Abstract
The large chromosomal deletions frequently observed in cancer genomes are often thought to arise as a “two-hit” mechanism in the process of tumor-suppressor gene (TSG) inactivation. Using a murine model system of hepatocellular carcinoma (HCC) and in vivo RNAi, we test an alternative hypothesis, that such deletions can arise from selective pressure to attenuate the activity of multiple genes. By targeting the mouse orthologs of genes frequently deleted on human 8p22 and adjacent regions, which are lost in approximately half of several other major epithelial cancers, we provide evidence suggesting that multiple genes on chromosome 8p can cooperatively inhibit tumorigenesis in mice, and that their cosuppression can synergistically promote tumor growth. In addition, in human HCC patients, the combined down-regulation of functionally validated 8p TSGs is associated with poor survival, in contrast to the down-regulation of any individual gene. Our data imply that large cancer-associated deletions can produce phenotypes distinct from those arising through loss of a single TSG, and as such should be considered and studied as distinct mutational events.
Keywords: cancer genomics, chromosome 8p deletion, RNAi screen
Most cancer genomes contain large heterozygous deletions of uncertain biological significance. Early studies on the tumor-suppressor genes (TSGs) RB and TP53 suggested that such deletions can arise as a single mechanism for loss of heterozygosity and, consequently, it is often assumed that they provide a “second-hit” event to inactivate a single TSG (1). However, genomic approaches have not conclusively identified a definitive TSG within some cancer-associated deletions, raising the possibility that they occur through genomic instability or selection for the reduced activity of multiple genes. Even in chromosomal regions where a bona fide “two-hit” TSG has been identified, the large deletions often associated with loss of heterozygosity reduce the dosages of neighboring genes, which could in principle contribute to tumorigenesis in a haploinsufficient manner.
Large deletions encompassing regions of chromosome 8p are very common in human tumors (2, 3) and often occur together with 8q gains encompassing MYC (4). Previously, we validated the 8p gene DLC1, encoding a Rho GAP, as a TSG using a mouse model of hepatocellular carcinoma (HCC), confirming that its attenuation can serve as a driving oncogenic event (3). Although DLC1 is at an epicenter of deletions, these deletions are frequently much larger and reduce the dosages of tens or hundreds of genes, often encompassing the entire 8p22 cytoband and beyond (2, 5, 6). Indeed, multiple candidate TSGs have been proposed in the region (5–8). Here we explore the hypothesis that chromosome 8p deletions arise owing to selection for the attenuation of multiple genes.
Results
Chromosome 8p Deletions Are Frequently Large and Co-Occur with 8q Gains and 17p Loss.
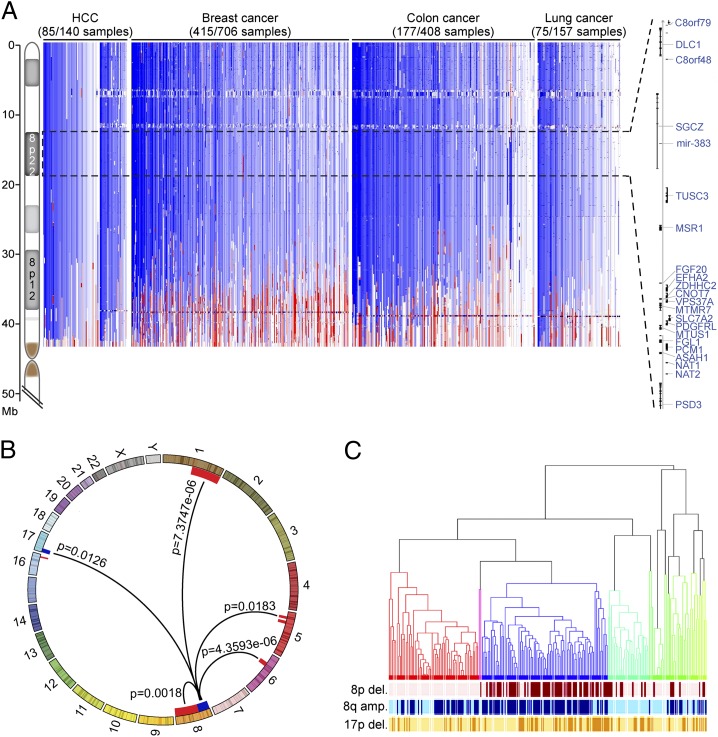
To better define regions affected by the 8p deletions frequently occurring in human cancers, we determined the extent of chromosome 8p deletions from cancer genome datasets derived from array-based comparative genomic hybridization (aCGH) performed at Cold Spring Harbor Laboratory and the Cancer Genome Atlas (TCGA) project, totaling 1411 primary tumor samples and cell lines of HCC and breast, colon, and lung cancers (Fig. 1A and Materials and Methods). According to these data, approximately half of these tumors harbor heterozygous deletions of human chromosome 8p, often encompassing a large portion of or even the entire chromosome arm (Fig. 1A). Focusing on 8p deletions in HCC, we noted that the most frequently deleted region on 8p centered around the DLC1 gene (Fig. 1A), that in HCC these deletions occur more frequently than those on chromosome 17p encompassing TP53 (3). However, this chromosome arm contains other candidate TSGs (5–8), and indeed, most deletions encompass regions adjacent to DLC1, including the whole 8p22 cytoband or even the whole chromosome 8p arm (Fig. 1A).
Fig. 1.
Chromosome 8p deletion characteristics and co-occurring genomic aberrations. (A) Size and extent of chromosome 8p deletions (in blue) and amplifications (in red) from individual HCCs, breast cancers, colon cancers, and lung adenocarcinomas based on aCGH data analysis (Materials and Methods). The 8p22 cytoband is highlighted by a dashed line, with the organization of the 8p22 genes indicated on the right. (B) Chromosome 8p deletions co-occur with genomic aberrations in HCC, including amplifications (red) of 1q, 5p, 6p, and 8q and deletions (blue) of 17p. Fisher’s exact test was used for statistical calculations. (C) Unsupervised hierarchical clustering of genomic aberrations indicates 12 groups within the HCC dataset (n = 197). Occurrence of 8p deletion (dark red), 8q amplification (dark blue), and 17p deletion (dark orange) within the individual samples is highlighted below the dendrogram.
To identify a relevant genetic context in which to study 8p loss, we analyzed 197 primary HCCs (3, 9, 10) for copy number aberrations associated with 8p deletions (Fig. 1B). Amplifications of chromosome 1q, 5p, 6p, and 8q (involving MYC) and losses including TP53 on 17p were significantly associated with 8p deletions (Fig. 1B). In addition, unsupervised hierarchical linkage clustering of 197 primary HCCs revealed that they fell within 12 groups, and that the 8p loss, 8q gain, and 17p loss cancers were clustered mainly within one subgroup that represents ∼40% of all HCCs (Fig. 1C). These data confirm that genotypes involving MYC overexpression and TP53 loss are a valid genetic context in which to study candidate 8p TSGs.
Chromosome 8p Harbors Multiple Genes That Inhibit Tumorigenesis in Mice.
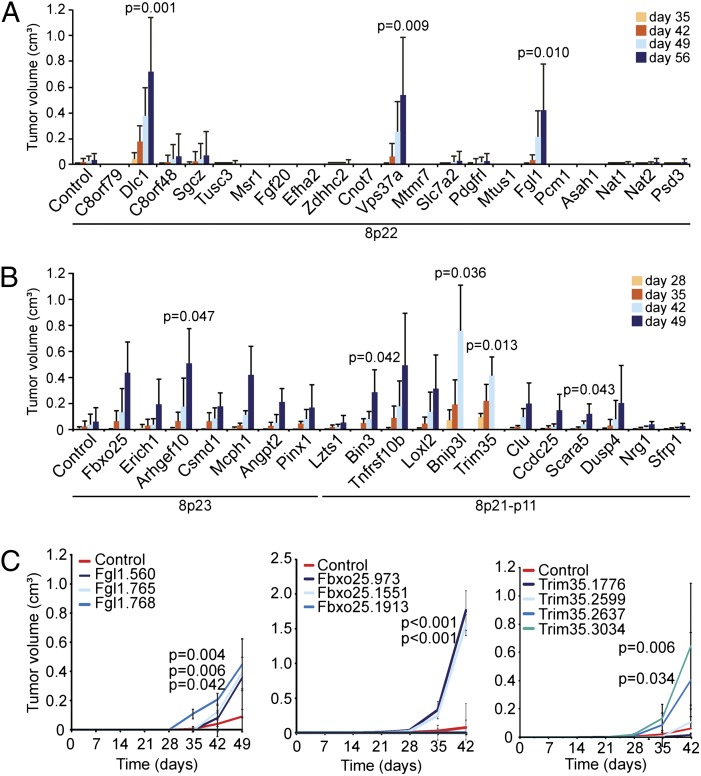
To identify TSGs located on 8p, we tested whether RNAi-mediated suppression of various 8p genes would promote tumorigenesis in a mouse HCC model used previously for TSG discovery (11). Initially focusing in an unbiased approach on the 8p22 region surrounding DLC1, we transduced pools of three shRNAs individually targeting each mouse ortholog of all 21 annotated 8p22 protein-coding genes into p53−/− liver progenitors overexpressing Myc, thereby approximating a relevant genetic context in human HCC progression. The resulting cell populations were then assessed for their tumorigenic potential (Fig. 2A). Whereas the parental cells transduced with control shRNA were only weakly tumorigenic, cells harboring three of the 8p22 pools, including one pool containing shRNAs targeting Dlc1, substantially promoted tumorigenesis above background (Fig. 2A). The two other scoring shRNA pools targeted fibrinogen-like 1 (Fgl1), a secreted protein of the fibrinogen family that is a candidate TSG in human HCC (12), and vacuolar protein sorting 37 homolog a (Vps37a), a component of the ESCRT-I complex mediating endosome sorting whose underexpression is associated with poor survival in HCC patients (13).
Fig. 2.
Chromosome 8p deletions target multiple TSGs. (A and B) Average volume of tumors derived from s.c. injected p53−/−;Myc immortalized liver cells infected with indicated shRNA pools. Error bars denote SD (n = 6). The Student t test comparing normalized samples at the time when mice were killed relative to controls was used for statistical calculations. (C) Average tumor volumes of s.c. injected p53−/−;Myc immortalized liver cells infected with indicated individual shRNAs. Error bars denote SD (n = 8). The Student t test comparing normalized samples at day 42 relative to control was used to calculate P values.
Although chromosome 8p22 is at a deletion epicenter in HCC, most 8p deletions span even larger regions (Fig. 1A). Thus, we questioned whether yet other candidate TSGs lay in these adjacent regions. Because there were too many genes to test individually, we used selection criteria based on high deletion frequency, underexpression in human HCC, and potential tumor-suppressive function, in accordance with the literature (Figs. S1 and S2 and Table S1). We then repeated our experiments using shRNA pools targeting the mouse orthologs of 19 genes from 8p23 and 8p21-p11. Surprisingly, shRNAs targeting many of these genes (14 of 19) promoted tumorigenesis over background, although with substantial variability in tumor incidence and size (Fig. 2B and Table S2). Five of these 14 genes demonstrated a statistically significant increase over background at the time of tumor harvest (Fig. 2B and Table S2).
For further validation of the original candidates, we subsequently tested the individual hairpins against the genes that showed significant tumor acceleration (i.e., Fgl1, Vps37a, Arhgef10, Bin3, Bnip3l, Scara5, and Trim35) plus one more (Fbxo25) that, although not statistically significant, yielded large tumors compared with control in a subset of mice (Table S2). Multiple shRNAs against Fgl1 or Vps37a that inhibited their corresponding targets promoted tumorigenesis in mice (Fig. 2C and Fig. S3); however, shRNA pools targeting Vps37a did not score consistently in all experiments, suggesting that it is a weak tumor suppressor or that its action is susceptible to subtle variations in experimental conditions. For most hits examined (e.g., Fbxo25, Fgl1, Trim35), the tumor-promoting effect of single shRNAs correlated closely with the observed level of knockdown (Fig. 2C and Fig. S3); however, this was not always the case (e.g., Arhgef10), perhaps indicating that some of the scoring shRNAs suppress translation, or that more complete gene suppression impacts an essential function, as we have described for Rad17 (14).
In addition to Fgl1 and Vps37a, among the genes with tumor-suppressive function validated in this way were the autophagy regulator Bnip3l (15), the F-box protein encoding Fbxo25 (16), and the ring finger, B-box, coiled-coil family gene Trim35 (17), which has been recently reported as an HCC TSG candidate (18) (Fig. 2C and Fig. S3). Also validated in these experiments were the candidate TSGs Scara5, which negatively regulates focal adhesion kinase signaling (19), and Arhgef10, previously implicated in breast and urothelial carcinomas (5, 6) (Fig. S3). Our screening assay was not without limitations, however; for example, none of the single hairpins against Bin3 accelerated tumorigenesis. Although this might indicate that the screening result was spurious, Bin3 shRNAs were also identified in a screen for TSGs in lymphoma (20), and Bin3 knockout mice are tumor-prone (21). Regardless, most hits identified in the screen were validated in follow-up assays.
To control for the validity and specificity of our gene selection criteria and screening approach, we repeated our experiments using shRNA pools targeting randomly selected genes on chromosome 8p, in contrast to the selection criteria (Figs. S1 and S2) that we applied earlier to enrich for potential candidate genes. In contrast to the high frequency at which candidates scored in our initial assay (Fig. 2B), only 1 of 17 shRNA pools targeting randomly selected chromosome 8p genes promoted tumorigenesis above background (Fig. S4A). We also tested shRNA pools targeting genes corresponding to human 5q31, a frequently deleted region in acute myelogenous leukemia, but not in HCC. Only 1 of these 23 targets scored in our assay, and suppression of this target had only weak tumor-promoting effects (Fig. S4B). The specificity of these results reinforces the biological relevance of our selection criteria to enrich for genes with tumor-suppressive function in our experimental system.
New TSGs Apparently Are Haploinsufficient.
Several of our observations imply that most of the validated genes that we identified are relatively weak tumor suppressors that function through a haploinsufficient mode of action. First, none of the new TSGs was as potent as the well-established TSG APC (Fig. S3A). Second, none of the scoring shRNAs tested was capable of complete knockdown, and the resulting tumors retained residual mRNA levels (Fig. S3B). Finally, and consistent with a previous report analyzing mutations of 8p21 genes (22), exon sequencing and methylation analysis of 13 matched tumor/normal HCC samples indicated that although 9 of these tumors harbored 8p deletions, neither DLC1 nor FGL1 showed evidence of somatic mutation or promoter hypermethylation. Except for ARHGEF10, DLC1, and SCARA5, none of these genes showed evidence of somatic point mutations in public databases (Table S3), and none approached the frequency of 8p deletion. Thus, it seems unlikely that any of these biologically active TSGs are subject to a two-hit mutational mechanism in HCC.
Individual 8p Genes Cooperate to Suppress Tumorigenesis.
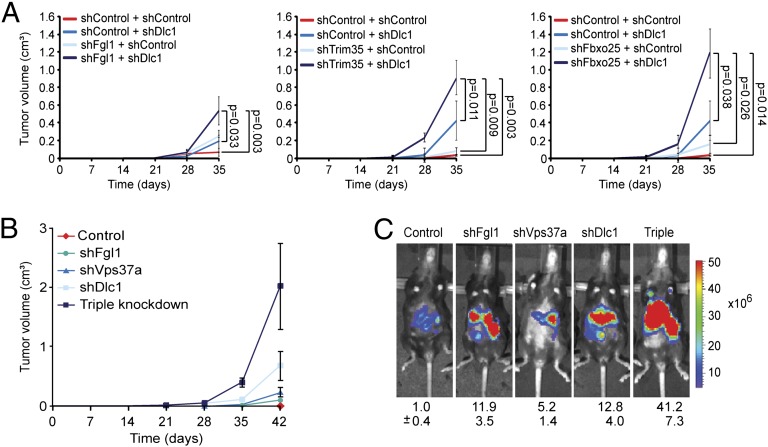
Our data imply that the impact of 8p deletions goes beyond the effects of DLC1 mutation or, for that matter, the attenuation of any individual gene. To determine whether the coattenuation of multiple genes with tumor-suppressive function produced additive or cooperative effects, we examined the impact of coattenuating DLC1—which is the only functionally validated 8p TSG in HCC (3) and at a deletion epicenter (Fig. 1A)—and the best-validated other TSGs from 8p22 (Fgl1) or adjacent regions (Fbxo25 and Trim35). Indeed, coattenuation of the 8p22 gene Fgl1, the 8p21 gene Trim35, or the 8p23 gene Fbxo25 with Dlc1 by cotransduction of shRNA vectors cooperated to promote tumor formation in mice (Fig. 3A). Consistently, cosuppression of the three scoring 8p22 genes (Dlc1, Fgl1, and Vps37a) synergistically accelerated tumor growth compared with single gene knockdown (Fig. 3 B and C). In addition, copy number loss was highly correlated with DLC1, TRIM35, and FBXO25 mRNA underexpression in primary HCC and invasive breast cancer, which is consistent with their codeletion in most epithelial tumors (Fig. S5 A and B). Collectively, these data imply that coattenuation of physically linked TSGs can cooperate during malignant transformation.
Fig. 3.
Cooperativity of 8p TSGs. (A) Average tumor volumes (n = 4) of s.c. injected p53−/−;Myc immortalized liver cells infected or coinfected with indicated shRNAs. Hairpins targeting Renilla served as controls. Error bars denote SD. Significance was calculated using the Student t test comparing normalized samples on day 35 relative to controls. (B) Average tumor volumes of s.c. injected p53−/−;Myc immortalized liver cells infected with indicated single shRNAs or coinfected with all three shRNAs (Fgl1, Vps37a, and Dlc1). Error bars denote SD (n = 4). Of note, given the experimental organization, the individual contribution of each TSGs in the triple-gene knockdown could not be determined. (C) Representative bioluminescence images from five mice with in situ liver tumors from intrasplenically injected p53−/−;Myc liver progenitor cells infected with indicated single shRNAs or triple-infected with Fgl1, Vps37a, and Dlc1 shRNAs. Numbers shown indicate mean ± SD intensities of luciferase signals (n = 5).
Down-Regulation of Multiple 8p TSGs Predicts Poor Survival.
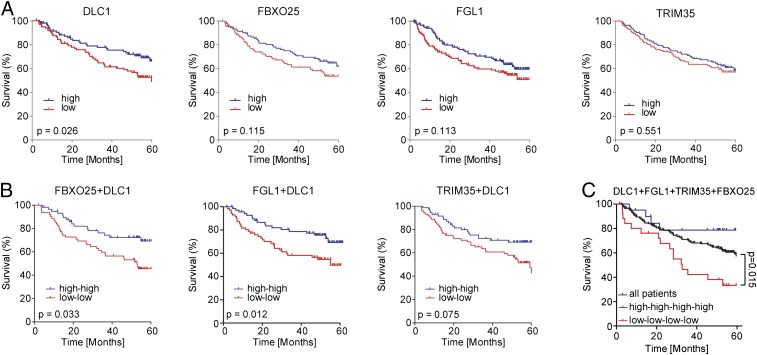
To substantiate our findings in human cancer, we next asked whether down-regulation of the validated individual 8p genes is associated with survival outcome in human HCC. We analyzed gene expression data from a cohort of 195 HCC patients with available survival data (23, 24). We found that although diminished expression of single genes or two genes together had no or only moderate association with survival outcome (Fig. 4 A and B), diminished expression of all four validated cooperating TSGs (DLC1, FGL1, FBXO25, and TRIM35) was significantly correlated with poor survival (Fig. 4C) and thus might predict more aggressive disease progression. The reduced copy number status of each of these four validated 8p genes was significantly associated with survival (Fig. S6A), which is consistent with previous reports examining the collective impact of 8p loss on survival (4, 25) and the fact each gene is invariably codeleted (Fig. S5A). Taken together, these data underscore the fact that chromosome 8p contains multiple genes—likely more than have been identified here—whose attenuated activity can promote tumorigenesis (Fig. S6B). Although each gene contributes only a modest effect, their combined attenuation may rival the impact of inactivating potent TSGs, such as APC, RB, and TP53.
Fig. 4.
Association of survival and 8p gene expression in patients with HCC. (A and B) Survival curves comparing high and low expression of indicated single genes (A) and combinations of two genes (B). (C) Survival association in patients with HCC (n = 192) for the entire cohort (all patients, black line) versus combined low expression (red line) or high expression (blue line) of DLC1, FGL1, TRIM35, and FBXO25. Statistical testing was performed as described previously (24).
Discussion
Our data suggest that some recurrent cancer-associated deletions reflect the selective advantage of simultaneously targeting multiple two-hit and/or haploinsufficient TSGs (1). Such a situation was described previously on chromosome 9p, although it was attributed mainly to the unique organization of the INK4a/ARF tumor-suppressor locus (26). Nonetheless, in other in vivo RNAi screens, we identified additional situations in which multiple genes with tumor-suppressive functions could be validated within the same genomic region (11, 20) and further analysis of our HCC data revealed that large deletions surrounding well-characterized TSG loci often encompass additional validated and/or candidate TSGs (Fig. S7), supporting our idea that the biology mediated by these large deletions goes beyond the effects of individual genes.
The extent and complexity of chromosome 8p deletions suggests that various candidate TSGs are targeted (5, 27, 28). Here we have shown experimentally that cosuppression of linked 8p TSGs promotes tumor formation more potently than any individual gene. Our data suggest that along with the frequency 8p deletions occurring in most epithelial tumors, 8p deletions specifically arise from selective pressure to attenuate the activity of multiple genes that may be separated by large distances, which explains the observed extent of the deletions at the copy number level. Although the possibility exists that genomic instability can fuel the losses that we observed on chromosome 8p and in other genomic regions, our data imply that these large events are selected for during tumor progression because of the presence of a discrete number of linked TSGs whose complete or partial attenuation individually may have only a modest effect on tumor growth. This provides an important nuance to the two prevailing views that a single gene in the region is the “driver” or that aneuploidy (chromosome imbalance) per se is crucial. The frequent large deletions on other chromosomes (e.g., 3p, 5q, 9p, 17p) suggest that deletion of linked cancer genes may play a broad role in cancer phenotypes.
None of the TSGs that we functionally identified showed evidence of somatic inactivation of the remaining allele at a frequency approaching that of their deletion, suggesting that these genes do not fit the canonical view of a TSG as defined based on studies of RB1 and TP53. Nonetheless, our functional data suggest that the expression of the 8p genes that we have identified is reduced in tumors with 8p deletions, and that the forced attenuation of these genes promotes malignant growth in an in vivo experimental system in a relevant cell type and genetic context. These results raise the possibility that large-scale genomic lesions can act through their effects on an opportunistic collection of linked genes rather than through disruption of a single resident gene. Given the extremely high frequency of these lesions in human cancers, this hypothesis warrants further investigation.
Although this study has addressed the origin of large somatic deletions occurring in human tumors, the notion that copy number alterations may frequently target multiple drivers likely extends to oncogenes located in common regions of amplification as well. Thus, work from our group and others has functionally validated multiple drivers in common amplicons that in some cases cooperate to produce more aggressive features and contribute to the maintenance of disease (29–32). Collectively, these data raise the possibility that methods integrating the complexity of copy number aberrations in tumors might be more accurate in predicting and delineating tumor behavior compared with methods that focus on individual genes (33, 34).
Whether there is a biological rationale underlying the physical linkage of some TSGs, and how their molecular function synergizes, are not clear. However, the fact that these genes can cooperate to suppress tumorigenesis implies that concomitant loss of multiple genes may create unexpected vulnerabilities not easily revealed through the study of single genes. Thus, codeletion of the 8p TSGs not only might create dependency on Rho signaling (3), but also might deregulate autophagy (15), ubiquitination (16), and other processes. Although the relevant biological effects remain to be determined and are the focus of ongoing work, our results demonstrate that cancer-associated deletions can create phenotypes unique from those arising through loss of a single TSG, and thus should be considered and studied as distinct mutational events.
Materials and Methods
Genomic Data Analysis.
We analyzed the aCGH data produced using representational oligonucleotide microarray analysis for the frequency and size of deletions in a series of human HCCs and breast, colon, and lung cancers available at Cold Spring Harbor Laboratory (3, 11, 33). We used this method to study gene dosage alterations in human HCC as described recently (33). Copy number aberrations (CNAs) were visualized from the individual representational oligonucleotide microarray analysis aCGH plots of the specific HCC samples using Integrated Genomics Viewer software (Broad Institute; http://www.broadinstitute.org/igv/home). In addition, available CNA from SNP6 arrays from the Cancer Genome Atlas (http://cancergenome.nih.gov/) for HCC and breast, colon, and lung adenocarcinomas were visualized using Integrated Genomics Viewer software and analyzed for the occurrence of chromosome 8p deletion.
Co-occurrence of gene deletions and amplifications in HCC was performed as described previously (34) by analyzing the aCGH dataset available at Cold Spring Harbor Laboratory combined with two previously reported HCC aCGH datasets (9, 10) publicly available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession nos. GSE19399 and GSE9845), totaling 197 primary HCCs and 12 HCC cell lines. In brief, statistically significant CNAs in HCCs were analyzed for frequency and co-occurrence in individual samples, and Fisher’s exact test was used to calculate P values for co-occurrence with chromosome 8p deletion.
Hierarchical clustering was performed by analyzing the combined HCC dataset (see above) using Nexus Copy Number software 5.1 (BioDiscovery), adding the significant CNAs as individual factors for each sample and using the complete linkage hierarchical cluster tool to group the samples based on the overall genomic aberrations. Subsequently, the annotated significant CNAs for individual samples were highlighted to visualize their occurrence within the clusters. In addition, Nexus Copy Number software 5.1 (BioDiscovery) was used to determine deletion frequencies of the 8p genes outside of 8p22 (Table S1).
Gene expression analysis was performed using the Oncomine database (www.oncomine.org), comparing multiple available HCC gene expression datasets. Comparison of copy number aberrations and gene expression was based on available TCGA datasets for HCC (53 samples) and invasive breast cancer (320 samples). Cancer genome datasets and bioinformatic tools for visualizing different parameters for analysis of genomic data are accessible through the MSKCC cBio Core homepage (www.cbioportal.org).
shRNA Design, Cloning, and Vector Construction.
miR30-shRNAs targeting murine orthologs of human 8p genes, which show synteny to mouse chromosomes 8A4-B2 (human 8p22), 8A1 (human 8p23), and 14D1 (8p21), were designed as described previously (35). miR30 design shRNAs were PCR-amplified from 97-mer oligonucleotides and cloned into MSCV-miR30-SV40-GFP or MSCV-miR30-PGK-Puromycin-IRES-GFP retroviral vectors (36) and sequence-verified. Myc was expressed using murine stem cell virus (MSCV) retroviral vectors (11). For double-cooperativity experiments, shRNAs were subcloned into MSCV-miR30-PGK-Neo-GFP or MSCV-miR30-PGK-Neo-mCherry.
Generation of Liver Carcinomas and Tumor Imaging.
Isolation, culture, and retroviral infection of murine hepatoblasts were described previously (32, 37). Liver progenitor cells from ED = 18 p53−/− fetal livers were immortalized with MSCV-based retroviruses expressing Myc–IRES-GFP or Myc-IRES-luciferase (11). To generate liver carcinomas, 2 × 106 ED = 13.5 liver progenitors were retrovirally transduced and transplanted into livers of female C57/B6 (age 6–8 wk) by intrasplenic injection or injected s.c. into NCR nu/nu mice. Hairpins targeting Renilla or an unspecific shRNA against human pRb were used as controls. Validation of single hairpin experiments (Fig. S3) were performed in newly derived liver progenitor cells from ED = 18 p53−/− fetal livers immortalized with MSCV-based retroviruses expressing Myc.
For double-cooperativity experiments, liver carcinomas were generated using liver progenitor cells immortalized with MSCV-based retroviruses expressing Myc by retroviral cotransduction of the two shRNAs in MSCV-based vectors containing either GFP or mCherry and subsequent s.c. injection of 1 × 106 cells into NCR nu/nu mice. The following shRNAs were used: shRen.713, shDlc1.3163, shTrim35.3034, shFbxo25.1551, and shFgl1.560. Triple-infection knockdown experiments were performed accordingly, except that 3× control virus was used, and single shRNA virus was mixed with 2× control virus to achieve comparable virus titers among the samples. Cells were injected either s.c. or intrasplenically. The tumor volume of s.c. tumors were calculated based on caliper measurements by the modified ellipsoidal formula: 0.5 × (length × width2). To address tumor penetrance, the number of tumors per injected site was counted (Table S2).
Intrasplenic injections and bioluminescence imaging for triple-knockdown experiments were performed as described previously (37–39).
Gene Expression and Survival Analysis from Human Samples.
The gene expression data of the HCC cohort have been published previously (23, 24). In brief, gene expression profiling was performed using the National Cancer Institute’s Human Array-Ready Oligo Set microarray platform and Affymetrix GeneChip HG-U133A 2.0 arrays, respectively. The microarray data are publicly available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession nos. GSE5975 and GSE14520). The gene expression of DLC1 and FGL1 was obtained from the Affymetrix arrays. TRIM35 and FBXO25 were not available on the Affymetrix platform and therefore the gene expression data of the National Cancer Institute’s Human Array-Ready Oligo Set microarray was used. For 195 patients, gene expression data were available on both microarray platforms, and for 192 patients, survival data as well as cause of death were available. Statistical analysis was performed as described previously (24).
Tissue Culture and Quantitative RT-PCR.
Retroviral-mediated gene transfer was performed using Phoenix packaging cells (provided by G. Nolan, Stanford University, Stanford, CA) as described previously (40). RNA purification and quanrititive RT-PCR (qRT-PCR) were performed as described previously (3). qRT-PCR reactions were done in triplicate using gene-specific primers. The expression level of each gene was normalized to β-actin or GAPDH. qRT-PCR primers were designed using PrimerBank (http://pga.mgh.harvard.edu/primerbank/) and are listed in SI Text.
Supplementary Material
Acknowledgments
We thank B. Ma, A. Shroff, and S. Muller for excellent technical assistance; Z. Xuan for help with statistics; L. Dow and M. Saborowski for critically reading the manuscript; and all members of the S.W.L. laboratory for critical discussions throughout the course of the study. T.K. is a recipient of German Research Foundation Postdoctoral Fellowship KI1605/1-1, W.X. is a recipient of an American Association for Cancer Research Centennial Predoctoral Fellowship, J. Zuber is a Seligson clinical fellow, S.W.L. is a Howard Hughes Medical Institute Investigator, and M.W. is an American Cancer Society Research Professor. This work was supported by a program project grant from the National Cancer Institute, a Cancer Target Discovery and Development consortium grant, and the Don Monti Memorial Research Foundation. M.W.’s support for this work comes from the Department of the Army (W81XWH04-1-0477) and the Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206062109/-/DCSupplemental.
References
- 1.Baker SJ, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum D, et al. Chromosome arm 8p and cancer: A fragile hypothesis. Lancet Oncol. 2003;4:639–642. doi: 10.1016/s1470-2045(03)01225-7. [DOI] [PubMed] [Google Scholar]
- 3.Xue W, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Gammal AT, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 5.Cooke SL, et al. High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer. 2008;8:288. doi: 10.1186/1471-2407-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams SV, et al. High-resolution analysis of genomic alteration on chromosome arm 8p in urothelial carcinoma. Genes Chromosomes Cancer. 2010;49:642–659. doi: 10.1002/gcc.20775. [DOI] [PubMed] [Google Scholar]
- 7.Berger AH, et al. Identification of DOK genes as lung tumor suppressors. Nat Genet. 2010;42:216–223. doi: 10.1038/ng.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorringe KL, et al. Are there any more ovarian tumor suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer. 2009;48:931–942. doi: 10.1002/gcc.20694. [DOI] [PubMed] [Google Scholar]
- 9.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang DY, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu HT, et al. Specific expression and regulation of hepassocin in the liver and down-regulation of the correlation of HNF1α with decreased levels of hepassocin in human hepatocellular carcinoma. J Biol Chem. 2009;284:13335–13347. doi: 10.1074/jbc.M806393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MW, et al. Expression of the HCRP1 mRNA in HCC as an independent predictor of disease-free survival after surgical resection. Hepatol Res. 2009;39:164–176. doi: 10.1111/j.1872-034X.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 14.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagens O, Minina E, Schweiger S, Ropers HH, Kalscheuer V. Characterization of FBX25, encoding a novel brain-expressed F-box protein. Biochim Biophys Acta. 2006;1760:110–118. doi: 10.1016/j.bbagen.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Kimura F, et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J Biol Chem. 2003;278:25046–25054. doi: 10.1074/jbc.M303438200. [DOI] [PubMed] [Google Scholar]
- 18.Jia D, et al. Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. Hepatology. 2011;54:1227–1236. doi: 10.1002/hep.24495. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, et al. Genetic and epigenetic silencing of SCARA5 may contribute to human hepatocellular carcinoma by activating FAK signaling. J Clin Invest. 2010;120:223–241. doi: 10.1172/JCI38012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scuoppo C, et al. A tumor suppressor network relying on the polyamine-hypusine axis. Nature. 2012 doi: 10.1038/nature11126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramalingam A, et al. Bin3 deletion causes cataracts and increased susceptibility to lymphoma during aging. Cancer Res. 2008;68:1683–1690. doi: 10.1158/0008-5472.CAN-07-6072. [DOI] [PubMed] [Google Scholar]
- 22.Mourra N, et al. High-resolution genotyping of chromosome 8 in colon adenocarcinomas reveals recurrent break point but no gene mutation in the 8p21 region. Diagn Mol Pathol. 2008;17:90–93. doi: 10.1097/PDM.0b013e31815bb5eb. [DOI] [PubMed] [Google Scholar]
- 23.Jia HL, et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 24.Roessler S, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren N, et al. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132:399–407. doi: 10.1007/s00432-005-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krimpenfort P, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 27.Pole JC, et al. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene. 2006;25:5693–5706. doi: 10.1038/sj.onc.1209570. [DOI] [PubMed] [Google Scholar]
- 28.Roessler S, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2011;142:957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan Y, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 30.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 37.Zender L, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.