Abstract
Over-expression of the transcriptional regulator Myc is thought to be the cause or a contributing factor in the development of a large number of human lymphomas and certain other cancers. Apoptotic cell death constitutes a tumor suppressive mechanism, particularly in the context of Myc over-expression. Accordingly, lymphoma development in Eμ-Myc transgenic mice, which mimic the Myc/IgH chromosomal translocation that causes Burkitt Lymphoma, is accelerated by concomitant over-expression of anti-apoptotic Bcl-2 family members or loss of proapoptotic BH3 only proteins, such as Bim. Bim binds with high affinity to all prosurvival Bcl-2-like proteins and can also interact with Bax/Bak, but it remains unclear which of these interactions are critical for its tumor suppressive function.
We have previously generated knock-in mutant mice in which the BH3 region of Bim has been exchanged with that for Bad, Noxa or Puma so that it can only bind to select pro-survival Bcl-2-like proteins: BimBad binding to Bcl-2, Bcl-xL and Bcl-w but not Mcl-1 or A1; BimNoxa binding only to Mcl-1 and A1 and as a control, BimPuma, which can still bind all pro-survival Bcl-2-like proteins. We have now inter-crossed these Bim mutant mice with Eμ-Myc transgenic mice and found that both the BimBad and the BimNoxa mutations but not the BimPuma mutation greatly accelerate Myc-induced lymphoma development and increase leukemic burden. These results demonstrate that for optimal tumor suppressive activity, Bim must be able to interact with all and not just select pro-survival Bcl-2 family members.
Keywords: Myc, Bim, tumor suppression, lymphoma, apoptosis
Introduction
The oncogene Myc is over-expressed in ~70% of human cancers and the Myc/IgH chromosomal translocation is the cause of Burkitt Lymphoma (Pelengaris et al., 2002). Eμ-Myc transgenic mice over-express Myc under control of the immunoglobulin heavy chain gene enhancer (Eμ), mimicking the Myc/IgH chromosomal translocation found in human Burkitt lymphomas (Adams et al., 1985). Myc over-expression causes abnormally increased proliferation of B lymphoid cells (Langdon et al., 1986) and acquisition of additional oncogenic lesions precipitates malignant clonal pre-B or B-cell lymphomas with a median latency of ~100 days (on a C57BL/6 background) (Michalak et al., 2009).
In addition to promoting abnormally increased cell proliferation, Myc over-expression also enhances apoptotic cell death under conditions of stress, such as limited growth factor supply (Pelengaris et al., 2002; Strasser et al., 1996). This apoptosis imposes a barrier against Myc-induced neoplastic transformation (Pelengaris et al., 2002) and, accordingly, concomitant over-expression of pro-survival Bcl-2-like proteins (Strasser et al., 1990) or loss of pro-apoptotic relatives, such as Bim (Egle et al., 2004) or Puma (Garrison et al., 2008; Hemann et al., 2004; Michalak et al., 2009), greatly accelerates lymphomagenesis in Eμ-Myc mice. Proteins of the Bcl-2 family, which comprises three subgroups with distinct functions, are major regulators of apoptosis (Youle and Strasser, 2008). The pro-survival members (Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1) are essential for cell survival, the BH3-only proteins (e.g. Bim, Puma, Bad, Noxa) initiate apoptosis signaling and Bax/Bak are required for mitochondrial outer membrane permeabilization (MOMP) and activation of the caspase cascade that dismantles the cells (Strasser et al., 2011). The molecular mechanisms for Bax/Bak activation are not fully resolved but appear to involve both direct activation by BH3-only proteins as well as indirect activation by BH3-only protein mediated blockade of the pro-survival Bcl-2 family members (Chipuk and Green, 2008; Merino et al., 2009; Strasser et al., 2011). BH3-only proteins bind with their BH3 region to a groove on the surface of pro-survival Bcl-2 proteins, but individual members of this subgroup differ substantially in their binding specificity. Bim and Puma interact with all pro-survival proteins and this accounts (at least in part) for their potent pro-apoptotic activity (Chen et al., 2005; Kuwana et al., 2005). Conversely, Bad and Noxa appear to be only weak killers (at least when over-expressed), and this has been attributed to their limited binding to Bcl-2, Bcl-xL and Bcl-w or Mcl-1 and A1, respectively (Chen et al., 2005; Kuwana et al., 2005). The binding specificity of BH3-only proteins is determined by their BH3 region (Chen et al., 2005; Kuwana et al., 2005). Therefore the importance of Bim’s ability to bind all pro-survival Bcl-2-like proteins could be investigated by generating Bim BH3 region replacement mutant knock-in mice (Merino et al., 2009), with BimBad binding only to Bcl-2, Bcl-xL and Bcl-w but not Mcl-1 or A1, BimNoxa only binding Mcl-1 and A1 and, as a control, BimPuma still binding all pro-survival family members (Figure 1a). BimBad as well as BimNoxa mutant mice had abnormally increased numbers of leukocytes (Merino et al., 2009), although this phenotype was clearly less pronounced compared to Bim-deficient (Bim−/− (Bouillet et al., 1999)) animals. Thus, for optimal induction of developmentally programmed cell death (at least in the hematopoietic system), Bim must be able to interact with all pro-survival Bcl-2 family members. We have exploited these Bim BH3 region replacement mutant mice to investigate the role of Bim’s binding specificity in tumor suppression, using the Eμ-Myc mouse lymphoma model. These studies demonstrate that for optimal suppression of Myc-induced lymphomagenesis, Bim must be able to bind all Bcl-2-like pro-survival proteins.
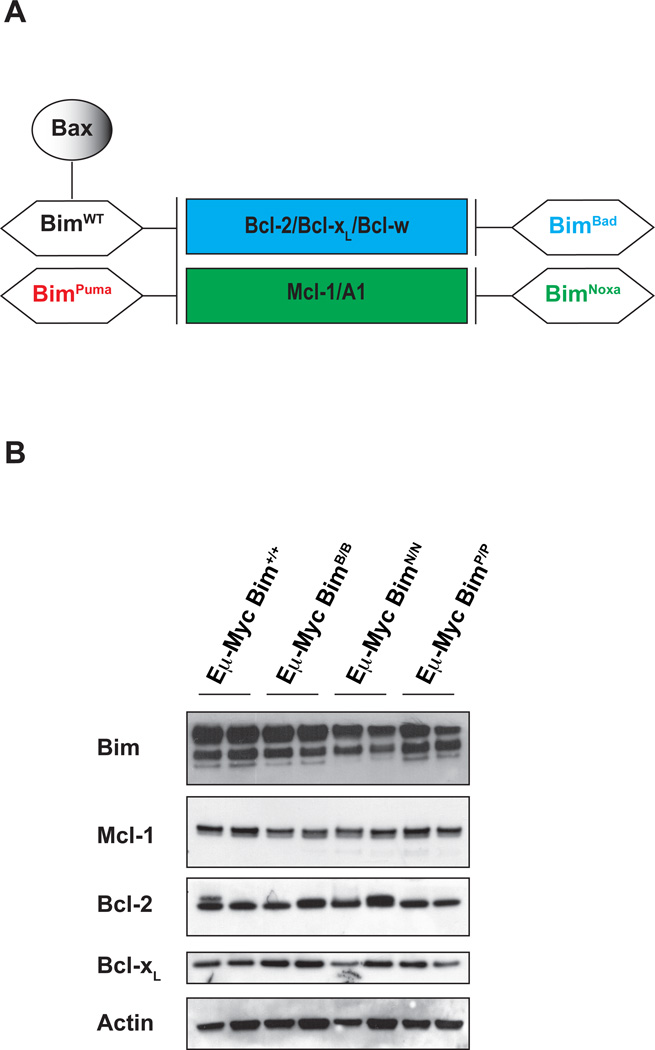
Figure 1. Expression of mutant Bim proteins in Eμ-Myc tumors.
(a) Schematic representation of specificity of binding of BimBH3 mutants to pro-survival Bcl-2 family members. (b) Western blot analysis to determine expression of BimBH3 mutants, Mcl-1, Bcl-2, Bcl-xL and Actin (used as loading control) in Eμ-Myc lymphomas (B220+IgM−) from control Eμ-Myc as well as Eμ-Myc/BimBad/Bad, Eμ-Myc/BimNoxa/Noxa and Eμ-Myc/BimPuma/Puma mice (extracts from two different mice for each genotype). Western blotting was carried out by standard procedures, using protein extracts generated by lysis of lymphoma cell suspensions in a buffer containing (20 mM Tris-pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1mM EGTA, 10% glycerol, 1% Triton-X 100, protease inhibitors MiniComplete, EDTA free, Roche). Proteins were detected using antibodies to Bim (14A8 or 3C5, ENZO), Bcl-2 (BD Pharmingen), Bcl-xL (BD Bioscience), Mcl-1 (Rockland) or Actin (Sigma). Secondary antibodies were HRP-conjugated and detection used Enhanced Chemoluminescence (ECL, Healthcare).
Results and Discussion
Bim functions as a tumor suppressor in both mice (Egle et al., 2004) and humans, with loss of both Bim alleles or suppression of Bim protein expression detected in mantle cell lymphoma (Tagawa et al., 2005) and certain other B lymphoid malignancies, such as Burkitt Lymphoma (Anderton et al., 2008). The tumor suppressor activity of Bim was first demonstrated in Eμ-Myc mice (Egle et al., 2004) where loss of one Bim allele shortened median survival from the normal ~100 days to ~77 days and loss of both alleles to 57 days. Although loss of Puma, Bmf, Bad or Noxa can also accelerate Myc-induced lymphomagenesis (Frenzel et al., 2010; Michalak et al., 2009), loss of one allele of these genes did not have significant impact, indicating these BH3-only proteins are less potent tumor suppressors in this context than Bim. These differences between BH3-only proteins could be due to their level of expression, their specific regulation and/or their ability to bind pro-survival Bcl-2 proteins and hence their efficacy to trigger apoptosis.
The BimBad and BimNoxa mutations both accelerated lymphoma development in Eμ-Myc transgenic mice
Although the tumor suppressive activity of Bim has been established, it is not clear whether this requires its ability to bind to all or only a limited subset of the pro-survival Bcl-2 family members. This question could be addressed by crossing Eμ-Myc mice with our mutant strains of mice in which Bim has been altered to restrict its binding to Bcl-2, Bcl-xL and Bcl-w (BimBad) or Mcl-1 plus A1 (BimNoxa). Regardless of the Bim genotype, all Eμ-Myc transgenic mice developed lymphoma. Western blot analysis demonstrated that BimBad, BimNoxa and BimPuma mutant proteins were expressed in such tumors at levels comparable to wt Bim (Figure 1b). Moreover, expression of the Bim BH3 exchange mutant proteins did not significantly affect the expression of pro-survival Mcl-1, Bcl-2 and Bcl-xL in Eμ-Myc lymphomas arising in these mutant animals (Figure 1b).
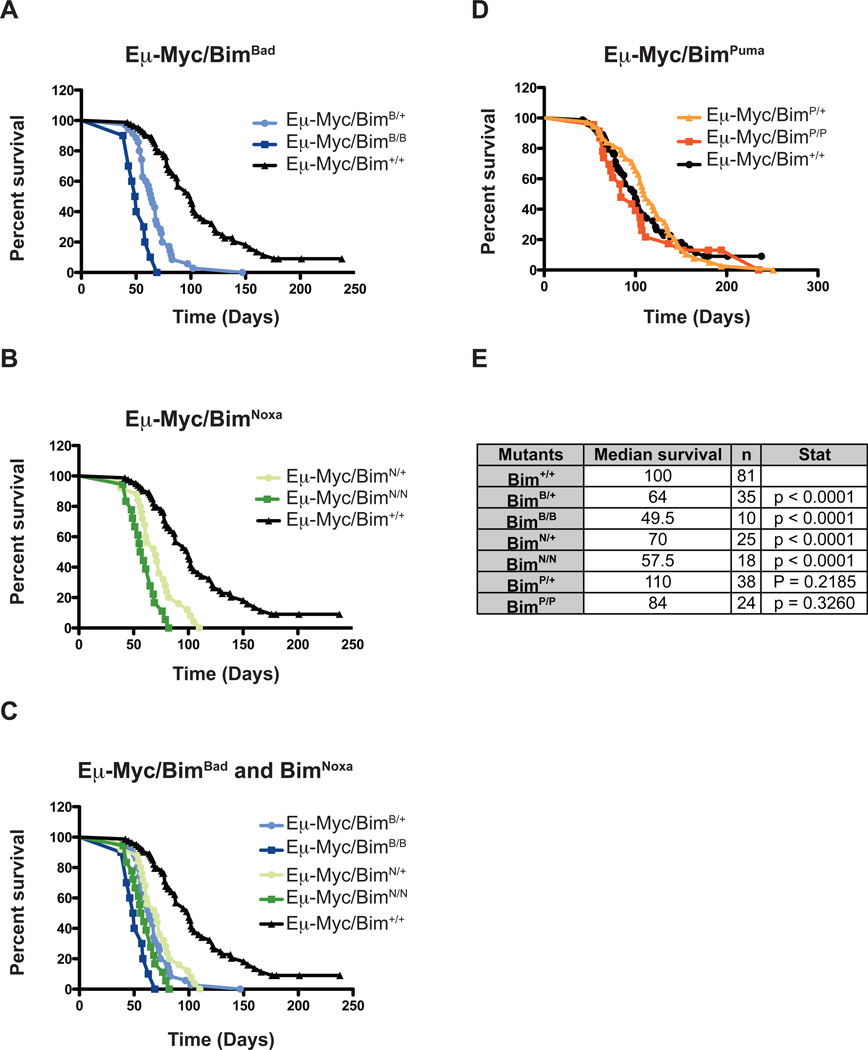
The survival of Eμ-Myc/BimBH3 mutant mice was compared with the survival of control Eμ-Myc mice. Remarkably, a single allele mutation of Bim into either BimBad or BimNoxa significantly decreased the survival of Eμ-Myc mice from ~100 to 64 or 70 days, respectively (Figure 2a and c). This median lifespan is similar to that previously reported for Eμ-Myc/Bim+/− mice (77 days; (Egle et al., 2004)), and consistent with the notion that Bim functions as a haplo-insufficient tumor suppressor. The Eμ-Myc/BimBad/Bad and Eμ-Myc/BimNoxa/Noxa mice became sick even more rapidly, with a median survival of 50 and 58 days, respectively (Figure 2c). This reduction in median survival is remarkably similar to the 57 days reported for Eμ-Myc/Bim−/− mice (Egle et al., 2004). Importantly, all the mice used in the present study were of the same genetic background (C57BL/6) as the mice used in our former study (Egle et al., 2004). Thus, in this model of tumorigenesis, BimBad and BimNoxa proteins behaved as complete loss-of-function mutants of Bim, indicating that in B lymphoid cells undergoing Eμ-Myc induced neoplastic transformation, the life-versus-death decision must be very tightly balanced. Since lymphomagenesis is accelerated both by disabling Bim from binding to either Bcl-2, Bcl-xL and Bcl-w (BimNoxa) or by preventing its interaction with Mcl-1 plus A1 (BimBad), we hypothesize that both of these subgroups of pro-survival Bcl-2 proteins must be critical to sustain pre-leukemic Eμ-Myc B lymphoid cells undergoing neoplastic transformation. Consistent with a role for Mcl-1 in such a process, its haplo-insufficiency protects mice from Myc induced AML development (Xiang et al., 2010). As for the relative importance of endogenous expression of Bcl-2, Bcl-xL or Bcl-w in Eμ-Myc induced lymphomagenesis, we predict that Bcl-xL is most critical, given that loss of Bcl-2 had no impact on this disease (Kelly et al., 2007) and that Bcl-xL but not Bcl-w is expressed at readily detectable levels in pre-malignant Eμ-Myc B lymphoid cells (Michalak et al., 2009).
Figure 2. The BimBad and BimNoxa mutations accelerated Eμ-Myc induced lymphomagenesis but the BimPuma mutation had no significant impact.
Analysis of lymphoma-free survival was carried out using Graph Pad Prism (version 5.0a). (a) Lymphoma-free survival of control Eμ-Myc as well as Eμ-Myc/BimBad/+, Eμ-Myc/BimBad/Bad, Eμ-Myc/BimNoxa/+ and Eμ-Myc/BimNoxa/Noxa mice. (b) Lymphoma-free survival of control Eμ-Myc as well as Eμ-Myc/BimPuma/+ and Eμ-Myc/BimPuma/Puma mice. (c) Summary of data presented in (a) and (b), showing for each genotype of mice the median survival, the number of mice analyzed and statistical analysis of difference in survival in comparison to control Eμ-Myc mice. Statistical evaluation of tumor-free survival data was undertaken using a log-rank test (Mantel-Cox) and 1-way ANOVA test on Kaplan-Meier curves.
The BimPuma mutation had no significant impact on lymphoma development in Eμ-Myc mice
We have previously shown that the replacement of the BH3 domain of Bim with the BH3 domain of Puma did not impair its ability to bind to all pro-survival Bcl-2-like proteins (Merino et al., 2009). This mutation did, however, cause a significant, albeit relatively minor, increase in leukocytes, indicating that actions of Bim that are independent of its ability to bind to pro-survival Bcl-2 family, such as direct interaction with Bax/Bak, must also be critical for optimal activity in developmentally programmed cell death (at least in the hematopoietic system). Interestingly, the median lifespans of Eμ-Myc/BimPuma/+ (110 days) and (despite a trend towards accelerated disease) even of the Eμ-Myc/BimPuma/Puma mice (84 days) was not significantly different from the median survival of control Eμ-Myc mice (Figure 2b and c). This indicates that the actions of Bim that are independent of its ability to bind pro-survival Bcl-2-like proteins are not essential for its tumor suppressive function, at least in Eμ-Myc mice. This apparent discrepancy with our previous conclusions (Merino et al., 2009) might simply be due to a difference of experimental setting. None of the cells considered in our former study were malignant, whereas they are in this study. This does, however, not exclude that direct activation of Bax/Bak can play a role in killing of Eμ-Myc B lymphoid cells undergoing transformation (Letai et al., 2004), as this function might be accomplished by another BH3-only protein present in these cells (e.g. Puma or Noxa) as long as Bim (i.e. BimPuma in this case) can neutralize all pro-survival Bcl-2 proteins.
The BimBad and BimNoxa but not the BimPuma mutation enhanced the severity of Eμ-Myc lymphoma
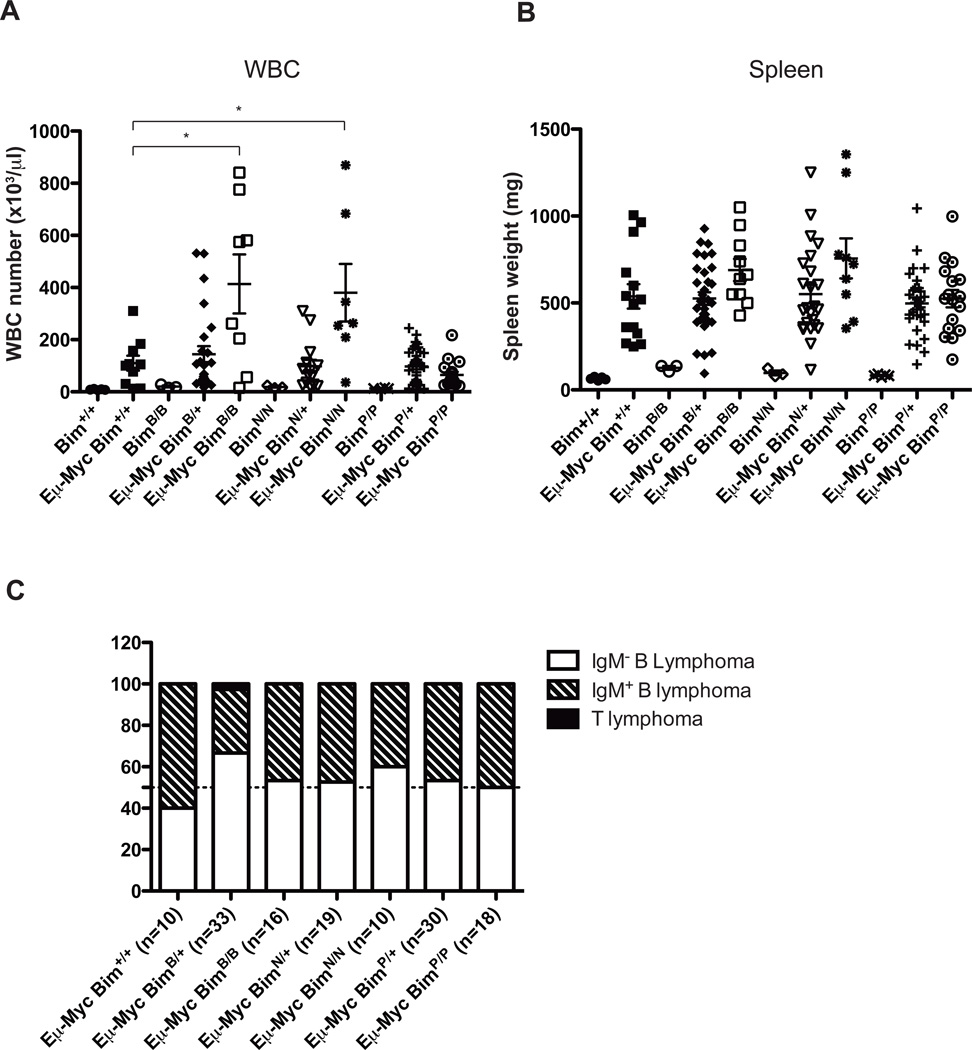
At autopsy, the spleen weights of all Eμ-Myc/BimBH3 exchange mutant mice were similar to those of sick control Eμ-Myc littermates (Figure 3a). Remarkably, however, the Eμ-Myc/BimBad/Bad and Eμ-Myc/BimNoxa/Noxa mice had significantly higher white blood cell (WBC) counts (Figure 3b). Conversely, the Eμ-Myc/BimPuma/Puma mice showed similar leukemic burden compared to control Eμ-Myc mice (Figure 3b). Finally, immuno-phenotyping did not reveal significant differences between lymphomas from the various genotypes of mice. B220+IgM+ B lymphomas were slightly more abundant in the control Eμ-Myc mice, whereas B220+sIgM− pre-B lymphomas predominated in all other genotypes (Figure 3c).
Figure 3. The BimBad and BimNoxa mutations but not the BimPuma mutation increased the leukemic burden in sick Eμ-Myc transgenic mice.
(a) Number of WBC and (b) spleen weight of wt (Bim+/+), control Eμ-Myc (Eμ-Myc/Bim+/+), BimBad/Bad, Eμ-Myc/BimBad/+, Eμ-Myc/BimBad/Bad, BimNoxa/Noxa, Eμ-Myc/BimNoxa/+, Eμ-Myc/BimNoxa/Noxa, BimPuma/Puma, Eμ-Myc/BimPuma/+ and Eμ-Myc/BimPuma/Puma mice at autopsy. Peripheral blood was analyzed using the ADVIA hematology analyzer (Bayer). Data represent means+/−SEM (n=numbers of animals analyzed for each genotype). Statistical analysis was performed using the Student t test. Statistically significant differences in comparison to Eμ-Myc/Bim+/+ mice are indicated (*P<0.05). (c) Diagrammatic representation of the relative frequencies of lymphoma types (sIg−pre-B lymphoma, sIg+ B lymphoma or Thy1+ T lymphoma) observed in mice of the indicated genotypes (n= numbers of mice analyzed for each genotype). Sick mice were sacrificed, lymphomas harvested and single cell suspensions prepared for FACS (FacsCalibur2; Becton Dickinson) analysis using fluorochrome-conjugated antibodies against IgM (5.1 or 333.12), IgD (11-26C) and B220 (RA3-6B2).
In conclusion, this study demonstrates that the binding to pro-survival Bcl-2 family members plays an important role in the tumor suppressor activity of the BH3-only protein Bim. The data also indicate that Mcl-1 and Bcl-xL may play critical roles in sustaining the survival of pre-leukemic Eμ-Myc B lymphoid cells whilst they are acquiring additional oncogenic lesions that promote their progression to full malignancy. This hypothesis could be tested by generating Eμ-Myc mice in which Mcl-1 or Bcl-xL can be deleted specifically in B lymphoid cells. If proven correct, one may consider to target Bcl-xL and/or Mcl-1 as a strategy for early intervention to prevent or delay Myc-induced tumorigenesis if early detection of pre-malignant lesions is possible or if patients are known to be predestined to develop such tumors.
Acknowledgments
We thank Mikara Robati and Owen Siggs for help with experiments, Jerry Adams, Suzanne Cory, Clare Scott and Cyril Clybouw for insightful discussions and Eμ-Myc mice, Bruno Helbert and Carley Young for mouse genotyping, Emily Sutherland and Giovanni Siciliano for mouse husbandry and Jason Corbin for blood sample analysis. This work was supported by the Australian National Health and Medical Research Council (program grant 461221, Australia Fellowship (AS) and Career Development Award (PB)), the National Cancer Institute (CA43540), the Australian Research Council (DM), the Leukemia and Lymphoma Society (LLS Specialized Center of Research grant 7015) and operational infrastructure grants through the Australian Government (IRISS) and the Victorian State Government (OIS).
Footnotes
Conflict of interest statement
The authors have no conflict of interest to disclose.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene. 2008;27:421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Köntgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A, Labi V, Chmelewskij W, Ploner C, Geley S, Fiegl H, et al. Suppression of B-cell lymphomagenesis by the BH3-only proteins Bmf and Bad. Blood. 2010;115:995–1005. doi: 10.1182/blood-2009-03-212670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci U S A. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Puthalakath H, Adams JM, Strasser A. Endogenous bcl-2 is not required for the development of Eμ-myc-induced B-cell lymphoma. Blood. 2007;109:4907–4913. doi: 10.1182/blood-2006-10-051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 Domains of BH3-Only Proteins Differentially Regulate Bax-Mediated Mitochondrial Membrane Permeabilization Both Directly and Indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in Eμ-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- Letai A, Sorcinelli MD, Beard C, Korsmeyer SJ. Antiapoptotic BCL-2 is required for maintenance of a model leukemia. Cancer Cell. 2004;6:241–249. doi: 10.1016/j.ccr.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O'Reilly LA, et al. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol. 2009;186:355–362. doi: 10.1083/jcb.200905153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nature Reviews Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Elefanty AG, Harris AW, Cory S. Progenitor tumours from Em-bcl-2-myc transgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO J. 1996;15:3823–3834. [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, et al. Genomewide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, et al. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest. 2010;120:2109–2118. doi: 10.1172/JCI39964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]