. Detailed information for cell culture and for identification of deleted probands and controls is given in SI Materials and Methods. For the screening of samples included in Table 1, we used PCR assays (Fig. S2), except for SSC samples, where Illumina arrays (36) were also used; for Toronto and the Wellcome Trust Case–Control Consortium, where Affymetrix 6.0 arrays were used; and for Paris and New York, where qPCR assays were used. All deletions in patients with autism were confirmed using custom arrays for the TMLHE region (Fig. 2A).
PCR and Sanger dideoxy-sequencing of TMLHE exons 1–8 was performed for 536 SSC male probands, 98 affected AGRE males from male-male multiplex families (brothers or half-brothers with the same mother), and 443 NIMH male controls (primers provided in Table S7).
CGH Array.
All arrays used in this study were designed and analyzed based on University of California, Santa Cruz (UCSC) Genome Browser hg18 (National Center for Biotechnology Information Build 36, March 2006). The coordinates found in tables and figures are converted to hg19 (Genome Reference Consortium: human GRCh Build 37, February 2009). An Agilent CGH custom array of design ID 028249 was used to confirm TMLHE deletions originally found by PCR assay or those that were detected by the 1M Illumina SNP array through a collaborative study of SSC families (36). The custom array design is available on the Agilent’s eArray website (www.agilent.com/genomics/earray). Analysis of CNVs was done using Agilent’s DNA Analytics software (version 4.0.76) with the following settings: aberration algorithm ADM-2, a minimum of three consecutive probes per region, and a minimum absolute average log2 ratio of 0.25 for any given region.
The protocol for DNA digestion, labeling, purification, and hybridization to the arrays followed the manufacturer’s instructions with some modifications, as described previously (37). Genomic DNA (800 ng) from the SSC individual and from a single male reference was used in the digestion. Each slide was scanned into an image file using the Agilent G2565 DNA Microarray Scanner at a 3-μm scan resolution. Each image file was quantified using Agilent Feature Extraction software (version 10.7.3.1). The Agilent custom-focused validation files were uploaded into the DNA Analytics software for analysis.
Enzyme Assays and Metabolite Determinations.
All individuals tested for TMLD enzyme activity were assayed for the presence or absence of exon 2 of TMLHE by PCR or CGH array. These included BPR controls, AGRE and SSC individuals, and a Centre d’Étude du Polymorphisme Humain control (NA12003) (38). TML was obtained from Sigma–Aldrich. [2H9]TML and [2H3]γBB were synthesized as described previously (11). [2H9]HTML was prepared enzymatically by incubating [2H9]TML with Neurospora crassa TLMD, heterologously expressed in Saccharomyces cerevisiae as described previously (39). The resulting mixture of [2H9]HTML and [2H9]TML was applied to Amicon Ultra 30-kDa filters (Millipore), and the deproteinized filtrate was used as an internal standard for TML and HTML. All other reagents were of analytical grade.
Lymphoblast pellets were homogenized in 10 mM Mops buffer containing 0.9% (wt/vol) NaCl, 10% (wt/vol) glycerol, and 5 mM DTT (pH 7.4). The protein concentration was determined by the method of Bradford (40) using human serum albumin as a standard. For measurement of TMLD and γBBD activities, the reaction mixture consisted of 20 mM potassium phosphate buffer containing 50 mM KCl, 3 mM 2-oxoglutarate, 10 mM sodium ascorbate, 0.5 mM DTT, 0.5 mM ammonium iron sulfate, 2.5 mg/mL BSA, 2 mM TML, and 0.2 mM [2H3] γBB at pH 7.4, with a final volume of 250 μL. The reaction was started by adding 50 μL of homogenate (target final protein concentration of 0.2 mg/mL for lymphoblast homogenates) to the reaction mixture and was incubated at 37 °C for 30 min. The reaction was terminated by the addition of ZnCl2 to a final concentration of 1 mM, and the reaction mixtures were placed on ice. The ZnCl2 solution also contained the following internal standards: 50 pmol of [2H9]HTML, 140 pmol of [2H9]TML, 140 pmol of [2H3]γ-BB, and 550 pmol of [2H3]carnitine. Subsequently, the reaction mixture was loaded onto an Amicon Ultra 30-kDa filter and centrifuged at 14,000 × g for 20 min to separate the metabolites (TML, HTML, γ-BB, and carnitine) from the enzymes and remove most of the proteins. The filtrate (100 μL) was derivatized with methylchloroformate, and the produced HTML was quantified using ion-pair ultra performance liquid chromatography (UPLC)-tandem MS essentially as previously described (11).
For determination of carnitine biosynthesis metabolites in plasma and urine, internal standards were added to each homogenate and derivatization was performed as described above. Plasma samples were deproteinized using an Amicon Ultra 30-kDa filter. Urine samples were directly derivatized, and TML, HTML, carnitine, and γ-BB were quantified using ion-pair UPLC-tandem MS as previously described (11). For immunoblot analysis, a Multiphor II Nova Blot electrophoretic transfer unit (Amersham Pharmacia Biotech) was used to transfer proteins onto a Protran nitrocellulose membrane (Whatman) as described by the manufacturer. After blocking of nonspecific binding sites with 50 g/L Protifar (Nutricia) and 10 g/L BSA in PBS with Tween 20 (1 g/L) for 1 h, the membrane was incubated for 2 h in the same buffer without Protifar with 1:3,000 dilution of rabbit polyclonal antibodies raised against human recombinant TMLD fused to maltose-binding protein (41). Detection was performed with IRDye 800-conjugated goat anti-rabbit antibody (LI-COR Biosciences) according to the manufacturer’s instructions. Membranes were then dried and scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences).
TDT, Metaanalysis, and Penetrance Calculations.
After 7 of 909 probands from multiplex male-male families were identified, the P value favoring a risk relationship of TMLHE deficiency to autism was 0.022 based on a one-sided Fisher’s exact test. This outcome could have occurred by chance or could have occurred because there is indeed a risk relationship. If the result occurred by chance, 3.5 of the seven siblings would be expected statistically to have the deletion, if all mothers are assumed to be carriers. If the result reflects a true risk relationship, a higher proportion, but not necessarily all, of the autistic siblings should have the deletion. Seven of the eight siblings carried the deletion. Statistical analysis of the sibling data was performed by implementing the TDT (42–45). Because the X chromosome is being analyzed, only transmissions from the mother are informative. We examined whether or not the deletion had been transmitted to the affected male sibling from his mother and applied McNemar’s χ2 (46) to the resulting 2 × 2 table to guard against significant results attributable to population substructure/admixture. To combine the results from the comparison of multiplex probands to controls (P = 0.023) and from the TDT analysis of affected male siblings (P = 0.012), metaanalysis was performed using Stouffer’s method (47). The metaanalysis resulted in a Z-score of 2.90 and a P value of 0.0037.
For estimating penetrance, we used a hypothetical population of 2 million individuals at risk with an equal number of males and females, as shown in Table S6. We assumed a frequency of autism of 1 in 100 with a 4:1 male/female ratio. We assumed that 1 in 350 normal males carried deletion of exon 2 of TMLHE. We then calculated the penetrance assuming that either 1 in 250 or 1 in 150 males with autism carries the deletion.
Supplementary Material
Acknowledgments
We thank all the families and the clinicians at the participating Simons Foundation Autism Research Initiative (SFARI) SSC sites and collaborators within the SSC Genetic Consortium; the principal investigators of the SSC Genetic Consortium are listed elsewhere (36). We also thank many other clinicians and families from around the world for making samples available. We are grateful for the participating AGRE Consortium families and all the resources provided by AGRE. We especially acknowledge access to the Illumina AGRE genotype data submitted by Dr. Hakon Hakonarson at the Center for Applied Genomics at The Children’s Hospital of Philadelphia. We thank Dr. Joachim Hallmayer for help in recruiting Autism Genome Project investigators. This study makes use of data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the data is available at www.wtccc.org.uk. J.S.S. acknowledges the assistance of Melissa Potter in the Computational Genomics Core for managing family, sample, genetic, and phenotypic data for samples examined at Vanderbilt University. A.L.B. thanks Dr. Karen Wang for help in working directly with a family, Drs. James Lupski and Huda Zoghbi for helpful discussions and critical reading of the manuscript, and Dr. John Belmont and Gladys Zapata for providing LCLs and DNA from the BPR collection. F.M.V. acknowledges assistance from the Metabolite/Mass Spectroscopy section of the laboratory Genetic Metabolic Diseases for technical assistance and L. IJlst for helpful discussions. C.B. acknowledges all the families and the clinicians participating in the Paris Autism Research International Sibpair study, particularly Christopher Gillberg, Marion Leboyer, Mary Coleman, Maria Rastam, Gudrun Nygren, and Richard Delorme. The AGRE is a program of Autism Speaks and is supported, in part, by Grant 1U24MH081810 from the National Institute of Mental Health (to Clara M. Lajonchere). Part of this work was supported by Grant SFARI 124827 from the Simons Foundation (to the investigators of the SSC Genetic Consortium) and Grant HD-37283 (to A.L.B) and Grant P30HD-0240640 from the National Institutes of Health. Part of this work was financially supported by the Fundação para a Ciência e Tecnologia, Lisbon, Portugal, by Grant SFRH/BD/38074/2007 (to. S.V.). Part of this work was supported by National Institutes of Health Grants R01 MH061009 and R01 NS049261 (to J.S.S.). Funding for part of this work was provided by the Wellcome Trust under Award 076113 and by Grant 077014/Z/05/Z. Funding for the Paris Autism Research International Sibpair study was provided, in part, by the Institut National de la Santé et de la Recherche Médicale, Fondation de France, Fondation Orange, Fondation pour la Recherche Médicale, Assistance Publique–Hôpitaux de Paris, and the Swedish Science Council.
Footnotes
Conflict of interest statement: Multiple authors are based in the Department of Molecular and Human Genetics at Baylor College of Medicine, which offers extensive genetic laboratory testing, and Baylor College of Medicine derives revenue from this activity. The department offers biochemical and molecular diagnostic testing for trimethyllysine hydroxylase, epsilon gene deficiency. P.B.S.C.-S., S.V., F.M.V., and A.L.B. have filed a patent related to some of the work reported.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120210109/-/DCSupplemental.
References
- 1. Conference Proceedings (2004) Carnitine. The science behind a conditionally essential nutrient. Proceedings of a conference. March 25–26, 2004. Bethesda, MD. Ann N Y Acad Sci 1033:1–197. [PubMed]
- 2.Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. Role of carnitine in disease. Nutr Metab (Lond) 2010;7:30. doi: 10.1186/1743-7075-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361:417–429. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slonim AE, et al. Dietary-dependent carnitine deficiency as a cause of nonketotic hypoglycemia in an infant. J Pediatr. 1981;99:551–555. doi: 10.1016/s0022-3476(81)80252-1. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt-Sommerfeld E, Penn D. Carnitine and total parenteral nutrition of the neonate. Biol Neonate. 1990;58(Suppl 1):81–88. doi: 10.1159/000243302. [DOI] [PubMed] [Google Scholar]
- 6.Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet. 2006;142C(2):77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strijbis K, Vaz FM, Distel B. Enzymology of the carnitine biosynthesis pathway. IUBMB Life. 2010;62:357–362. doi: 10.1002/iub.323. [DOI] [PubMed] [Google Scholar]
- 8.Evans JD, Jacobs TF, Evans EW. Role of acetyl-L-carnitine in the treatment of diabetic peripheral neuropathy. Ann Pharmacother. 2008;42:1686–1691. doi: 10.1345/aph.1L201. [DOI] [PubMed] [Google Scholar]
- 9.Sarma S, Gheorghiade M. Nutritional assessment and support of the patient with acute heart failure. Curr Opin Crit Care. 2010;16:413–418. doi: 10.1097/MCC.0b013e32833e10d4. [DOI] [PubMed] [Google Scholar]
- 10.Celestino-Soper PBS, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum Mol Genet. 2011;20:4360–4370. doi: 10.1093/hmg/ddr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaz FM, et al. Analysis of carnitine biosynthesis metabolites in urine by HPLC-electrospray tandem mass spectrometry. Clin Chem. 2002;48:826–834. [PubMed] [Google Scholar]
- 12.Miles JH, et al. Development and validation of a measure of dysmorphology: Useful for autism subgroup classification. Am J Med Genet A. 2008;146A:1101–1116. doi: 10.1002/ajmg.a.32244. [DOI] [PubMed] [Google Scholar]
- 13.Jacquemont M-L, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bon BW, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: One disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinawi M, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott FJ, Baron-Cohen S, Bolton P, Brayne C. Brief report: Prevalence of autism spectrum conditions in children aged 5-11 years in Cambridgeshire, UK. Autism. 2002;6:231–237. doi: 10.1177/1362361302006003002. [DOI] [PubMed] [Google Scholar]
- 22.Kalra V, Seth R, Sapra S. Autism—Experiences in a tertiary care hospital. Indian J Pediatr. 2005;72:227–230. [PubMed] [Google Scholar]
- 23.Boone PM, et al. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monfregola J, et al. Functional characterization of the TMLH gene: Promoter analysis, in situ hybridization, identification and mapping of alternative splicing variants. Gene. 2007;395(1–2):86–97. doi: 10.1016/j.gene.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. American Journal of Biochemistry and Biotechnology. 2008;4(2):198–207. [Google Scholar]
- 27.Mostafa GA, El-Gamal HA, El-Wakkad ASE, El-Shorbagy OE, Hamza MM. Polyunsaturated fatty acids, carnitine and lactate as biological markers of brain energy in autistic children. International Journal of Child Neuropsychiatry. 2005;2(2):179–188. [Google Scholar]
- 28.Lombard J. Autism: A mitochondrial disorder? Med Hypotheses. 1998;50:497–500. doi: 10.1016/s0306-9877(98)90270-5. [DOI] [PubMed] [Google Scholar]
- 29.Filipek PA, Juranek J, Nguyen MT, Cummings C, Gargus JJ. Relative carnitine deficiency in autism. J Autism Dev Disord. 2004;34:615–623. doi: 10.1007/s10803-004-5283-1. [DOI] [PubMed] [Google Scholar]
- 30.Rubio JC, et al. Cerebrospinal fluid carnitine levels in patients with Alzheimer’s disease. J Neurol Sci. 1998;155(2):192–195. doi: 10.1016/s0022-510x(97)00314-6. [DOI] [PubMed] [Google Scholar]
- 31.Shinawi M, Gruener N, Lerner A. CSF levels of carnitine in children with meningitis, neurologic disorders, acute gastroenteritis, and seizure. Neurology. 1998;50:1869–1871. doi: 10.1212/wnl.50.6.1869. [DOI] [PubMed] [Google Scholar]
- 32.Fujita M, et al. Hepatic uptake of gamma-butyrobetaine, a precursor of carnitine biosynthesis, in rats. Am J Physiol Gastrointest Liver Physiol. 2009;297:G681–G686. doi: 10.1152/ajpgi.00238.2009. [DOI] [PubMed] [Google Scholar]
- 33.Wong VC, Hui SL. Epidemiological study of autism spectrum disorder in China. J Child Neurol. 2008;23(1):67–72. doi: 10.1177/0883073807308702. [DOI] [PubMed] [Google Scholar]
- 34.Kim YS, et al. Prevalence of autism spectrum disorders in a total population sample. Am J Psychiatry. 2011;168:904–912. doi: 10.1176/appi.ajp.2011.10101532. [DOI] [PubMed] [Google Scholar]
- 35.Geier DA, et al. A prospective double-blind, randomized clinical trial of levocarnitine to treat autism spectrum disorders. Med Sci Monit. 2011;17:PI15–PI23. doi: 10.12659/MSM.881792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders SJ, et al. Multiple recurrent de novo copy number variations (CNVs), including duplications of the 7q11.23 Williams-Beuren syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou Z, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarroll SA, et al. International HapMap Consortium Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 39.Swiegers JH, Vaz FM, Pretorius IS, Wanders RJ, Bauer FF. Carnitine biosynthesis in Neurospora crassa: Identification of a cDNA coding for epsilon-N-trimethyllysine hydroxylase and its functional expression in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2002;210(1):19–23. doi: 10.1111/j.1574-6968.2002.tb11154.x. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Vaz FM, Ofman R, Westinga K, Back JW, Wanders RJ. Molecular and Biochemical Characterization of Rat epsilon-N-Trimethyllysine Hydroxylase, the First Enzyme of Carnitine Biosynthesis. J Biol Chem. 2001;276:33512–33517. doi: 10.1074/jbc.M105929200. [DOI] [PubMed] [Google Scholar]
- 42.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 43.Ewens WJ, Spielman RS. The transmission/disequilibrium test: History, subdivision, and admixture. Am J Hum Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- 44.McGinnis RE, Ewens WJ, Spielman RS. The TDT reveals linkage and linkage disequilibrium in a rare disease. Genet Epidemiol. 1995;12:637–640. doi: 10.1002/gepi.1370120619. [DOI] [PubMed] [Google Scholar]
- 45.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 46.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12(2):153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- 47.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM., Jr . Adjustment During Army Life. Princeton: Princeton Univ Press; 1949. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.