Abstract
Cell differentiation requires integration of gene expression controls with dynamic changes in cell morphology, function, and control. Post-transcriptional mRNA regulation and signaling systems are important to this process but their mechanisms and connections are unclear. During C. elegans oogenesis, we find that two groups of PUF RNA binding proteins (RNABPs), PUF-3/11 and PUF-5/6/7, control different specific aspects of oocyte formation. PUF-3/11 limits oocyte growth, while PUF-5/6/7 promotes oocyte organization and formation. These two PUF groups repress mRNA translation through overlapping but distinct sets of 3’ untranslated regions (3’UTRs). Several PUF-dependent mRNAs encode other mRNA regulators suggesting both PUF groups control developmental patterning of mRNA regulation circuits. Furthermore, we find that the Ras-MapKinase/ERK pathway functions with PUF-5/6/7 to repress specific mRNAs and control oocyte organization and growth. These results suggest that diversification of PUF proteins and their integration with Ras-MAPK signaling modulates oocyte differentiation. Together with other studies, these findings suggest positive and negative interactions between the Ras-MAPK system and PUF RNA-binding proteins likely occur at multiple levels. Changes in these interactions over time can influence spatiotemporal patterning of tissue development.
Keywords: Translational control, PUF protein, Oogenesis, RNA-binding protein, Ras, Map Kinase, ERK, translation
INTRODUCTION
During development, cell differentiation depends on the coordination of many levels of gene expression with dynamic changes in numerous cell processes. Gene expression controls include not only transcriptional regulation but also control of specific mRNAs and their protein products in the cytoplasm. RNA regulation systems are critical to numerous developmental events, but how they integrate with each other and with signaling pathways to control differentiation is poorly understood.
Early metazoan development requires coordination of mRNA translation and localization with germ cell, oocyte, and early embryo development (Bettegowda and Smith, 2007; de Moor et al., 2005; Lasko, 2009). One example is the C. elegans germ line, where oocytes develop in a precise temporal and spatial pattern that depends on mRNA regulation. Mitotic germline stem cells at the gonad distal tip produce nuclei that enter meiosis, progress through early meiotic prophase, and differentiate into oocytes at the proximal end (Fig. 1A). This developmental program depends on specific patterns of translational control (Crittenden et al., 2002; D’Agostino et al., 2006; Ghosh and Seydoux, 2008; Jadhav et al., 2008; Jones and Schedl, 1995; Lublin and Evans, 2007; Mootz et al., 2004; Zhang et al., 1997). For many maternal mRNAs, specific 3’UTR elements and some 5’UTR elements are bound by sequence-specific RNA-binding proteins that repress translation at specific stages (Jadhav et al., 2008; Kershner and Kimble, 2010; Lee and Schedl, 2001; Lublin and Evans, 2007; Marin and Evans, 2003; Merritt et al., 2008; Mootz et al., 2004). For example, the KH protein GLD-1 functions in early meiosis and disappears upon pachytene exit, whereas the PUF protein PUF-5 is made at the beginning of pachytene exit and controls late steps of oogenesis (Hansen et al., 2004; Jones et al., 1996; Lublin and Evans, 2007; Marin and Evans, 2003). As a consequence, the fate of a single mRNA depends both on the combination if its UTR elements and the developmental pattern of RNA regulators that bind these specific RNA elements.
Fig. 1. PUF-3/11 and PUF-5/6/7 have different functions during late oogenesis.
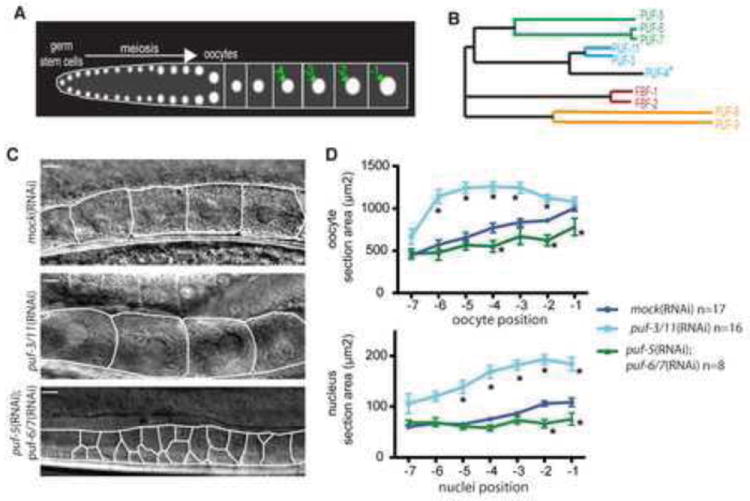
(A) Schematic of a C. elegans gonad. Distal tip to proximal end is left to right; oogonia and oocytes arise in the proximal arm. The oocyte in position -1 is the most proximal, fully differentiated oocyte prior to ovulation. (B) PUF proteins comprise four paralogous subgroups. PUF-4(asterisk) lacks two RNA-binding PUF repeats and likely has unique function. The puf-10 gene (not shown) of the PUF-5/6/7 group is a likely pseudogene (Lublin and Evans, 2007). (C) Normarski images of proximal gonad arms of mock(RNAi), puf-5;puf-6/7(RNAi) and puf-3/11(RNAi) animals. White lines delimit oocytes. In puf-3/11 animals, oocytes and nuclei were enlarged when compared to control. In puf-5(RNAi);puf-6/7(RNAi) animals, oocytes reside in multiple rows and vary in size. Scale bars are 10 um. (D) Quantification of oocyte cell and nucleus size: oocyte or nucleus area was plotted as a function of the oocyte position along the proximal gonad (as in Fig. 1A). Error bars represent the s.e.m for one representative experiment. *Denotes statistically significant differences from mock RNAi controls (p<0.01-0.001).
PUF proteins comprise a large, conserved family of RNA-binding proteins that control mRNA translation and stability from yeast to humans (Quenault et al., 2011; Wickens et al., 2002). PUF proteins control stem cells, cell fate patterning, differentiation, and neuron function (Kaye et al., 2009; Kimble and Crittenden, 2007; Kohlmaier and Edgar, 2008; Lublin and Evans, 2007; Quenault et al., 2011; Vessey et al., 2010; Walser et al., 2006; Wharton and Aggarwal, 2006). C. elegans has ten members, which can be clustered into 4 main branches: the PUF-8/9, FBF-1/2, PUF-3/11, and PUF-5/6/7 subfamilies (Fig. 1B) (Stumpf et al., 2008). Several PUFs function during the female phase of germline development in adult hermaphrodites. PUF-8 is expressed in the mitotic germ cells where it functions redundantly with MEX-3 to promote mitosis (Ariz et al., 2009). The FBF-1/2 group plays a key role in the maintenance of stem cell pools and their specification (Bernstein et al., 2005; Crittenden et al., 2002; Crittenden et al., 2003; Lamont et al., 2004; Suh et al., 2009; Wang et al., 2009). The PUF-5/6/7 subfamily controls later stages of oocyte differentiation (Lublin and Evans, 2007). The PUF-3/11 group, closest in homology to PUF-5/6/7, is the only branch for which no biological function has been described so far. Most PUF proteins contain 8 repeats of a three helix motif, each repeat binding a nucleotide in a sequence specific manner (Edwards et al., 2001; Wang et al., 2002; Wang et al., 2001; Wang et al., 2009). Each subgroup of the C. elegans PUF family binds related but distinct sequences (Bernstein et al., 2005; Koh et al., 2009; Opperman et al., 2005; Stumpf et al., 2008). However, their numerous mRNA substrates in vivo are only beginning to be uncovered (Crittenden et al., 2002; Kershner and Kimble, 2010; Lamont et al., 2004; Lublin and Evans, 2007; Merritt and Seydoux, 2010; Zhang et al., 1997).
The many functions of PUF proteins suggest coordination with signaling pathways that govern tissue specification and differentiation. In diverse eukaryotes, PUF proteins control mRNAs from several developmental signaling or patterning pathways (Kershner and Kimble, 2010; Kimble and Crittenden, 2007; Lasko, 2009; Lublin and Evans, 2007; Quenault et al., 2011; Walser et al., 2006; Wharton and Aggarwal, 2006). For example, PUF proteins in yeast, worm, and human cells bind and repress mRNAs for the Ras-MapKinase (Ras-MAPK) signaling pathway, leading in general to suppression of Ras-MAPK function (Lee et al., 2007a; Prinz et al., 2007; Whelan et al., 2011). In C. elegans germline stem cells, FBF-mediated repression of mpk-1 (MAPK) mRNA helps prevent premature activation of Ras-MAPK activity (Lee et al., 2007a). PUF functions must also themselves be regulated, and PUFs respond to signaling systems that control stem cell development, oocyte to embryo transitions, and early polarity (Johnstone and Lasko, 2001; Kimble and Crittenden, 2007; Kohlmaier and Edgar, 2008; Li et al., 2009). However, the mechanisms of PUF regulation are incompletely understood.
The Ras-MAPK pathway is not only a target but also a possible regulator of mRNA control systems. Ras is a small GTPase that induces a conserved pathway to activate the ERK (Map) protein kinases, which in turn phosphorylate downstream substrates (Chang and Karin, 2001; Moghal and Sternberg, 2003; Sundaram, 2006). Ras-MAPK is best known to control transcription, but also modulates post-transcriptional controls including several translation factors (Mahoney et al., 2009; Proud, 2007). In C. elegans, the Ras-MAPK pathway regulates multiple events during C. elegans oogenesis (Church et al., 1995; Lee et al., 2007b). Starting in late meiotic pachytene, MPK-1 undergoes waves of activation to promote meiotic progression, oocyte growth control, membrane and oocyte organization, and oocyte maturation (Church et al., 1995; Lee et al., 2007b; Ohmachi et al., 2002). These pleiotropic functions result from the coordinated phosphorylation of numerous substrates by MPK-1 during later stages of oogenesis (Arur et al., 2009). Most substrates likely control post-transcriptional events because transcription is inhibited in late oogenesis, an idea strongly supported by the substrates identified so far (Arur et al., 2009). Given the dominant role of translational control systems in oogenesis, Ras-MAPK may modulate specific RNA-binding complexes during this process. This idea is supported by a recent study showing direct inhibition of the translational control factor NOS-3 by MPK-1 (Arur et al., 2011).
In this study, we find that the PUF proteins PUF-3/11 and Ras-MAPK signaling combine with PUF-5/6/7 to control C. elegans oocyte differentiation. PUF-3/11 controls a similar stage of oogenesis as PUF-5/6/7, but these two PUF groups control distinct and specific aspects of oocyte formation. Furthermore, we find that PUF-3/11 and PUF-5/6/7 control overlapping but different sets of mRNAs. Remarkably, the Ras-MAPK pathway modulates oocyte growth and organization in part through promotion of translational repression by PUF-5/6/7. Ras and MPK-1 control PUF-5 protein modification or isoforms, and function with PUF-5 to regulate oocyte differentiation. Therefore, a network of mRNA repression pathways that are modified by Ras-MAPK signaling allows coordination of cellular processes essential to oocyte formation. This and other studies suggest that PUF proteins both regulate and are controlled by the Ras-MAPK pathway at different developmental stages. These interactions modulate the spatiotemporal pattern of germline development.
MATERIALS AND METHODS
Plasmids and strains
Strains were maintained by standard methods (Brenner, 1974). The following strains and alleles were used: N2 (Bristol wild-type strain); SD939, mpk-1(ga111) unc-79(e1068) III; SD551, let-60(ga89) IV; JH2270, Ppie-1gfp∷H2B∷UTRfbf-1; JH2296, Ppie-1gfp∷H2B∷UTRfbf-2; JH2207, Ppie-1gfp∷H2B∷UTRfog-2; JH2423, Ppie-1gfp∷H2B∷UTRfog-1; JH2436, Ppie-1gfp∷H2B∷UTRgld-1; JH2252, Ppie-1gfp∷H2B∷UTRglp-1; JH2377, Ppie-1gfp∷H2B∷UTRmes-3; JH2221, Ppie-1gfp∷H2B∷UTRmex-5; JH2200, Ppie-1gfp∷H2B∷UTRnos-3; JH2320, Ppie-1gfp∷H2B∷UTRpgl-1; JH2349, Ppie-1gfp∷H2B∷UTRpgl-3; JH2427, Ppie-1gfp∷H2B∷UTRpos-1; JH2311, Ppie-1gfp∷H2B∷UTRspn-4, as described (Merritt et al., 2008). A puf-5(RNAi) feeding vector (pTE7.30) was generated by cloning the full length open reading frame (ORF) of puf-5 cDNA into L4440. For simultaneous RNAi of puf-5 and puf-6/7, a puf-5 cDNA fragment (32-580 of puf-5 ORF) and a puf-6 cDNA fragment (847-1440 of the ORF) were cloned together into L4440 to make pTE7.31. To deplete PUF-3/11, two alternative feeding vectors were used. The first vector was generated by the fusion of a fragment from puf-3 cDNA (15-610 of puf-3 ORF) and a fragment from puf-11 cDNA (685- 1378 of puf-11 ORF). The second vector, as well as the let-60(RNAi) feeding vector, come from the C. elegans RNAi genome library (Kamath and Ahringer, 2003). All DNAs were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Center.
RNAi, microscopy, and fluorescence imaging analysis
RNAi was performed by feeding as described previously (Barbee and Evans, 2006; Timmons et al., 2001). For each experiment, L4440 with no insert (empty vector; “mock” RNAi) was used as a control. For RNAi experiments conducted with the temperature sensitive strains mpk-1(ga111ts) and let-60(ga89gf) and matching N2 controls, L4 worms were grown on feeding plates at 15°C for 36 h, then transferred as adults at 25°C for 12h. Otherwise, L4 worms were grown at 20°C for 30 h, then at 25°C for 18h. An RNAi screen for new genes that control RNP granules, which led to discovery of PUF-3/11, will be described elsewhere (A. Hubstenberger, C. Cameron, S. Noble, T.C. Evans, unpublished observations).
Epifluorescence images were acquired at room temperature with an Axioskop microscope (Carl Zeiss, Inc.) using a 40× 1.4 NA Plan Apochromat objective, imaged with Axiocam MRm (Carl Zeiss, Inc.), and quantified with Axiovision 4.6.3 software (Carl Zeiss, Inc.). For each experiment, mock and RNAi depleted animals were processed in parallel, and UV intensity, exposure times, and display adjustments were matched. For imaging and quantification of GFP reporter transgenes, worms were transferred onto 2% agarose pads with M9 buffer plus 30 mM NaN3. The GFP maximum intensity was determined in each nucleus by drawing a profile line across the nucleus. For immunofluorescence (IF), animals were dissected, fixed, and stained with antibodies as described previously (Barbee et al., 2002). Antibodies used include rabbit anti–GLP-1 (gift from J. Kimble, University of Wisconsin–Madison, Madison, WI; (Evans et al., 1994)), rabbit anti-GLD-1 (gift from T. Schedl, Washington University School of Medicine at St. Louis, St. Louis, MO; (Jones et al., 1996)), rabbit polyclonal anti-PUF-5 (Lublin and Evans, 2007). To quantify GLP-1 expression in each gonad, the pixel intensity mean was measured by drawing a 50 μm diameter circle in the most intense proximal oocyte and in the distal tip. PUF-5 boundaries were determined by drawing a profile line to measure PUF-5 intensity along the gonad long axis. Position of the PUF-5 boundary was determined by counting number of DAPI stained nuclei between PUF-5 boundary and the last nucleus localized in the gonad cortex.
Oocyte and nucleus area was measured either in intact worms or in extruded gonads. Both approaches gave similar results. For dissection, worms were picked onto a coverslip into a drop of M9 buffer containing 30 mM NaN3 (Fisher Scientific), and cut behind the pharynx to extrude gonads. The coverslip was inverted onto an agarose pad. The area of nucleus sections was determined by drawing a circle superimposed with the nuclear envelope, and the area of the oocyte sections was determined by drawing a line along the oocyte plasma membrane.
RNA extraction and RT-PCR
One hundred worms were washed twice with M9 and once with H2O. RNA was extracted using Trizol LS (Invitrogen). cDNAs were generated using random hexamers with the Superscript III First-Strand Synthesis System (Invitrogen). Real-Time PCR was performed in triplicate using SYBR Green Reagent on a StepOnePlus Real-Time PCR System (Applied Biosystems). Primers were designed around exon-exon junctions to avoid amplification of contaminant genomic DNA using the Primer Express 3.0 software. For gfp, we used primers 5′-AGGTGATGCAACATACGGAAA-3’ and 5’-AAGCATTGAACACCATAACAG-3’. For act-1, we used primers 5’- TTGCCCCATCAACCATGAA-3’ and 5’-CCGATCCAGACGGAGTACTTG-3’. Results were analyzed using the StepOne Software v2.1 (Applied Biosystems). Relative mRNA levels were calculated using the Quantitation Comparative CT (ΔΔCT) method and normalized to actin mRNA levels. Results were confirmed for at least two independent RNAi experiments. Semi-quantitative PCR was performed as described previously (Noble et al., 2008).
Western blot
Hermaphrodites were bleached and the remaining newly hatched L1 worms were starved for 24 h at 15°C on unseeded plates to synchronize the population. Worms were then grown at 15°C on NGM plates until reaching the L4 stage, washed 4 times in M9 buffer Carbenicillin 25 μg/mL, and transferred onto RNAi plates. Worms were grown for an additional 36 h at 15°C and then 12 h at 25°C. Worm extracts were prepared by boiling worms in a Laemli buffer. GLP-1 proteins were separated on a 7% SDS-PAGE and visualized by western blot using 1:2000 affinity purified anti-GLP-1 (Rabbit) and 1:10 000 HRP-conjugated anti-rabbit antibodies. PUF-5 proteins were separated on a 8.5% SDS-PAGE and visualized by western blot using 1:6000 affinity purified anti-PUF-5 (Rabbit) and 1:10 000 HRP-conjugated anti-rabbit antibodies.
Electrophoretic mobility shift assay (EMSA)
MBP∷PUF proteins were purified as described (Lublin and Evans, 2007). EMSA reactions were incubated at room temperature by mixing 32P end-labeled RNA oligos at 10nM with MBP∷PUF proteins at the indicated concentrations in binding buffer (20 mM HEPES pH 7.3, 50 mM KCl, 1 mM EDTA, 2 mM DTT, 0.1 mg/ml BSA, 0.02% Tween 20) for 1.5 h. Samples were mixed with loading buffer (5% glycerol final), and loaded onto 5% native gel. Gels were run at 4 watts for 1 h, fixed (30% Ethanol, 10% acetic acid) for 10 min, dried, exposed to a storage phosphor screen for 30 min, and scanned on a Typhoon 9400 (Amersham Biosciences). Experiments were repeated three times and linear regressions were generated using Prism 5 to estimate Kd values (GraphPad Software).
RESULTS
PUF-3/11 and PUF-5 regulate distinct biological processes during late stages of oogenesis
From an RNAi screen for genes that control RNA-Protein (RNP) complexes, PUF-3 was identified as a new RNP regulator during late oogenesis (A. Hubstenberger, C. Cameron, S. Noble, T.C. Evans, unpublished observations). PUF-5 and its close paralogs PUF-6/7 function redundantly at a similar stage to control oocyte differentiation and organization (Lublin and Evans, 2007). Additionally, among the PUF subfamilies, the PUF-5 and PUF-6/7 group is most homologous to the PUF-3/11 subfamily (Fig. 1B). A sixth PUF, PUF-4, also resides within the PUF-3/11 subgroup, but PUF-4 lacks two PUF repeats critical to RNA-binding and is thus unlikely to perform a similar function. We therefore analyzed the role of PUF-3/11 in oogenesis and compared it to PUF-5/6/7, after RNAi depletion under identical conditions. We will refer to puf-3(RNAi) as puf-3/11(RNAi) because of 90.2% nucleotide identity between puf-3 and puf-11, and because no differences were seen among puf-3(RNAi), puf-11(RNAi) and puf-3;puf-11(RNAi) phenotypes. As was seen previously, puf-5;puf-6/7(RNAi) induced smaller disorganized oogonia and oocytes that failed to enlarge their nuclei (Fig. 1C,D). Most puf-5(RNAi) animals have wild type gonads and viable progeny, and a low percentage show oocyte misorganization (Fig. 2; Fig. 6C,G). By contrast, puf-3/11(RNAi) caused premature enlargement of oocyte cell size and nuclear size while oocyte organization was maintained (Fig. 1C,D). Nuclei in puf-3/11(RNAi) oocytes were larger than RNAi control oocytes but contained diakinetic condensed chromosomes (Fig. 2L). In addition, puf-3/11(RNAi) caused 100% embryonic arrest (n=154), as did depletion of puf-11 (n>50). Some phenotype differences could result from differences in RNAi depletion. However, RNAi of PUF-5 alone caused oocyte misorganization at low penetrance but “large oocyte” phenotypes were never seen, whereas PUF-3/11 depletion cause highly penetrant “large oocyte” phenotype but never mis-organization characteristic of puf-5;puf-6/7 RNAi. These results suggest that PUF-3/11 and PUF-5/6/7 control a similar stage of oogenesis but influence distinct aspects of oocyte differentiation. PUF-3/11 limits or delays oocyte and nuclear growth while PUF-5/6/7 is essential for oocyte growth, nucleus transformations, and cell organization.
Fig. 2. PUF-3/11 and PUF-5/6/7 repress translation through common and distinct 3’UTRs.
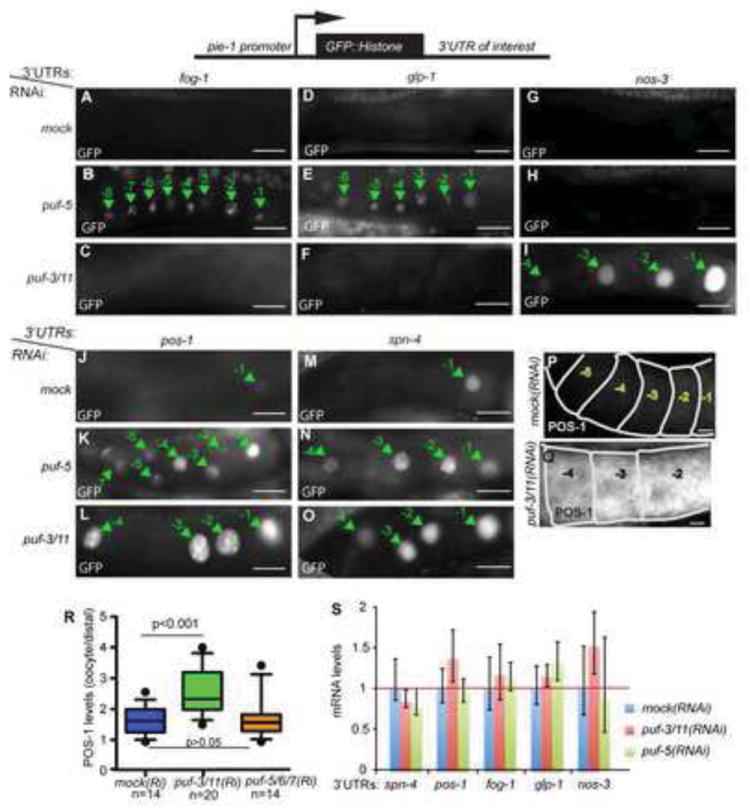
(A-O): Fluorescence micrographs of GFP∷H2B expressed from reporter transgenes under the control of the indicated 3’UTRs are shown for proximal gonad arms of live worms; oocyte nuclei (green arrows) are numbered as in Fig. 1A. Scale bars are 20 μm. Bottom diagram shows summary for RNAi effects on 3’UTR reporters, in which the top row summarizes GFP expression in control worms (white=no expression, grey=mild, black=strong) and bottom rows with colored boxes summarize expression of different reporters after RNAi depletions (at left); green=no change, red=increased expression. (P-Q) POS-1 protein staining after empty vector (mock) RNAi (P) or puf-3/1(RNAi) (Q). (R) Whisker plot (10-90th percentile) quantifying POS-1 fluorescence staining; puf-3/11(RNAi) levels were significantly higher but puf-5;puf-6/7(RNAi) levels were not. (S) mRNA levels were quantified after different RNAi depletions for the indicated 3’UTR reporters by real time quantitative RT-PCR. Each bar represents an average of three replicates. No significant increases in relative mRNA levels of puf-5(RNAi) or puf-3/11(RNAi) compared to mock RNAi were detected for all reporters (p>0.05). The experiment was repeated at least twice with similar results.
Fig. 6. Ras/MPK-1 and PUF-5 functionally interact to control oocyte organization and growth.
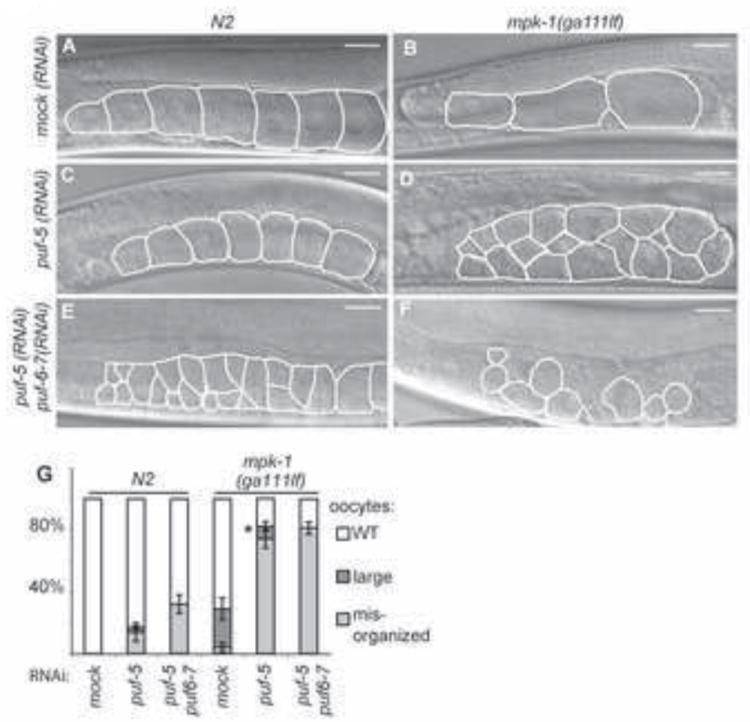
(A-F) Normarski images of hermaphrodites of the indicated genotypes and RNAi conditions are shown. White outlines mark oocytes. Scale bars are 20 μm. (G) Penetrance of phenotypes were quantified. Each value represents an average of at least 3 independent experiments. *Indicates statistically significant difference of mpk-1(ga111ts);puf-5(RNAi) from either puf-5(RNAi) or mpk-1(ga111) single mutant/RNAi (p<0.001). “Oocyte misorganization” in mpk-1(ga111ts);puf-5;puf-6/7 gonads was different and more severe than all other strains (as shown in F); oocytes were smaller and individual cells were difficult to define.
PUF-3/11 and PUF-5 repress overlapping but distinct mRNAs
The different roles of PUF-3/11 and PUF-5/6/7 in oogenesis suggest that these PUF proteins have at least some different mRNA targets. To test this prediction, PUF-3/11 and PUF-5 were depleted in transgenic strains that express reporter mRNAs encoding a GFP:HIS(Histone2B) fusion protein under control of different 3’UTRs (Merritt et al., 2008). Each reporter transgene was driven by the pie-1 promoter. The HIS tag targets GFP to nuclei (Fig. 2). Loss of PUF-5 alone disrupted repression by the glp-1 and fog-1 3’UTRs (Fig. 2B,E; Fig. 4D, Fig. 5C). PUF-5 repression of these 3’UTRs was predicted by previous studies, which showed PUF-5/6/7 repression of endogenous glp-1 translation and PUF-5 binding in vitro to an element from the fog-1 3’UTR (Lublin and Evans, 2007; Stumpf et al., 2008). By contrast, puf-3/11(RNAi) caused strong de-repression of reporter mRNA carrying the nos-3 3’UTR, but did not perturb repression by either the glp-1 or fog-1 3’UTRs (Fig. 2C,F,I; Fig. 4D,G). Repression by the nos-3 3’UTR was not altered in all gonads after either puf-5(RNAi) or puf-5;puf-6/7(RNAi)(Fig. 2H; Fig. 4G; n>50 for both conditions). Together with their phenotype differences, the opposite effects of PUF loss on fog-1 (and glp-1) compared to nos-3 reporters suggest that PUF-3/11 and PUF-5/6/7 regulate distinct sets of mRNA targets (compare Fig. 2A-F with 2G-I; and Fig. 4D to 4G). This conclusion is further supported by differences in RNA-binding specificity seen in vitro (Koh et al., 2009). Surprisingly, both PUF-3/11 and PUF-5 were required for full repression by the pos-1 and spn-4 3’UTRs (Fig. 2K,L,N,O). Thus, these related PUF proteins may co-regulate some mRNAs, directly or indirectly. The levels of all reporter mRNAs tested were not significantly altered following PUF-3/11 or PUF-5 depletion (Fig. 2S). Localization of these mRNAs was not investigated, although glp-1 and pos-1 mRNAs are distributed throughout wild type gonads and disruption of glp-1 3’UTR elements does not alter distribution (Crittenden et al., 1994; Evans et al., 1994; Noble et al., 2008). Therefore, these data support PUF-mediated repression of mRNA translation.
Fig. 4. Ras functions with PUF-5 to repress specific 3’UTRs.
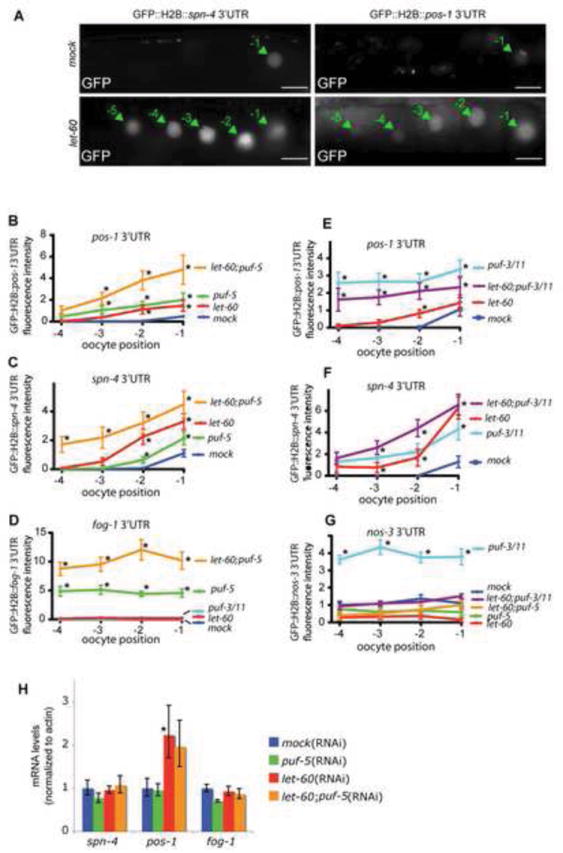
(A) Fluorescence imaging of GFP∷H2B reporters under the control of the indicated 3’UTRs in the proximal gonad of live worms. Green arrows point to oocyte nuclei. Numbers refer to their position as defined in Fig. 1A. Note that, in pos-1 and spn-4 control worms, the GFP is weakly detected in the nucleus of the last oocyte in some gonads. Scale bars are 20 μm. (B-G) Quantification of the let-60/Ras(RNAi) effects on different 3’UTRs. Each graph plots GFP maximum intensity in nuclei against oocyte position for the indicated single or double RNAi conditions for one experiment (n=8-24 gonads for all graphs; Error bars are s.e.m.). Similar results were obtained for at least two independent experiments. *Denotes statistically significant differences (p<0.01-0.001) from empty vector (mock) RNAi controls. (H) Quantification of mRNA levels for the 3’UTR reporters as determined by real time quantitative RT-PCR as in Fig. 2S; pos-1 reporter mRNA levels were significantly increased (*, p<0.01).
Fig. 5. The Ras-MAPK pathway functions with PUF-5 to repress endogenous glp-1 mRNA.
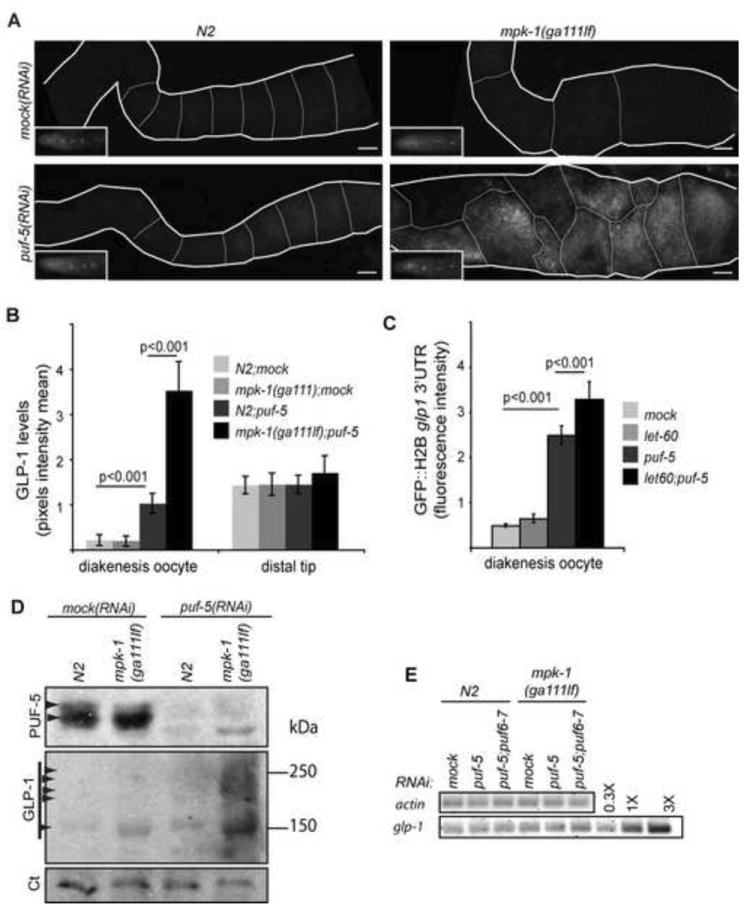
(A) Dissected gonads were stained by immunoflorescence with α-GLP-1. Only the proximal part of gonads is shown. Gonads are outlined by thick white lines, oocytes by thin white lines. Gonads were stained in parallel from the same experiment with matched exposure times and adjustments. Insets: GLP-1 staining in the tip of the distal gonad was used as an internal control. Scale bare: 50 μm. (B) GLP-1 protein expression was quantified by measuring GLP-1 pixel intensity mean in a 50 μm diameter circle of the most intense proximal oocyte and normalized to the distal tip. Strains were N2 or mpk-1(ga111ts), with empty vector RNAi (mock) or puf-5(RNAi) (puf-5) (n=8-24 for each strain/RNAi). Error bars represent s.d. for one experiment. Similar results were obtained with three independent experiments. (C) Gonads after the indicated single or double RNAi were dissected and GFP levels of the glp-1 3’UTR reporter were measured as in Fig. 4 (n=10-18 for each strain/RNAi). Error bars reflect s.d. between animals for one experiment. Similar results were obtained for 4 independent experiments. (D) GLP-1 and PUF-5 expression was assessed by western blot in the indicated strains. Total protein was extracted from the same number of synchronized adult hermaphrodites. puf-5 RNAi strongly reduced both of two PUF-5 bands (arrowheads). GLP-1 runs as multiple diffuse bands (arrowheads) due to glycosylation (Crittenden et al., 1994). Ct: loading control from same blot. (E) Semi quantitative RT-PCR of endogenous glp-1 mRNA extracted from equal numbers of whole worms showed no change in glp-1 mRNA levels in puf-5(RNAi);mpk-1(ga111) animals. Primers distinguished spliced (shown) from unspliced mRNA and DNA products, which were not detected. actin mRNA levels were similar in both samples (data not shown).
To test whether PUF-3/11 controls endogenous mRNAs, we examined POS-1 protein in puf-3/11(RNAi) gonads and found significant ectopic POS-1 protein in puf-3/11 oocytes compared to control oocytes (Fig. 2P-R). Our previous work showed that endogenous POS-1 protein levels were unaffected by loss of PUF-5/6/7, which we confirmed in this study (Fig. 2R)(Lublin and Evans, 2007). One possibility is that pos-1 mRNA is repressed to different extents by all of these PUFs, with PUF-3/11 being the dominant regulator. The GFP:HIS reporter may be a more sensitive assay of PUF function. Alternatively, endogenous pos-1 may be repressed by redundant mechanisms including other RNA regulators, other mRNA regions, or protein stability control. In addition, de-repression of multiple mRNAs that themselves encode translational or post-translational regulators (as several PUF targets are) could lead to complex protein expression patterns (see Discussion). Nonetheless, our data support an important role for PUF-3/11 repression of pos-1 translation through 3’UTR elements. We do not know the extent to which other PUF-sensitive 3’UTRs control their endogenous mRNAs. Furthermore, incomplete PUF depletion could hide some additional overlap of PUF-3/11 and PUF-5/6/7. Regardless, these results support distinct translational repression activities for PUF-3/11 and PUF-5/6/7 for different subsets of mRNAs, which is consistent with their different roles in oogenesis.
PUF-3 binds conserved elements in regulated 3’UTRs
PUF-3/11 and PUF-5/6/7 could control these mRNA targets directly or indirectly. Analysis of spn-4 and pos-1 DNA sequence suggested elements related to PUF binding elements (PBE) in both 3’UTRs that are strongly conserved in related nematodes C. remanei and C. briggsae (Fig. 3A; Fig. S1A,B) (Koh et al., 2009; Stumpf et al., 2008). To test if PUF proteins could bind directly to these elements, recombinant protein binding to synthetic RNAs was tested by gel mobility shift assays. Because PUF-5 fusion proteins were labile and bound RNAs inconsistently, we focused on a MBP:PUF-3 fusion protein. Four conserved regions with possible PBE-like motifs were detected in the spn-4 3’UTR and tested by gel shift assay (Fig. S1B,C). However, only one bound with high affinity (Kd~5nM) to the MBP:PUF-3 fusion (Fig. 3B; Fig. S1C). Similarly, the conserved potential PBE region of the pos-1 3’UTR also bound MBP:PUF-3 with high affinity (Kd~23nM) (Fig. 3B). To test the specificity of MBP:PUF-3 for these RNAs, the core UGU motifs were mutated to ACA. MBP:PUF-3 failed to bind both pos-1 and spn-4 mutant RNAs (Kd>1μm) (Fig. 3B). Thus, PUF-3 binds specifically and with high affinity to specific elements within the pos-1 and spn-4 3’UTRs, consistent with direct PUF-3/11-mediated repression of these mRNAs. Both sequences contain a conserved PUF-3/11 binding consensus (P3BE) (Fig. 3A; Fig. S1) (Koh et al., 2009). The nos-3 3’UTR also has potential P3BE variants, but these have not been tested. Although direct binding of PUF-5 to pos-1 and spn-4 RNAs was inconclusive (unpublished data), variants of PUF-5 elements (P5BEs) lie within them (Fig. 3A) (Stumpf et al., 2008). In addition, PUF-5 can bind a P5BE variant within the fog-1 3’UTR in the yeast 3-hybrid assay (Stumpf et al., 2008). Thus, these mRNAs could be direct PUF-5/6/7 targets. The glp-1 3’UTR also has a potential P5BE within the region most critical for oocyte repression but it has not been tested and is not conserved among nematodes, leaving open direct or indirect control of this mRNA (Lublin and Evans, 2007).
Fig. 3. PUF-3 specifically binds high affinity sites in pos-1 and spn-4 3’UTRs.
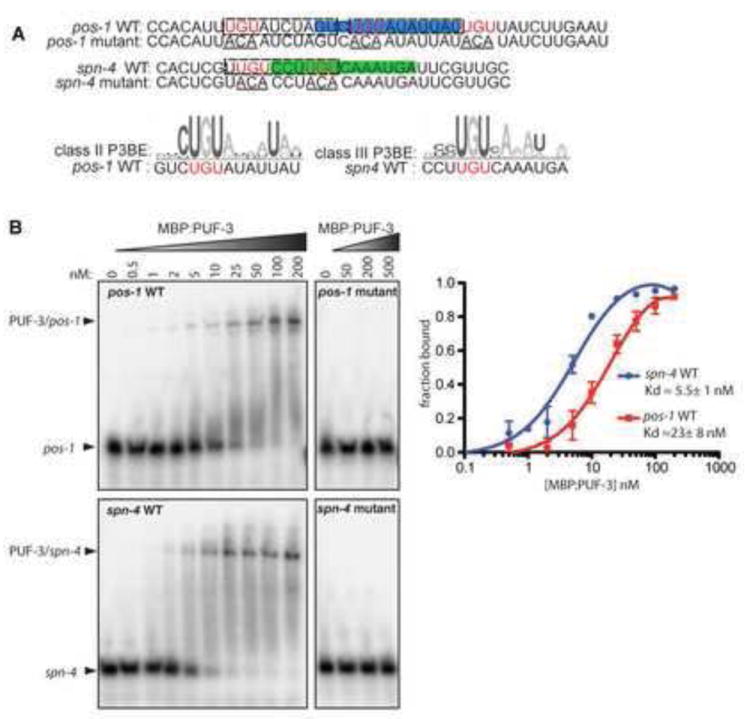
(A) pos-1 and spn-4 3’UTR fragments contain potential PUF-3/11 consensus PBE sequences (P3BEs) consistent with class II (blue) or class III (green) sites, as defined by (Koh et al., 2009). Potential PUF-5 element (P5BE) variants reside in same RNAs (dashed boxes). PBEs in both RNAs are highly conserved (Fig. S1A,B) (B) Gel shift assays of MBP:PUF-3 binding to pos-1 and spn-4 RNAs shown at top of (A). Wild type RNA probes (pos-1 WT and spn-4 WT) contained UGU motifs (red in A) that were mutated to ACA (underlined in A) in pos-1 mutant and spn-4 mutant RNA probes. Arrows indicate unbound RNAs and RNA/PUF-3 complexes. The fraction of bound RNA was plotted against protein concentration to obtain an apparent Kd values.
Ras and MPK-1 function with PUF-5 to repress translation of specific mRNAs
The stage when PUF-5/6/7 and PUF-3/11 regulate oogenesis overlaps with activation of the Ras-MAPK pathway (Lee et al., 2007b). Therefore, we wondered if Ras-MAPK function interacts with either of these PUF pathways. To test this idea, Ras was disrupted in the same 3’UTR reporter strains that were regulated by PUF-5/6/7 or PUF-3/11. Strong loss of the Ras-MAPK pathway function causes germ cells to arrest at the mid/late pachytene transition (Church et al., 1995; Lee et al., 2007b). To examine Ras-MAPK function in later oogenesis stages, we used two approaches: (1) an RNAi protocol that allows bypass of pachytene arrest, and (2) an mpk-1(ga111ts) temperature-sensitive allele that strongly reduces MPK-1 function after temperature up-shift (see Materials and Methods)(Lee et al., 2007b). As with loss of PUF-5 or PUF-3/11, RNAi of let-60/Ras induced ectopic expression of reporters carrying the pos-1 and spn-4 3’UTRs (Fig. 4A,B,C). Ectopic expression of GFP was detected in the most proximal two to five oocytes (position -2 up to -5) (Fig. 4A-C). Both 3’UTR reporters remained repressed in earlier meiotic stages suggesting that Ras promotes mRNA repression specifically during the diplotene to diakinesis period of oogenesis. Under these RNAi conditions, oocytes were enlarged in let-60(RNAi) animals but entered diakinesis and had normal organization, as seen previously with partial loss of Ras-MAPK function (Church et al., 1995; Gutch et al., 1998; Lee et al., 2007b). Thus, loss of mRNA repression is not likely a consequence of meiotic progression failure, or severe differentiation defects. Ras could promote repression specifically through certain 3’UTRs or have general effects on translation or other aspects of gene expression. However, loss of Ras function did not induce GFP expression from the strongly silenced fbf-1 3’UTR reporter, nor did it increase GFP expression from reporters with either low (gld-1 3’UTR) or strong (mex-5 3’UTR) expression levels (Fig. S2). LET-60/Ras loss also did not alter reporter mRNA levels, with the exception of the pos-1 reporter (Fig. 4H). These results suggest that Ras promotes translational repression of at least some PUF-dependent mRNAs specifically and not general translation, transcription, or GFP stability. However, the effects of Ras on pos-1 reporter mRNA levels could reflect MPK-1 control of more than one process, which would not be surprising given the multiple pathways influenced by this signaling system (Arur et al., 2009).
These results suggest that the Ras-MAPK pathway might promote translational repression by PUF-5/6/7, PUF-3/11, or both. To further test this idea, we first asked if Ras loss could enhance defects of puf-5 single RNAi. Indeed, puf-5(RNAi);let-60(RNAi) gonads produced stronger de-repression of the spn-4 and pos-1 reporters than either puf-5 or let-60 single RNAi (Fig. 4B,C). For example, ectopic expression of spn-4 reporter in distal oogonia (-4 to -3 oocytes) was strong in puf-5(RNAi);let-60(RNAi) but almost undetectable in puf-5(RNAi) gonads or let-60(RNAi) gonads (Fig. 4C). Furthermore, while loss of LET-60 alone did not visibly alter repression by the glp-1 or fog-1 3’UTRs, let-60(RNAi) strongly enhanced puf-5(RNAi)-induced defects in repression of these mRNAs (Figs 4D, 5C). This enhancement was mRNA-specific; let-60(RNAi);puf-5(RNAi) did not activate the fbf-1 reporter, and actually suppressed expression of mex-5 and gld-1 reporters even though puf-5(RNAi) alone induced small expression increases (Fig. S2). To test if the Ras-MAPK pathway promotes repression of endogenous PUF-5-dependent mRNAs, we asked if the mpk-1(ga111ts) mutant could enhance puf-5(RNAi) disruption of endogenous GLP-1 repression. In wild type, GLP-1 protein was very weak to undetectable by western blot and immunofluorescence in oocytes (Fig. 5A,B,D). In mpk-1(ga111ts) gonads, GLP-1 staining in fixed gonads was not detectably increased (Fig. 5A,B). By western blot however, GLP-1 was detected at low levels in mpk-1(ga111ts) gonads, equivalent to puf-5(RNAi) animals (Fig. 5D). The mpk-1(ga111ts) mutation may also have caused a slight increase in GLP-1 gel mobility, although the diffuse glycosylated forms of GLP-1 complicate a firm conclusion (Fig. 5D) (Crittenden et al., 1994). Remarkably, RNAi of puf-5 in mpk-1(ga111ts) animals induced a large burst of GLP-1 expression in oocytes, as seen both by immunofluorescence and western blot (Fig. 5A,B,D). A similar synergistic defect on endogenous GLP-1 was observed with Ras(RNAi) (data not shown). No significant changes in RNA levels were seen for reporter mRNAs carrying the spn-4 or fog-1 3’UTRs after either let-60/Ras(RNAi), puf-5(RNAi), or puf-5;let-60(RNAi) (Fig. 4H). Similarly, RNA levels of endogenous glp-1 mRNA were not altered in mpk-1(ga111ts) or mpk-1(ga111ts);puf-5(RNAi) animals (Fig. 5E). Collectively, these results suggest that the Ras-MAPK pathway and puf-5/6/7 function together to repress translation of specific mRNAs. Ras-MAPK is probably not essential for PUF-5/6/7 function, but is more likely a positive modulator.
To test if Ras-MAPK also influences PUF-3/11 repression, we asked if LET-60/Ras depletion enhanced translational repression defects induced by puf-3/11(RNAi). In contrast to PUF-5-dependent reporters, let-60(RNAi) did not disrupt repression by the nos-3 3’UTR nor did let-60(RNAi) enhance puf-3/11(RNAi) in de-repressing the nos-3 reporter (Fig. 4G). Instead, let-60(RNAi) partially suppressed expression from the nos-3 reporter, and strongly suppressed puf-3/11(RNAi) de-repression of this mRNA. LET-60 depletion also did not enhance puf-3/11(RNAi) defects in pos-1 3’UTR repression (Fig. 4E). A small increase in translation from the spn-4 3’UTR was observed in let-60;puf-3/11(RNAi) gonads compared to puf-3/11(RNAi) or let-60(RNAi) alone (Fig. 4F). These results suggest that repression of at least some mRNAs by PUF-3/11 is insensitive to, or even inhibited by Ras-MAPK function, in contrast to this pathway’s positive modulation of repression by PUF-5/6/7. These findings could mean that Ras-MAPK specifically promotes function of the PUF-5/6/7 control system. Alternatively, strong loss of PUF-3/11 function in these experiments might prevent detection of MPK-1 modulation of PUF-3/11 activity.
The Ras-MAPK pathway selectively synergizes with PUF-5 to control oocyte organization
Our results suggest that Ras-MAPK promotes mRNA repression through direct or indirect interactions with PUF repression systems, and may be specific to PUF-5 and PUF-6/7. To further explore these interactions and how they may contribute to oogenesis, we examined developmental consequences of perturbation of both pathways. We first induced loss of MPK-1 in mpk-1(ga111ts) gonads with or without puf-5(RNAi). When mpk-1(ga111ts) animals were shifted to restrictive temperature after germ cells entered diplotene, oocytes remained in a single row but were enlarged compared to N2 control worms treated in parallel (Fig. 6B), as seen with let-60(RNAi) and in previous studies (Church et al., 1995; Gutch et al., 1998; Lee et al., 2007b). PUF-5 depletion alone in wild type animals produced wild type oocytes arranged in a single row in most gonads (Fig. 6C), as expected because of the redundant function of PUF-6/7 (Lublin and Evans, 2007). However, when PUF-5 was depleted in mpk-1(ga111ts), oocytes were severely disorganized forming multiple rows in 75% of gonads examined (Fig. 6D,G). A very similar synergistic phenotype was seen in let-60(RNAi);puf-5(RNAi) animals (data not shown). Thus, MPK-1 functions with PUF-5 to maintain oocyte organization, supporting the idea that puf-5/6/7 and mpk-1 promote translational repression of common mRNAs.
To further test interaction of Ras-MAPK with PUF-5-mediated mRNA control, we asked if PUF-5 loss could alter defects in a let-60 gain-of-function mutant. The let-60(ga89gf-ts) allele causes increased and prolonged MPK-1 activation, and inhibits oocyte growth at restrictive temperature (Eisenmann and Kim, 1997; Lee et al., 2007b). In our conditions, let-60(ga89) animals produced small oocytes arranged in multiple rows (Fig. 7B), as was seen previously (Lee et al., 2007b). Interestingly, puf-5 RNAi partially suppressed the oocyte growth defect in let-60(ga89) animals (Fig. 7D,E,F). This result is consistent with the idea that Ras-MPK controls some process(es) important to oogenesis through stimulation of PUF-5 activity, either in a dependent or parallel pathway. However, oocyte mis-organization in let-60(ga89) was not rescued by PUF-5 loss (Fig. 7B,D), and inactivation of PUF-5/6/7 function induces oogenesis defects different from either Ras/MPK loss or hyperactivation (compare Fig. 1C,6E to 6B and 7B). In addition, loss of MPK-1 and PUF-5/6/7 together produces a more dramatic oogenesis defect than either alone (Fig. 6F). Therefore, Ras/MPK and PUF-5 direct different sets of pleiotropic pathways, one or more of which interact functionally. Our data collectively support a model where Ras/MPK-1 directly or indirectly stimulate PUF-5 repression activity to fully repress some mRNA targets. Alternatively or in addition, PUF-5 control of MPK-1 activation or additional downstream substrates is also possible.
Fig. 7. PUF-5 loss alters oogenesis defects in Ras gain-of-function.
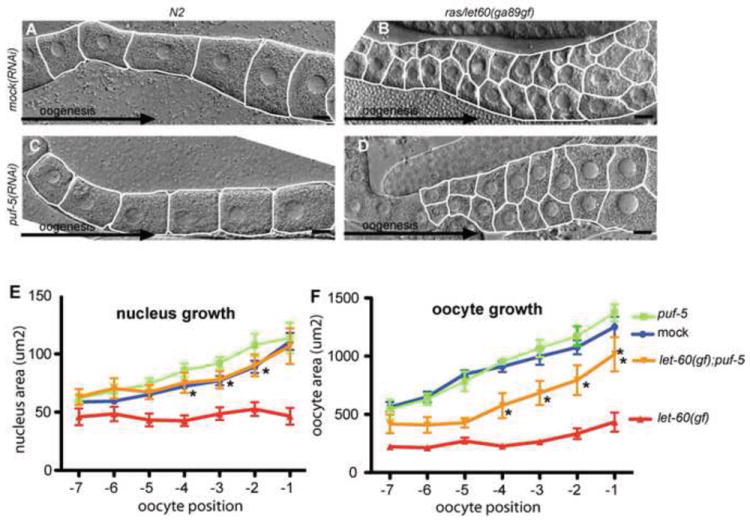
(A-D) Nomarski images of dissected gonads of the indicated genotypes or RNAi are shown. The black arrows represent the distal-proximal axis orientation. White outlines mark oocytes. Scale bars are 10 μm. (E and F) Oocyte and nucleus sizes were measured from dissected gonads (n=8-12) in indicated mutant and RNAi conditions, as defined in Fig. 1A. Strain key for the plots in E and F (right of F) denote empty vector RNAi (mock), puf-5 RNAi (puf-5), let-60(ga89gf) (let-60(gf)), and let-60(ga89gf);puf-5(RNAi) (let-60(gf);puf-5). Errors bars represent the s.e.m. for one experiment. *Denotes statistically significant differences from let-60(ga89gf);mock(RNAi) (p<0.001). The experiment was repeated three times with similar results.
PUF-3/11 interactions with the Ras pathway are distinct from PUF-5 and PUF-6/7. Curiously, the puf-3/11(RNAi) large-oocyte phenotype bears some resemblance to partial loss of MPK-1 and Ras (Figs 1C, 6A). PUF-3/11 depletion in mpk-1(ga111ts) animals produced enlarged oocytes that were similar in size to either single mutant or RNAi alone (Fig. S3). However, puf-3/11(RNAi) did not significantly alter the small oocyte defects of Ras gain-of-function in let-60(ga89) animals (Fig. S3). PUF-3/11 therefore may promote oocyte growth upstream or independently of Ras-MAPK. The different interaction of PUF-3/11 and PUF-5/6/7 with MPK-1 support the idea that PUF-3/11 has at least some distinct sets of mRNA targets from PUF-5/6/7, and are consistent with the possibility that Ras-MAPK may specifically influence the PUF-5/6/7 repression system.
MPK-1 controls PUF-5 proteins
If the Ras-MAPK pathway modulates mRNA repression through control of PUF-5 and PUF-6/7, it might control PUF-5 protein expression or activity. To test this, we asked if loss or gain of pathway function altered PUF-5 protein levels by both immunofluorescence and western blot. PUF-5 protein levels did not differ significantly between wild type, the mpk-1(ga111lf) mutant, or the let-60(ga89gf) mutant (Fig. 8A). Therefore, MPK-1 is not required for PUF-5 accumulation. However, on western blots, we detected two different PUF-5 bands from wild type (N2) extracts, suggesting PUF-5 exists in two forms (Fig. 8A; see also Fig. 5D). Both forms are PUF-5 specific because both disappeared after puf-5(RNAi) (Fig. 8A). Interestingly, MPK-1 activity controlled the relative amounts of these PUF-5 protein products. Extracts from mpk-1(ga111) worms had higher proportion of the faster migrating form compared to N2 extracts, whereas let-60(ga89) extracts had higher proportion of the slower migrating form (Fig. 8A). The nature of these alternative PUF-5 forms is not known. They could represent post-translational PUF-5 modifications induced directly or indirectly by MPK-1 (see Discussion). Alternatively, Ras/MPK-1 may alter production of PUF-5 isoforms. Regardless, together with our other results, these data are consistent with MPK-1 control of PUF-5 and PUF-6/7.
Fig. 8. MPK-1 controls PUF-5 protein forms and onset of expression.
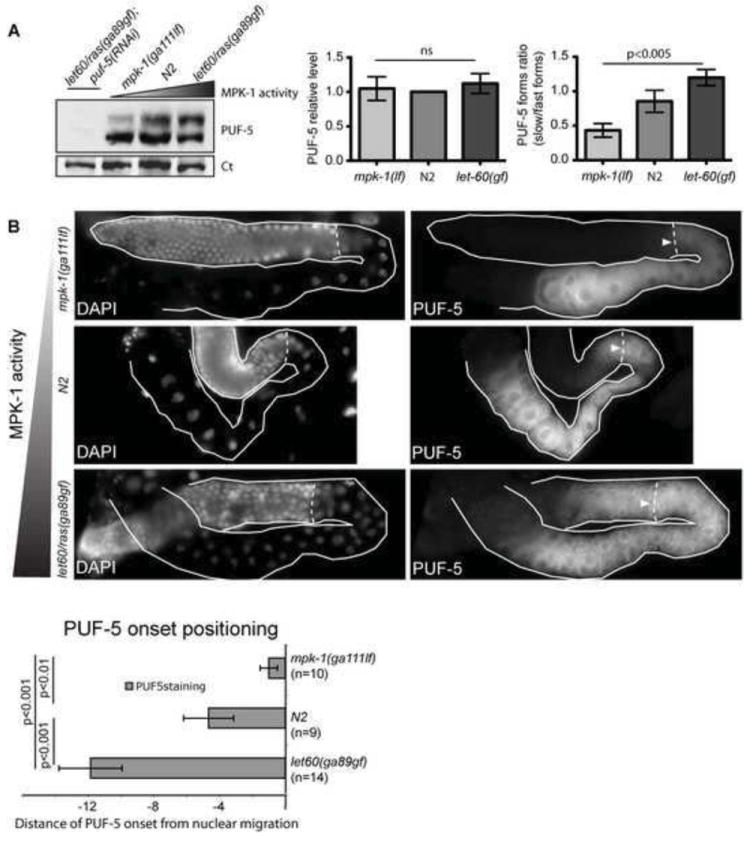
(A) PUF-5 western blot: protein was extracted from equal numbers of synchronized adult hermaphrodites after 12 h at restrictive temperature for the indicated genotypes. Two PUF-5 forms were observed, which were strongly reduced after puf-5(RNAi). In three experiments, the ratio of slower/faster PUF-5 forms increased with Ras-MAPK activity. Bar graphs show quantification of total PUF-5 levels (left graph) normalized to loading control (Ct) band in the same blot, and ratio of slow/fast forms (right graph) by densitometry; ns=not signficant. (B) MPK-1 controls PUF-5 onset. Dissected gonads of indicated genotypes were stained in parallel for PUF-5, and imaged with matched exposures. DAPI staining revealed nuclear positioning. Bottom panel: Quantification of PUF-5 onset relative to nuclear migration from periphery to gonad core. Values are nuclear diameters from this transition (negative values are distal to transition). Error bars are s.d. for one representative experiment.
We also detected subtle control of the spatial onset of PUF-5 expression by the Ras-MAPK pathway. PUF-5 expression normally begins as germ nuclei detach from the gonad periphery and progress from late pachytene to diplotene stages of meiosis (Lublin and Evans, 2007). At this same transition, the KH domain RBP GLD-1 disappears from its peak expression earlier in meiosis in the distal gonad (Jones et al., 1996). GLD-1 is likely a direct repressor of puf-5 translation in early meiosis (Lee and Schedl, 2001). MPK-1 is activated close to this “GLD-1/PUF-5” switch, during the mid-late pachytene transition (Lee et al., 2007b). In wild type gonads, PUF-5 expression was first detected before late pachytene germ nuclei moved from the gonad periphery to form “single file” diplotene stage in the center of the gonad core (Fig. 8B). In the mpk-1(ga111) gonads, PUF-5 expression reproducibly appeared to be delayed relative to this nuclear migration. By contrast, in let-60(ga89) gf gonads, PUF-5 expression appeared prematurely, prior to this nuclear transition (Fig. 8B). GLD-1 expression showed a reciprocal shift, disappearing prematurely in Ras(gf) and belatedly in mpk-1(lf) (Fig. S4). Therefore, the GLD-1/PUF-5 switch relative to the transition of nuclear localization depends on Ras-MAPK activity. This altered RNABP switch could be indirectly due to changed timing of meiosis transition relative to nuclear migration, or to more direct effects on RNP regulators that mediate the GLD-1/PUF-5 switch.
DISCUSSION
Oogenesis is an elaborate process that requires coordination of specific mRNA control systems with regulation of oocyte growth, cytoplasm and nucleus changes, membrane dynamics, and cell cycle. The results presented here show that the RNA-binding translation repressors PUF-3/11 and PUF-5/6/7 both control late C. elegans oogenesis, but repress distinct sets of mRNAs. Diversification of these mRNA repression systems allows execution of different cellular processes essential to oocyte formation. The mRNA targets identified so far suggest these PUFs control pathways of mRNA regulation and developmental signaling systems. Additional mRNA targets are also likely, perhaps including those that more directly influence cell growth and organization. Furthermore, we found that the Ras-MAPK pathway adds another layer of regulation by positive modulation of translational repression through specific 3’UTRs. Ras-MAPK works in part with PUF-5 to control mRNA subsets and oocyte differentiation. Ras-MAPK is not likely essential for PUF-5/6/7 activity, but rather optimizes repression of some mRNA targets. Likewise, PUF-5/6/7 represents only one of many parallel pathways influenced by the Ras-MAPK system. Taken together, this work suggests that oocyte growth and organization depend on regulation of multiple mRNAs by two related but distinct PUF protein control systems, and that the Ras-MAPK pathway modulates the output of at least one of these PUF systems to ensure proper oocyte differentiation (Fig. 9). Moreover, these findings combined with previous results suggest PUF proteins have both negative and positive interactions with the Ras-MAPK pathway at different levels and different times of germline development (Lee et al., 2006; Lee et al., 2007a). Together these interactions control the spatiotemporal organization of the germline stem cell to oocyte program (Fig. 9).
Fig. 9. Model for PUF control and function patterning during oogenesis.
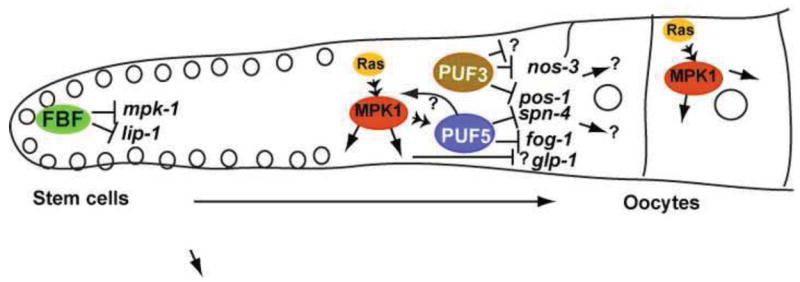
PUF proteins FBF-1/2 repress mpk-1 mRNA to suppress Ras-MAPK function in stem cells and early meiosis (Lee et al., 2006; Lee et al., 2007a). Ras-MAPK is activated in early pachytene and promotes PUF-5/6/7 activity among multiple other parallel activities. PUF-5/6/7 and PUF-3/11 repress both distinct and common mRNA targets. Additional unknown targets and indirect controls (?) of all pathways are probable, and interactions between Ras-MAPK and PUFs could include multiple levels and feedback controls. Several PUF mRNA targets (pos-1, spn-4, nos-3) encode mRNA control factors that likely influence downstream mRNAs. Other unknown mRNA targets (?) may more directly control cell organization and growth. Oogenesis may rely on concerted control of both types of mRNA targets, together with Ras/MAPK modulation of PUF-5/6/7 and other parallel pathways.
Role of the PUF protein family in mRNA repression and late oogenesis
In C. elegans, PUF proteins form a large family of ten paralogs that have diverged both in function and RNA-binding specificity. PUF-3/11 reside in one branch of this family (Fig. 1B) but their functions were previously unknown. We show that PUF-3/11 represses translation in vivo of several developmentally-important maternal mRNAs during late oogenesis. In addition, we show that PUF-3 can directly bind in vitro very specifically and with high affinity to specific elements in the pos-1 and spn-4 3’UTRs, elements that are highly conserved among related nematode species. This suggests PUF-3/11 control of pos-1 and spn-4 translation is direct. Most PUF proteins studied to date appear to bind with highest affinity to a single sequence consensus (PBEs), based on in vitro selection experiments (Bernstein et al., 2005; Opperman et al., 2005; Stumpf et al., 2008). PUF-11, however, was shown to bind to three distinct classes of PBEs (Koh et al., 2009). The spn-4 PUF-3/11 binding site contains a class III consensus, and the pos-1 PUF-3/11 binding RNA contains a class II consensus (Fig. 3A). As might be predicted, nos-3 also contains two potential PUF-3/11 binding sites that are conserved, suggesting nos-3 could also be a direct mRNA target. However, PUF-3/11 (or PUF-5/6/7) control of some mRNAs could be indirect through regulation of other RNA-binding regulators. POS-1, SPN-4, and NOS-3 are known regulators of mRNAs and embryonic proteins, and thus de-repression of these or other mRNAs after PUF depletion could lead to complex patterns of mRNA and protein mis-regulation (Farley et al., 2008; Huang et al., 2002; Kraemer et al., 1999; Ogura et al., 2003).
PUF-5 and PUF-6/7 comprises a branch of the PUF family that is closest in homology to PUF-3/11. Our results show that PUF-3/11 and PUF-5/6/7 control oogenesis and mRNA repression at a similar stage, suggesting that they function at a close and possibly coincident time of development. The dependence of pos-1 and spn-4 3’UTR control on both PUF-3/11 and PUF-5/6/7 suggests these PUF proteins may co-regulate some mRNA targets. However, several results argue that PUF-5/6/7 and PUF-3/11 also control distinct sets of mRNAs. First, some 3’UTRs were specifically controlled by PUF-5/6/7 (fog-1, glp-1) or PUF-3/11 (nos-3). Even co-regulated mRNAs may differ in their sensitivity to PUF-3/11 and PUF-5/6/7, which is supported by the differential control of endogenous pos-1 by these PUFs. Second, loss of these PUF groups produces dramatically distinct phenotypes. Finally, PUF-5 and PUF-3 interact in different ways with the Ras-MAPK signaling system. In support of these conclusions, in vitro RNA binding site selection experiments suggest PUF-3/11 and PUF-5/6/7 bind different RNA sequences (Koh et al., 2009; Stumpf et al., 2008). Taken together, our results suggest the diversification of these PUF proteins within the PUF family allowed for intricate and independent modulation of different factors and processes.
How do PUF-3 and PUF-5 subgroups control the intricacies of oocyte development? As mentioned above, several mRNA targets identified so far encode other RNA-binding factors that control hierarchies of translational control in the embryo and/or germ line. These PUFs therefore may partly act as master regulators of these mRNA control circuits, restricting their activities to the proper developmental times. However, it is very likely that both PUF groups have many other mRNA targets, some of which may encode direct mediators of various cell processes. Indeed, PUF-5 can bind several other PBEs in target mRNAs with diverse functions (Stumpf et al., 2008). These considerations lead to two models for how these PUF proteins modulate oogenesis (Fig. 9). In one model, PUF-3/11 and PUF-5/6/7 regulate mRNA control cascades, which in turn control mRNAs for factors that limit or promote cell organization and growth. Another model would be that these PUFs control multiple pathways, and direct control of membrane and cell dynamics/growth regulators promotes oocyte formation. The models are not mutually exclusive.
The Ras-MAPK pathway and PUF protein interactions during oogenesis
Our results show that Ras and its downstream effector MPK-1 add a layer of control to maternal mRNA repression by PUF proteins. The Ras-MAPK pathway controls multiple aspects of oogenesis, including meiotic progression, oogonia and nuclear organization, oocyte growth, and oocyte maturation (Lee et al., 2007b). Recent work revealed that MPK-1 phosphorylates many substrates that likely act post-transcriptionally in the C. elegans germ line (Arur et al., 2009). These MPK-1 substrates support a model where MPK-1 activates or suppresses multiple parallel pathways, which together control various post-nuclear events in the cytoplasm during oocyte development (Arur et al., 2009). Our results suggest that one of these parallel pathways positively support PUF-5/6/7 repression of oocyte mRNAs. First, Ras-MAPK promotes repression by several PUF-5-dependent 3’UTRs, but does not influence regulatory properties of other 3’UTRs. Second, PUF-5 and MPK-1 synergistically cooperate to control oocyte organization. Finally, PUF-5 protein is modified by Ras-MAPK activity. The nature of PUF-5 modification is not known. It is unlikely that MPK-1 directly phosphorylates PUF-5 since no consensus MAPK/ERK phosphorylation sites reside in PUF-5 or PUF-6/7. More likely, PUF-5 is modified indirectly through one or more MPK-1 substrates. Regardless, taken together, these several findings suggest that Ras-MAPK promotes PUF-5/6/7 activity or controls a parallel pathway that supports PUF-5/6/7 function (Fig. 9). A simple model is that PUF-5 modification by a MPK-1 activated pathway alters RNA binding or association with co-repressive factors. Alternatively, MPK-1 could modulate other RNA-associated factors that influence repression of PUF mRNA targets. For example, MPK-1 control of general translation factors might indirectly assist silencing of PUF-5 mRNPs. Another intriguing possibility is the recently discovered direct inactivation of NOS-3 by MPK-1 (Arur et al., 2011). NOS-3 is an RNABP that belongs to the Nanos family, some of which partner with PUF proteins (de Moor et al., 2005; Wilhelm and Smibert, 2005). Perhaps inactivation of NOS-3 alters PUF-5/6/7 activity for certain mRNAs. Alternatively, MPK-1 inactivation of NOS-3 protein may have independent functions, perhaps working in concert with PUF-3/11 repression of nos-3 mRNA. These models are not mutually exclusive given the pleiotropic nature of Ras-MAPK regulation. Moreover, PUF proteins are known to also modulate MAPK pathway mRNAs in both invertebrates and vertebrates (Lee et al., 2007a; Prinz et al., 2007; Whelan et al., 2011). Therefore, there could be additional feedback control of PUF-5/67 or PUF-3/11 on this signaling pathway in late oogenesis (Fig. 9).
This and previous studies suggest the Ras-MAPK pathway has complex interactions with PUF proteins and other translational control factors in diverse organisms. Within the stem cell compartment of the C. elegans gonad, the PUF proteins FBF-1/2 repress mpk-1 translation, which functions with other mechanisms to suppress MPK-1 activity in early-stage germ cells (Lee et al., 2006; Lee et al., 2007a). Our results suggest that once MPK-1 is activated in later stages, PUF-5/6/7-dependent mRNAs are indirect downstream targets that collectively mediate some of its many functions. Therefore, complex negative and positive interactions between PUFs and Ras-MAPK change over time during the stem cell to oocyte developmental program (Fig. 9). These dynamics likely contribute to spatial and temporal organization of this stem cell system. In other systems, MAPK promotes translation through several pathways, in oocyte development and many other tissue systems (MacNicol and MacNicol, 2010; Mahoney et al., 2009; Proud, 2007; Whelan et al., 2011). These pathways influence the general translation machinery and some mRNA-specific systems. It is likely that Ras-MAPK has multiple effects on mRNA control in diverse systems, including the C. elegans germ line. In support of this pleiotropy, direct targets of MPK-1 in C. elegans gonads include several RNA-associated factors (Arur et al., 2011; Arur et al., 2009). Therefore, this pathway likely influences mRNA regulation in diverse developmental and cellular contexts.
Supplementary Material
(A) Alignments of pos-1 and spn-4 3’UTRs from C. elegans (top) and C. briggsae (bottom). The first nucleotides follow stop codons, and likely polyadenylation signals (orange boxes) reside near 3’ ends. Lines link nucleotide identities, and red boxes denote alignment gaps and non-identities. Blue bars show sequences in RNA probes used for gel shift assays. The pos-1 3’UTR contains a highly conserved region with several putative PBEs, including a consensus PUF-3 PBE (P3BE) (boxed in blue). The spn-4 3’UTR contains four regions with conserved PBE-related motifs (hatched boxes); spn-4.2 RNA binds PUF-3 (shown in B; same as spn-4 WT in Fig. 3) and has a consensus P3BE (green box). More details of pos-1 WT and spn-4.2 (spn-4 WT) RNAs are shown in Fig. 3A. (B) Gel shift assay of MBP:PUF-3 binding to radiolabeled pos-1 WT RNA and four different spn-4 RNAs. Only spn-4.2 (spn-4 WT) RNA and pos-1 WT RNAs bound MBP:PUF-3 with high affinity, and were analyzed in more detail in Fig. 3B. MBP:PUF was used at 50 nM and 500 nM.
Expression levels of the GFP:HIS expressed from reporters under the control of the indicated 3’UTRs were measured in the proximal gonad of live worms following let-60/Ras(RNAi), puf-5 or puf-3/11 RNAi, or let-60;puf-5 double RNAi. Nuclear positions and GFP maximum intensity were determined as in Figs. 1 and 2. Error bars reflect the S.E.M. for one experiment, which was repeated at least twice for each reporter. Asterisks reflect statistically significant differences from the mock (L4440) RNAi control (p<0.01). PUF-5 depletion induced slight increases in mex-5 and gld-1 reporter expression, but let-60(RNAi) did not enhance but instead suppressed this expression.
Oocyte and nucleus size were quantified in puf-3(RNAi), let-60/Ras(RNAi) and let-60/Ras(ga89ts) and double RNAi/mutant animals (n=14-18 gonads for each condition). In all panels, puf-3 refers to puf-3(RNAi), mock refers to empty vector (L4440) RNAi; and let-60(gf) refers to let-60(ga89gf) mutant strain. Oocyte or nucleus size was plotted as a function of the oocyte position along the proximal gonad. **denotes p<0.001, and *denotes p<0.01 from mock RNAi (in A) or let-60(gf);puf-3 (in B and C).
GLD-1 and PUF-5 levels during nuclear migration show reciprocal sensitivity to Ras-MPK-1 activity. Dissected gonads were stained for endogenous GLD-1 or PUF-5, and staining levels were quantified at the transition of nuclei from periphery to gonad core in mpk-1(ga111ts), wild type (N2), and let-60Ras (ga89gf) fixed gonads processed and stained in parallel, using matched exposures settings.
Two PUF RNA-binding protein groups control different processes in oogenesis.
PUF-3/11 and PUF-5/6/7 repress translation of distinct and common mRNAs.
The Ras-MapKinase pathway promotes translational repression by PUF-5/6/7.
Ras-MapKinase regulates PUF-5 protein
Ras-MapKinase and PUF-5/6/7 cooperate to control oocyte organization.
Acknowledgments
We want to thank Judith Kimble and Tim Schedl for providing antibodies and mutant strains. For providing transgenic and mutant strains, we thank Jayne Squirrell, John White, Jim Priess and the Caenorhabditis Genetics Center (funded by the National Institutes of Health National Center for Research Resources); Steve Britt and Joan Hooper for use of equipment and materials. This work was funded by grants from the National Science Foundation (0725416) and the National Institutes of Health (R01 GM07968) to T.C. Evans.
Footnotes
Competing interest statement
The authors declare no competing financial interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Berkseth M, Nayak S, Hansen D, Zarkower D, Schedl T. MPK-1 ERK controls membrane organization in C. elegans oogenesis via a sex-determination module. Dev Cell. 2011;20:677–688. doi: 10.1016/j.devcel.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc Natl Acad Sci U S A. 2009;106:4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Evans TC. The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev Biol. 2006;291:132–143. doi: 10.1016/j.ydbio.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Lublin AL, Evans TC. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr Biol. 2002;12:1502–1506. doi: 10.1016/s0960-9822(02)01111-9. [DOI] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda A, Smith GW. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci. 2007;12:3713–3726. doi: 10.2741/2346. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Eckmann CR, Wang L, Bernstein DS, Wickens M, Kimble J. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc Lond B Biol Sci. 2003;358:1359–1362. doi: 10.1098/rstb.2003.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- D’Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3’ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Kim SK. Mechanism of activation of the Caenorhabditis elegans ras homologue let-60 by a novel, temperature-sensitive, gain-of-function mutation. Genetics. 1997;146:553–565. doi: 10.1093/genetics/146.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TC, Crittenden SL, Kodoyianni V, Kimble J. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Farley BM, Pagano JM, Ryder SP. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Seydoux G. Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics. 2008;178:235–243. doi: 10.1534/genetics.107.083212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutch MJ, Flint AJ, Keller J, Tonks NK, Hengartner MO. The Caenorhabditis elegans SH2 domain-containing protein tyrosine phosphatase PTP-2 participates in signal transduction during oogenesis and vulval development. Genes Dev. 1998;12:571–585. doi: 10.1101/gad.12.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- Huang NN, Mootz DE, Walhout AJ, Vidal M, Hunter CP. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Jones AR, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3’UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Edgar BA. Proliferative control in Drosophila stem cells. Curr Opin Cell Biol. 2008;20:699–706. doi: 10.1016/j.ceb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lasko P. Translational control during early development. Prog Mol Biol Transl Sci. 2009;90:211–254. doi: 10.1016/S1877-1173(09)90006-0. [DOI] [PubMed] [Google Scholar]
- Lee MH, Hook B, Lamont LB, Wickens M, Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007a;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007b;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- MacNicol MC, MacNicol AM. Developmental timing of mRNA translation--integration of distinct regulatory elements. Mol Reprod Dev. 2010;77:662–669. doi: 10.1002/mrd.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney SJ, Dempsey JM, Blenis J. Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog Mol Biol Transl Sci. 2009;90:53–107. doi: 10.1016/S1877-1173(09)90002-3. [DOI] [PubMed] [Google Scholar]
- Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3’ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 2010;137:1787–1798. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal N, Sternberg PW. The epidermal growth factor system in Caenorhabditis elegans. Exp Cell Res. 2003;284:150–159. doi: 10.1016/s0014-4827(02)00097-6. [DOI] [PubMed] [Google Scholar]
- Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- Noble SL, Allen BL, Goh LK, Nordick K, Evans TC. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J Cell Biol. 2008;182:559–572. doi: 10.1083/jcb.200802128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 2003;130:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- Ohmachi M, Rocheleau CE, Church D, Lambie E, Schedl T, Sundaram MV. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr Biol. 2002;12:427–433. doi: 10.1016/s0960-9822(02)00690-5. [DOI] [PubMed] [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Prinz S, Aldridge C, Ramsey SA, Taylor RJ, Galitski T. Control of signaling in a MAP-kinase pathway by an RNA-binding protein. PLoS One. 2007;2:e249. doi: 10.1371/journal.pone.0000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram MV. RTK/Ras/MAPK signaling. WormBook. 2006:1–19. [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci U S A. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Battu G, Hoier EF, Hajnal A. Distinct roles of the Pumilio and FBF translational repressors during C. elegans vulval development. Development. 2006;133:3461–3471. doi: 10.1242/dev.02496. [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci U S A. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- Whelan JT, Hollis SE, Cha DS, Asch AS, Lee MH. Post-transcriptional regulation of the Ras-ERK/MAPK signaling pathway. J Cell Physiol. 2011 doi: 10.1002/jcp.22899. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Smibert CA. Mechanisms of translational regulation in Drosophila. Biol Cell. 2005;97:235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Alignments of pos-1 and spn-4 3’UTRs from C. elegans (top) and C. briggsae (bottom). The first nucleotides follow stop codons, and likely polyadenylation signals (orange boxes) reside near 3’ ends. Lines link nucleotide identities, and red boxes denote alignment gaps and non-identities. Blue bars show sequences in RNA probes used for gel shift assays. The pos-1 3’UTR contains a highly conserved region with several putative PBEs, including a consensus PUF-3 PBE (P3BE) (boxed in blue). The spn-4 3’UTR contains four regions with conserved PBE-related motifs (hatched boxes); spn-4.2 RNA binds PUF-3 (shown in B; same as spn-4 WT in Fig. 3) and has a consensus P3BE (green box). More details of pos-1 WT and spn-4.2 (spn-4 WT) RNAs are shown in Fig. 3A. (B) Gel shift assay of MBP:PUF-3 binding to radiolabeled pos-1 WT RNA and four different spn-4 RNAs. Only spn-4.2 (spn-4 WT) RNA and pos-1 WT RNAs bound MBP:PUF-3 with high affinity, and were analyzed in more detail in Fig. 3B. MBP:PUF was used at 50 nM and 500 nM.
Expression levels of the GFP:HIS expressed from reporters under the control of the indicated 3’UTRs were measured in the proximal gonad of live worms following let-60/Ras(RNAi), puf-5 or puf-3/11 RNAi, or let-60;puf-5 double RNAi. Nuclear positions and GFP maximum intensity were determined as in Figs. 1 and 2. Error bars reflect the S.E.M. for one experiment, which was repeated at least twice for each reporter. Asterisks reflect statistically significant differences from the mock (L4440) RNAi control (p<0.01). PUF-5 depletion induced slight increases in mex-5 and gld-1 reporter expression, but let-60(RNAi) did not enhance but instead suppressed this expression.
Oocyte and nucleus size were quantified in puf-3(RNAi), let-60/Ras(RNAi) and let-60/Ras(ga89ts) and double RNAi/mutant animals (n=14-18 gonads for each condition). In all panels, puf-3 refers to puf-3(RNAi), mock refers to empty vector (L4440) RNAi; and let-60(gf) refers to let-60(ga89gf) mutant strain. Oocyte or nucleus size was plotted as a function of the oocyte position along the proximal gonad. **denotes p<0.001, and *denotes p<0.01 from mock RNAi (in A) or let-60(gf);puf-3 (in B and C).
GLD-1 and PUF-5 levels during nuclear migration show reciprocal sensitivity to Ras-MPK-1 activity. Dissected gonads were stained for endogenous GLD-1 or PUF-5, and staining levels were quantified at the transition of nuclei from periphery to gonad core in mpk-1(ga111ts), wild type (N2), and let-60Ras (ga89gf) fixed gonads processed and stained in parallel, using matched exposures settings.


