Summary
Mutations are typically perceived as random, independent events. We describe here non-random clustered mutations in yeast and in human cancers. Genome sequencing of yeast grown under chronic alkylation damage identified mutation clusters that extend up to 200 kb. A predominance of “strand-coordinated” changes of either cytosines or guanines in the same strand, mutation patterns and genetic controls indicated that simultaneous mutations were generated by base alkylation in abnormally long single-strand (ss)DNA formed at double-strand breaks (DSBs) and replication forks. Significantly, we found mutation clusters with analogous features in sequenced human cancers. Strand-coordinated clusters of mutated cytosines or guanines often resided near chromosome rearrangement breakpoints and were highly enriched with a motif targeted by APOBEC family cytosine-deaminases, which strongly prefer ssDNA. These data indicate that hyper-mutation via multiple simultaneous changes in randomly formed ssDNA is a general phenomenon that may be an important mechanism producing rapid genetic variation.
Introduction
Mutations drive disease and evolution. Along with frequency, location and timing are key parameters that define the biological outcomes of mutations. However, little is known about mechanisms that govern mutation distribution across the genome and across generations, especially those that would produce multiple simultaneous changes. While most mutations appear to be independent and distribute randomly, studies of mutations in mouse and human cells as well as tumor samples indicate that multiple mutations within genes can occur more frequently than predicted by chance (Drake, 2007 and refs within). This suggests that a portion of mutational events, up to 1% of the total, are non-random and could be chronologically linked (Wang et al., 2007). How these “mutation clusters” form is unclear, but acquisition of mutations within a single or few cell generations could enhance evolvability. As most mutations are neutral or deleterious, the production of multiple mutations in a locus increases the chances of inactivating a specific gene, but also allows for compensatory changes that can provide a selective advantage (Camps et al., 2007 and refs within). Furthermore, clustered mutations allow locally high mutation densities without overloading the rest of the genome, thereby reducing the chance of deleterious mutations that could obscure an advantageous change.
A proof of principle for the effectiveness of multiple, closely-timed mutations comes from somatic hyper-mutation (SHM) in activated B-cells. Following re-arrangement of the antigen receptor loci, lymphocytes express Activation Induced Cytosine Deaminase (AID) (reviewed in (Di Noia and Neuberger, 2007)). This protein deaminates cytosines to uracils primarily within immunoglobulin loci, directly causing C to T transitions or driving Pol eta-dependent mis-incorporations. Together, these result in multiple mutations clustered within the variable region of the antigen receptor. Subsequent selection allows expansion of cells containing variable domains with high antigen affinity. Beyond the specific example of SHM, studies of protein evolution (reviewed in (Camps et al., 2007)), as well as recent findings with regulatory sequences (Frankel et al., 2011)have shown that evolution often utilizes mutations that individually are neutral or even deleterious, but when combined cause a significant phenotypic change. Such multiple changes by random mutagenesis are highly improbable and in cases where “fitness valleys” must be overcome, either a duplication event followed by successive mutations, or linked, simultaneous changes are required (see (Smith, 1970)).
Here, we describe simultaneous induction of large numbers of clustered mutations and dissect the mechanisms that lead to their formation in yeast genomes and in human tumors. Mutation distributions and spectra, as well as genetic controls, implied that clustered mutations in yeast arose simultaneously through error-prone restoration of persistent ssDNA formed at DSBs or replication forks. Analysis of human cancer datasets also revealed clusters of mutations with characteristics strongly suggesting that their formation involves ssDNA. Moreover, many clusters, as well as a significant fraction of total mutations in cancers, carried a distinct mutation signature similar to those of APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like) cytosine deaminases.
Results
DNA damage induces clustered mutations
We hypothesized that mechanisms exist which can lead to the formation of mutation clusters at frequencies detectable in moderately sized mitotic populations. Therefore, we designed a strain of the yeast, Saccharomyces cerevisiae, that enabled selection of 2 or more closely spaced mutations induced by exposure to a DNA damaging agent. Specifically, we moved the URA3 and CAN1 coding sequences and endogenous promoters from their normal, widely separated positions in chromosome V to adjacent positions within the LYS2 gene on chromosome II (Figure 1). This chromosomal location allows the selection of events where URA3 and CAN1 are inactivated by multiple point mutations but not by gross chromosomal rearrangements (Chen and Kolodner, 1999), because the latter would eliminate surrounding essential genes. We reasoned that mutation clusters could originate from multiple unrepaired lesions in either double stranded (ds) or ssDNA. Regions around DSBs are prone to spontaneous mutation (Hicks et al., 2010; Ponder et al., 2005; Strathern et al., 1995; Yang et al., 2008), and when these regions are manipulated to be artificially long-lived, they can accumulate ssDNA with multiple DNA damages that each lead to a mutation (Burch et al., 2011; Yang et al., 2010; Yang et al., 2008). Clusters originating from lesions in dsDNA would invoke strong local inhibition of DNA repair by a yet unknown mechanism (See Discussion). To discriminate between ssDNA and dsDNA sources of clusters, we induced mutations by chronic, low dose exposure to methyl-methanesulfonate (MMS). This mutagen has a diagnostic ssDNA mutation signature (Yang et al., 2010) but also induces mutations through damage to dsDNA.
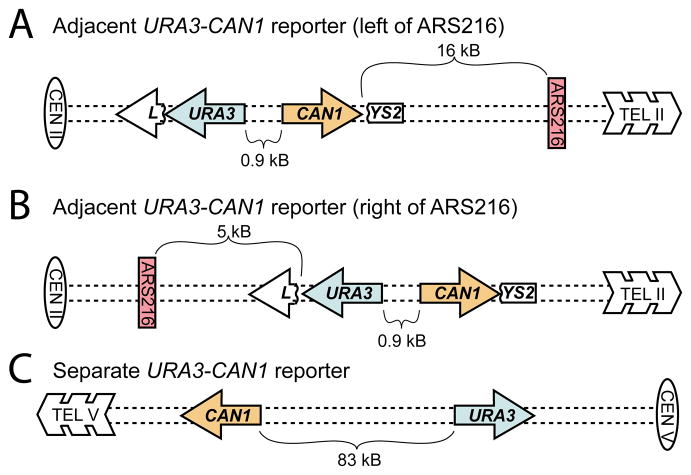
Figure 1.
Multiple Mutation Reporters. Presented are yeast strains with URA3 (blue) and CAN1 (orange) separated by either 0.9 kb inserted into LYS2 (adjacent reporters) to the left (A) or right (B) of ARS216 (red) on chromosome II or by 83 kb on the left arm of chromosome V (separated reporters; C). Isolates that were mutant in both genes were selected by resistance to the drugs 5-FOA and canavanine.
Both haploid and diploid versions of the URA3-CAN1 reporter strains were grown on rich media containing non-lethal concentrations of MMS (Table S1) for approximately 25 generations to induce DNA alkylation and mutations. Growth in the presence of MMS greatly elevated the frequencies of cells resistant to the drugs 5-FOA and canavanine (CAN) individually (ura3 and can1 selection, respectively; Table S2) as well as the frequency of isolates resistant to both drugs (referred to as FOA-CAN resistant; Figure 2A). Exposure of wild type haploid yeast carrying adjacent URA3-CAN1 genes to 2 mM and 4 mM MMS increased the estimated frequencies FOA-CAN resistance to 1.4 and 7.9 colonies/plate, respectively, over a background of 0.15 resistant colonies/plate resulting from spontaneous deletion of the reporter. After correcting for the Loss of Heterozygosity (LOH) (Table S2) required for expression of recessive phenotypes, the estimated frequency of mutations conferring FOA-CAN resistance for diploid versions of these strains treated with 4 mM MMS appeared even higher than that for similarly treated haploid cells, ranging from 57 to 91 resistant colonies/plate. In all cases, the frequency of dually resistant isolates from the adjacent URA3-CAN1 strains was much higher than expected from the accumulation of independent ura3 and can1 mutations as measured by FOA-CAN dual resistance of separated URA3-CAN1 strains (compare Adj and Sep in Figure 2A; Table S2). This suggested an association of the mutational events.
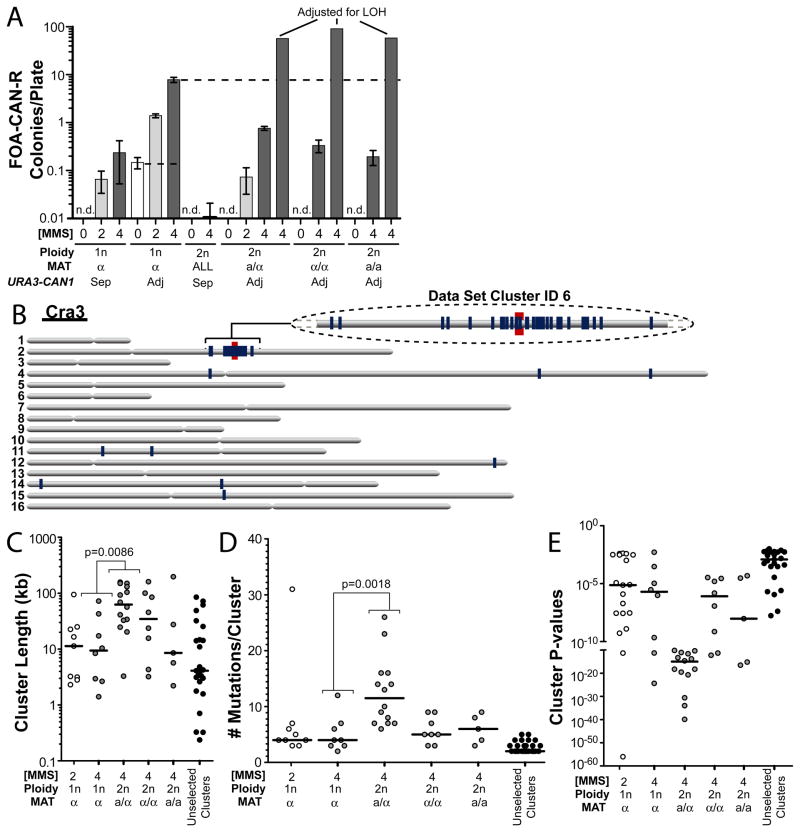
Figure 2.
Selection and Characterization of Mutation Clusters. FOA-CAN mutation frequencies for wild type haploid (1n) and diploid (2n) yeast carrying the adjacent (Adj) or separated (Sep) URA3-CAN1 reporters (see Figure 1 reporters A and C). Selected isolates were sequenced. (A) The median number of colonies/plate. Error bars indicate 95% confidence intervals (CI). “ALL” represents colonies from MAT a/α, α/α, and a/a combined. LOH adjusted values are indicated (see also Table S2, Supplemental text). (B) An example FOA-CAN resistant genome (named Cra3): gray bars represent each of the 16 yeast chromosomes, the red line indicates the location of URA3 and CAN1, and blue lines denote the position of mutations. A segment of chromosome II containing a mutation cluster is enlarged. Cluster lengths (C), the number of mutations per cluster (D), and Cluster P-values (E) of all sequenced chromosome II clusters arising from cells treated with 2 (white circles) or 4 mM (gray circles) are presented. Black circles indicate unselected clusters. Solid lines indicate median values.
Sanger sequencing of the URA3, CAN1, and LYS2 regions suggested mutation clusters may be large because the data contained an unusually high frequency of mutations in excess of the 2 minimally required for resistance (20% of sequenced clones treated with 2 mM and 50% treated with 4 mM; Table S3A). We therefore investigated the appearance of mutations throughout the genomes of selected isolates. Sixty-nine yeast clones were sequenced, chosen from 4 genotypes, 2 MMS concentrations, and 3 selection methods (i.e., unselected, 5-FOA selected, and FOA-CAN selected). As shown in Table S4A, 59–78% mutations occurred within genes, comparable to the percentage of the genome that is protein coding. Base pair substitutions accounted for 90% of all mutations; 7% were small insertions or deletions (termed Indels) and 3% were categorized as “complex,” consisting of 2 or more mutations with each separated by less than 10 bp from their nearest neighbor. Since complex mutations likely originate from trans-lesion synthesis past a single DNA lesion (Harfe and Jinks-Robertson, 2000), they were treated as a single event.
The median genome-wide mutation density in MMS treated yeast was low, at 12 mutations per haploid genome or ~1 mutation/Mb (Table S4). To address the possibility of clustering, we identified groups of mutations in which each mutation was no further than 100 kb from the next (chosen to be a 10 fold higher mutation density than genome-wide) and calculated the likelihood of observing the mutation distribution assuming all the mutations in the genome were independent and random. Groups of mutations with P-values <0.01 were classified as mutation clusters. Using this analysis, all ura3 and can1 mutations selected by FOA-CAN resistance comprised at least part of a mutation cluster (Table S4B). Surprisingly, FOA-CAN selected clusters ranged from 2 to a remarkable 30 mutations spanning more than 150 kb (Figure 2C and D, Table S4C, Figure S1). Importantly, the remaining 99% of the genome was relatively devoid of mutations (containing 1 to 52 mutations) (Table S4B, e.g., Figure 2B). P-values of FOA-CAN selected clusters ranged between 10−2 and 10−57 with P<10−5 for 55% of clusters (Figure 2E). Clusters were generally longer in diploid yeast than in haploid (Figure 2C), with all FOA-CAN selected clusters greater than 100 kB in length originating from diploid strains. This apparent impact of ploidy on cluster size, however, may be at least partly due to inhibition of non-homologous end-joining (NHEJ) in the a/alpha mating type (Valencia-Burton et al., 2006) and refs within), as NHEJ can compete with DSB end resection (see below). Indeed, no statistical difference in cluster size was observed when comparing diploids with homozygous mating types (clusters from alpha/alpha and a/a mating types combined) to similarly treated haploids (P=0.205 for length and P=0.36 for number of mutations comparing yeast treated with 4 mM MMS; Figure 2C and D). In addition to clusters containing mutations in URA3 and CAN1, we also identified 25 unselected clusters among 17 isolates with P-values ranging between 1.72X10−8 and 9.9X10−3 (Table S4D). These appear to be bona fide as they are not associated with subtelomeric regions (i.e., having mutations within 30 kb of a chromosome end) or other repeat regions (Long Terminal Repeats (LTRs) or retrotransposons) that can produce false clusters as a result of mapping artifacts. Unselected clusters displayed characteristics similar to those selected by FOA-CAN resistance (see below). The existence of unselected clusters indicates that while the occurrence of FOA-CAN resistant colonies is rare, cluster formation in other areas of the genome may be relatively frequent or that specific sub-populations may be particularly prone to mutation clusters.
MMS-induced mutations in clusters are strand-coordinated and simultaneous
The median density of MMS-induced mutations in clusters was 1 mutation per 3 kb, over 500 fold greater than in the rest of the genome. Furthermore, the probability of observing any individual cluster by the random coincidence of mutations was often incredibly low (e.g., P-values = 9.4x10−57 to 3.3x10−5 for clusters with >4 mutations). This suggested that the mutations within a cluster were not independent but instead were concerted in time and by mechanism. Supporting the non-random incidence of mutations in clusters, the estimated frequency of FOA-CAN resistance in the haploid strain with adjacent URA3 and CAN1 genes was 50 fold higher than when these genes were separated by 83 kb (Figure 2A, Table S2). Additional evidence confirming the simultaneous incidence of clustered mutations (i.e., occurring within one generation) emerged from the analysis of the MMS-induced mutation spectrum. While all types of mutation were present in clusters, substitutions at G:C base pairs predominated (Figure 3, Figure S2). Strikingly, when only clustered mutations in G:C pairs were displayed, a phenomenon that we defined as “strand-coordination” became evident. The substitutions of G:C pairs occurred almost exclusively as long series of either mutated cytosines or mutated guanines in the sequenced strand (Figure 4A and B). If DNA damage and subsequent mutations were accumulated through multiple cell divisions, mutation of cytosine (or guanine) would have the opportunity to occur on either DNA strand. Consequently, clusters formed with non-simultaneous mutations of G:C pairs (i.e. occurring in different cell generations) would be expected to show a random mix of guanines and cytosines mutated in the same strand. Thus, strand-coordinated stretches of mutations are likely generated simultaneously within a single cell cycle.
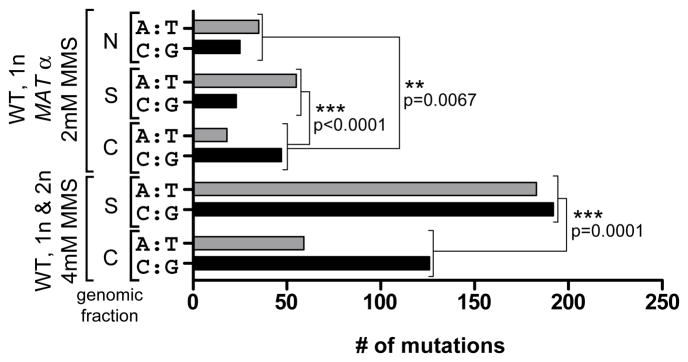
Figure 3.
MMS-induced Mutated Base Pairs. The total mutated A:T and C:G base pairs in FOA-CAN dually resistant wild type haploid and diploid yeast carrying the URA3-CAN1 reporter selected after 2 day exposure to 2 or 4 mM MMS were divided based upon whether they were clustered (C) or scattered (S). “N” indicates scattered mutations for isolates where no FOA-CAN selection was applied. Mutations in URA3 and CAN1 as well as annotated repeat regions including LTRs and retrotransposons were excluded from totals. P-values were determined with two-sided Fisher’s Exact Test (see also Figure S2).
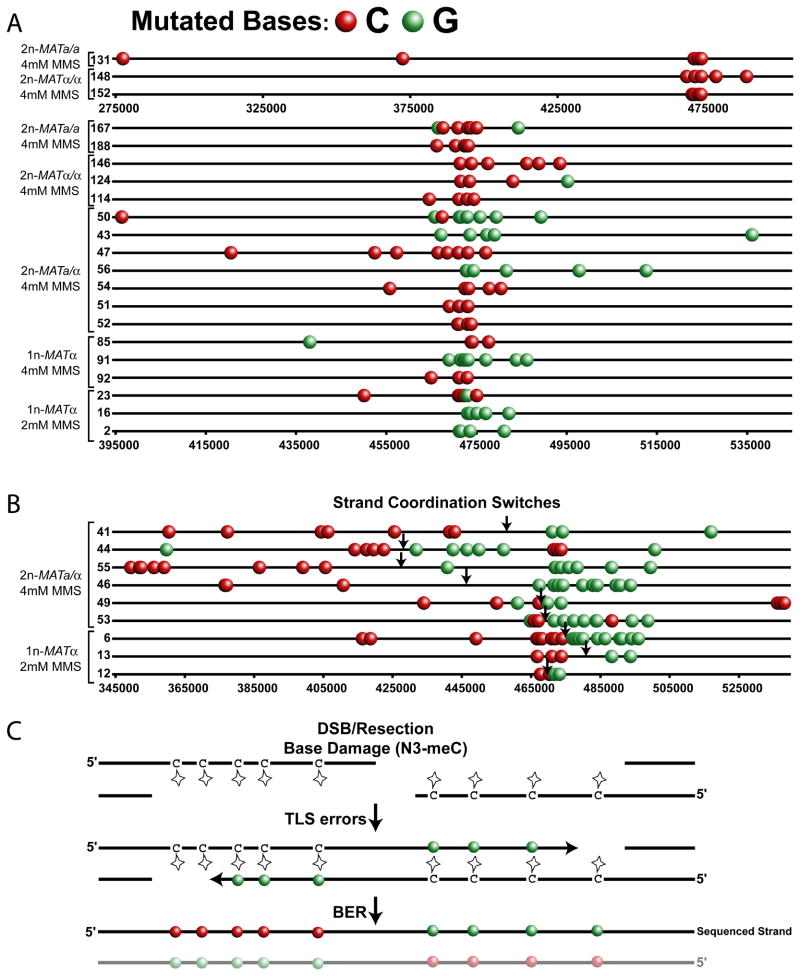
Figure 4.
Strand-Coordination of Clustered Mutations. Each line (numbers represent the Dataset Cluster ID from Table S4B) depicts the mutated cytosines (red circles) and guanines (green circles) in the sequenced strand of a FOA-CAN selected cluster containing more than 3 guanines and/or cytosines originating from the indicated wild type strains carrying the adjacent URA3-CAN1 reporter treated with either 2 or 4 mM MMS. Only mutations at C and G bases are depicted. The complete mutation data, including the mutations at A and T bases, is presented in Figure S1 and Table S4B. Clusters are separated into two categories (A) non-switching clusters and (B) switching clusters. A coordination switch is defined as 2 or more consecutive coordinated mutations followed by at least 2 consecutive coordinated mutations of the opposite base. Approximate break points are indicated by arrows. Within categories, clusters are sorted by genotype, MMS treatment and finally by cluster length. Clusters labeled with Dataset Cluster IDs 131,148 and 152 are presented as a subpanel within (A) since they require a significantly different scale than the remaining non-switching clusters. (C) Model of MMS induced clusters caused by N3-methyl cytosine adducts in the single-strand overhangs of resected DSBs.
Clustered mutations arise from alkylation damage to ssDNA
We next compared the MMS-induced mutation spectra of non-clustered (termed scattered) mutations distributed throughout the genome to that found in mutation clusters. Based on the concentration of MMS used, scattered mutations either occurred primarily at A:T bases (2 mM MMS = 71% A:T and 29% G:C) or showed no base preference (4 mM MMS = 49% A:T and 51% G:C). At either concentration of MMS, however, mutation clusters were enriched in mutated G:C pairs (Figure 3 and Figure S2) despite the higher prevalence of A:T bases in chromosome 2 (Figure S2) indicating the G:C preference was not due to the base composition in the region of selection. This enrichment suggests the contribution of unique mutagenic lesions in cytosine and/or guanine within the mutation cluster spectrum. The strand-coordination of clustered mutations requires that there be only one primary mutagenic lesion per base pair which, in the case of G:C pairs, could be a methylated cytosine or a methylated guanine. Based upon the known spectrum of MMS-induced alkylation (Beranek, 1990), the mutagenic capacity of MMS lesions, and our previous work describing the mutation spectrum of MMS damaged ssDNA (Yang et al., 2010), we suggest that the selected mutation clusters result largely from N3-methyl cytosine, a mutagenic single-strand specific MMS lesion. Consistent with this suggestion, the mutation spectrum in clusters showed a significant under-representation of C to G and G to C transversions (Table S4A), as previously reported for error-prone bypass of N3-methyl cytosine lesions in E.coli (Delaney and Essigmann, 2004). Thus, persistent ssDNA is apparently formed naturally, likely in response to DNA damage. Once formed, these regions can accumulate ssDNA-specific damage, presumably due the lack of repair capacity in ssDNA, ultimately resulting in error-prone trans-lesion synthesis (TLS) during the restoration of ssDNA to dsDNA.
Clustered mutations are formed during DSB repair
The identification of ssDNA as the target for MMS-induced mutation clusters suggests several cellular processes that may contribute to cluster formation, including resection of a DSB during homology directed repair. Resection from a DSB occurs only in the 5′ to 3′ direction (Mimitou and Symington, 2011). Consequently, N3-methyl cytosines accumulated in the ssDNA on opposite sides of a break would belong to opposite strands of the repaired double helix. Since sequencing only reports one strand of a chromosome oriented in the 5′ to 3′ direction, a cluster resulting from damage to bi-directional resection of a DSB should display mutated cytosines to the 5′ side of a DSB, while the 3′ side, which is complementary to the damaged ssDNA stretch, should display mutated guanines (Figure 4C). The position of the switch from C-coordination to G-coordination in the sequenced strand would indicate the approximate location of the break site. Among the 69 sequenced genomes, 9 clusters contained strand-coordination switches (Figure 4B). Seven of these clusters clearly demonstrated the expected pattern for N3-methyl cytosine lesions within bi-directional resection tracts: C-coordination always on the 5′-side and G-coordination on 3′-side. The 2 remaining switching clusters may also be associated with DSBs but appear to be more complex events (Figure 4B, Dataset Cluster IDs 44 and 49). Thus, recombinational DSB repair likely accounts for least 13% of the total clusters we sequenced. Interestingly, many switching clusters spanned further than 100 kb and would require resection tracts significantly longer than those previously estimated using direct measurements at site-specific (Chung et al., 2010 and refs within) or radiation-induced breaks (Westmoreland et al., 2009) in populations. Such unexpectedly long resection seems unnecessary for the repair of most DSBs, suggesting these clusters reflect a minor category of events where downstream steps of recombination were delayed. Remarkably, even these unusual repair intermediates are restored without deletion despite the presence of multiple lesions in the long stretches of ssDNA.
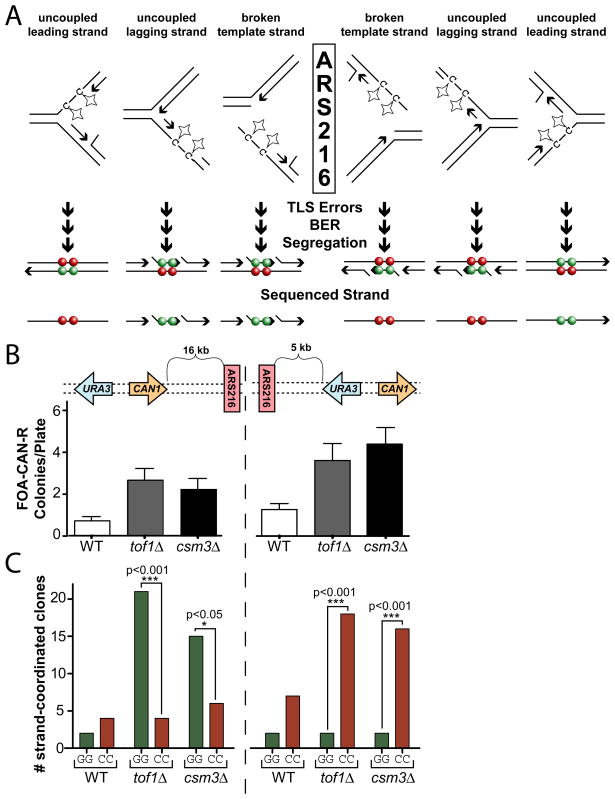
Dysfunctional replication forks result in mutation clusters
While long bi-directional resection of DSBs was the likely origin of large clusters with strand-coordination switches, clusters without switching could be generated by a variety of mechanisms producing ssDNA, one potential source being dysfunctional replication forks. Long ssDNA regions are produced when replicative polymerases stall at lesions but the MCM helicase continues to proceed (Lopes et al., 2006). To assess the contribution of DNA replication in the formation of non-switching clusters, we utilized genetic defects that exacerbate fork dysfunction. In Saccharomyces cerevisiae, deletion of TOF1, CSM3, or MRC1 which encode components of the replication fork protection complex, increases polymerase-helicase uncoupling during hydroxyurea treatment (Bando et al., 2009; Katou et al., 2003). We therefore disrupted TOF1 and CSM3 to increase the prevalence of replication-associated ssDNA and selected for MMS-induced mutation clusters using two versions of the deficient strains: one containing the URA3-CAN1 reporter to the left (5′) of the ARS216 origin of replication and a second with the reporter to the right (3′) of ARS216 (Figure 1A and B, respectively). Regardless of the location of the URA3-CAN1 reporter, disruption of either TOF1 or CSM3 increased the frequency of FOA-CAN resistant clones 3–4 fold (Figure 5B), indicating that 66 to 75% of the isolated clusters were caused by replication fork defects. Sanger Sequencing of the URA3 and CAN1 coding regions revealed that clusters from both the tof1Δ and csm3Δ backgrounds were strand-coordinated (i.e., having a higher frequency of C-C and G-G mutation pairs than the C-G and G-C mutation pairs) similar to clusters obtained from wild type strains (Table S3B). Moreover, the strand-coordinated clones displayed “strand bias” dependent on the position of the selective reporter relative to the origin of replication. Strand-coordinated clusters selected to the left (5′-side) of ARS216 in the tof1Δ and csm3Δ strains were composed primarily of mutated guanines whereas those selected to the right (3′-side) of the origin consisted almost exclusively of mutated cytosines (Figure 5C and Table S3C). This change in bias based on the position of the reporter confirms that the majority of strand-coordinated clusters in the tof1Δ and csm3Δ backgrounds are associated with the replication fork and suggests that ssDNA generated at stalled forks may be a significant contributor to cluster formation in wild type cells. Furthermore, the specific bias observed (guanines mutated on the left (5′) of origin and cytosines mutated to the right (3′)) implies that replication-associated ssDNA was formed by either specific uncoupling of lagging strand synthesis, possibly the result of increased Okazaki fragment length, or through damage of the ssDNA exposed during one-ended recombinational repair of a collapsed fork (one of the outcomes on Figure 5) by mechanism of break-induced replication (Llorente et al., 2008).
Figure 5.
Replication-Associated Clusters. (A) Possible causes of ssDNA at dysfunctional forks. Breakage of the leading strand template would produce a similar outcome as breakage of the lagging strand template and thus is omitted for simplicity. Expected biases of mutated cytosines (red circles) and/or guanines (green circles) in the sequenced strand depend on their location relative to the replication origin. (B) FOA-CAN mutation frequencies induced with 2 mM MMS. Error bars indicate 95% CI. (C) The URA3 and CAN1 open reading frames of selected clones from (B) were PCR amplified and Sanger sequenced. Isolates containing G-G or C-C strand coordinated mutations were tabulated. P-values were calculated by 2-tailed g test for goodness-of-fit to an expected 1:1 ratio.
Clusters of strand-coordinated mutations highlight mutagenic pathways in human cancers
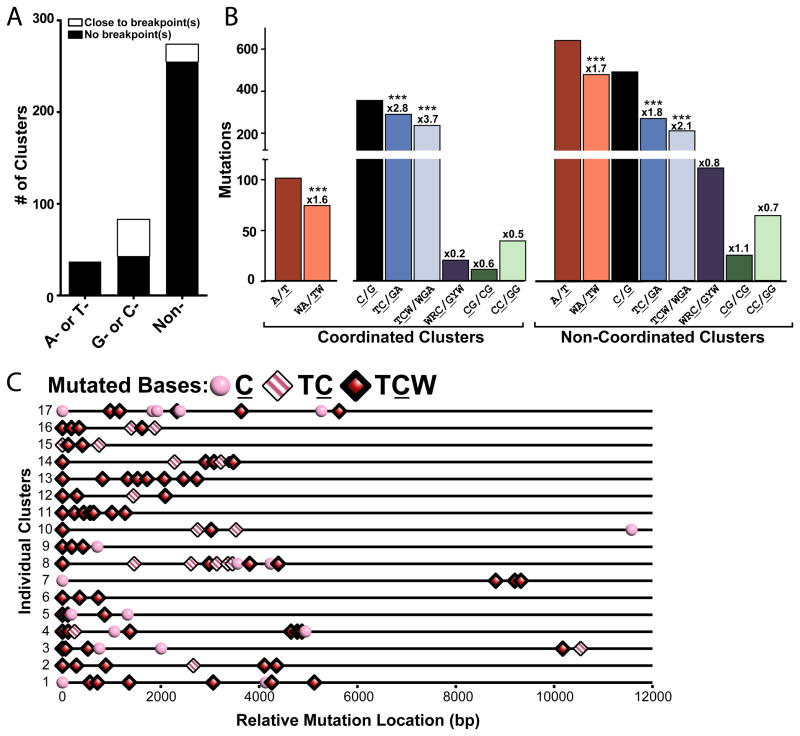
Do mutation clusters occur in other biological systems? To address this question, we searched for clusters among the 210,022 mutations identified in whole-genome sequencing of the following human malignant tumors: 23 multiple myelomas (Chapman et al., 2011), 7 prostate cancers (Berger et al., 2011), and 2 head and neck squamous cell carcinomas (HNSCC) (Stransky et al., 2011). We looked for mutation densities and clustering P-values similar to those of MMS-induced clusters in yeast. Specifically, we defined a cluster as 2 or more mutations in which: (i) all immediate neighbors were separated by no more than 10 kb and (ii) and whose cluster P-value was no greater than 10−4 (Supplemental text). To focus on clustered mutagenesis occurring at random genomic locations, we also excluded clusters identified in a small fraction of the genome containing known targets for AID (Table S5) as they are likely the result of SHM (see Introduction). In non-SHM regions, we found 394 clusters scattered among all chromosomes containing 1,625 mutations and ranging in size from 12 to 53,411 base pairs (Figure 6, Tables S6 and S7). Similar to our observations in yeast, clusters in cancers had an overall strong strand-coordinated component. This indicated that in the majority of clusters, most of the mutations occurred simultaneously in the same DNA molecule (Table S7B). This conclusion allowed us to apply cluster analysis to sequenced cancers in spite of the often heterogeneous cellular composition of tumors. As the coincidence of mutations is even more likely for clusters composed entirely of mutations of the same base in the sequenced strand, we classified clusters into 3 groups: A- or T-coordinated, C- or G-coordinated, or non-coordinated for further analysis aimed at identifying mechanisms leading to cluster formation. There were 8, 18 and 94 completely strand-coordinated clusters in the HNSCC, prostate, and myeloma datasets respectively, with the majority of these C- or G-coordinated (Figure S3A, Table S7C).
Figure 6.
Mutation Clusters in Sequenced Human Cancers. (A) Mutation clusters in non-SHM regions separated by type of strand-coordination (“A- or T-” coordinated, “G- or C-” coordinated, and “non-” coordinated). White bars indicate the number of clusters co-localized with breakpoint(s). Co-localization was registered when the region covered by the cluster plus left and right flanks of 20,000 nucleotides contained at least one breakpoint. Black bars depict the number of clusters not associated with a specific breakpoint. (B) Number of mutations from the coordination classes in (A) that occurred within specific sequence motifs. Numbers above bars indicate the fold enrichment of clustered mutations occurring at a motif over the frequency of the motif occurs in the chromosomal sequence that the cluster spans (See Cluster Sequence in Table S9). Asterisks demark motifs significantly enriched in mutation clusters (P-value <0.0001) as determined by Chi-square (See also Figures S3). (C) Distribution of mutations within 17 C-coordinated clusters with greater than 3 mutations. Mutated cytosines are categorized by their presence in a TC motif (pink and white striped diamonds), TCW motif (red diamonds), or no identified motif (pink circles).
Mutations in A- (or T-) coordinated clusters were enriched at a motif, WA/TW (where W is A or T; the mutated base is underlined). This motif is identified with mutations generated by TLS DNA polymerase eta in Ig-SHM regions (Rogozin et al., 2001). Consistent with Pol eta introducing the mutations, A- or T-coordinated clusters were composed primarily of A to G and T to C transitions (Table S7D), however, alternative ways of generating A- or T-coordinated clusters exist but are not distinguished in this study.
Nearly all mutations in C- or G-coordinated clusters were in TC or GA motifs, which in turn were strongly enriched at TCW or WGA in each type of cancer and in the total of the three datasets (Figures 6B, 6C and S3). This motif specificity exactly matches the signature of the RNA and DNA editing cytosine deaminases APOBEC1 (A1) (Beale et al., 2004), APOBEC3A (A3A) (Thielen et al., 2010), APOBEC3B (A3B), APOBEC3F (A3F) (Bishop et al., 2004) and APOBEC3H (A3H) (Henry et al., 2009) (hereafter referred to as TC-specific APOBECs). In agreement with uracils being formed by deamination, the most frequent substitution at cytosine was thymine, which could be due to the insertion of adenine across from uracils or AP sites generated by uracil DNA glycosylase. Guanine was the second-most frequent substitution at cytosine, which is also consistent with the mutation properties of AP sites in yeast as well as mammalian TLS (Table S7D) (Gibbs et al., 2005; Simonelli et al., 2005). Another striking feature of C- or G-coordinated clusters was that in all three cancer datasets, they often co-localized with one or more rearrangement breakpoints identified in those studies. In contrast, no A- or T-coordinated clusters co-localized with breakpoints (Figures 6A and S3A, Table S7C). The specific colocalization of C- or G-coordinated clusters with rearrangements suggests that they may be associated with ssDNA intermediates of DNA repair processes that generate rearrangements (Chen and Kolodner, 1999), and further strengthens our speculation about the APOBECs, which have a strong preference for ssDNA over dsDNA (Harris and Liddament, 2004). Altogether, the combination of strand-coordination, prominent enrichment with a motif targeted by a physiological ssDNA-specific mutagen and co-localization with rearrangement breakpoints strongly suggests that C- and G-strand coordinated clusters originated from ssDNA stretches formed by abnormal DSB processing or at uncoupled replication forks, similar to the mechanisms that we revealed in our yeast system.
Interestingly, non-coordinated clusters were also enriched in mutations at the WA/TW and TCW/WGA motifs suggesting that this class of clusters may arise from the combined action of the two mechanisms separately responsible for each category of coordinated clusters. Non-coordinated clusters, similar to C- or G-coordinated clusters, were not enriched in any other motifs analyzed (Figure 6B) (e.g., CC for APOBEC3G (Beale et al., 2004) or CpG) including the WRC/GYW (where R is T or C and Y is A or G) motif characteristic of AID activity (Rogozin and Kolchanov, 1992), suggesting that these clusters are not the result of inappropriately targeted SHM.
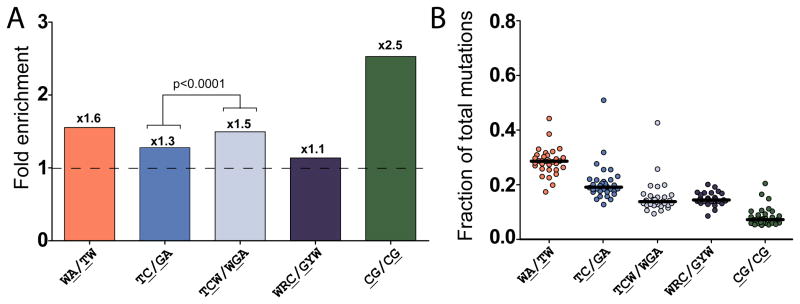
We proposed that mechanisms that lead to clustered mutations could have contributed significantly to the overall mutagenesis during the evolution of the sequenced tumors. Mutations in the CpG (CG/CG) motif are usually highly enriched in tumors (Greenman et al., 2007). Indeed, there was a statistically significant enrichment of CpGs among mutations as compared with the presence of these motifs in the immediate neighborhood of mutated bases (+/− 10 nt) across all of the tumors examined (Figure 7A, Figure S4, Table 8). However, since the presence of CpG motifs in genomes is generally reduced (Swartz et al., 1962), the fraction of mutations in CpG was usually small (Figure 7B, Figure S4, Table S8). Importantly, we observed enrichment of the 2 motifs that dominate clustered mutations, WA/TW and TC/GA. Similar to mutations in clusters, there was not a consistent genome-wide enrichment of mutations at the AID motif (WRC/GYW) in all tumor samples, even though potential genomic targets of AID were not excluded from this analysis. Mutations in TC/GA were enriched across all samples and comprised one of the most frequent classes of mutations, reaching ~50% of all mutations in one tumor (Figure 7B, Table S8B). Importantly, the enrichment with TCW or WGA, which define APOBEC A1/A3A/A3B/A3F/A3H specificity, was higher than the enrichment with the less stringent TC/GA motif across all cancer samples (Figure 7A, Table S8B) suggesting that these enzymes may be responsible for a fraction of TC/GA mutations and contribute significantly to the overall mutagenesis seen in tumors.
Figure 7.
Genome-wide Prevalence of Mutated Motifs in Sequenced Human Cancers. (A) Median fold enrichment of genome-wide mutations at various motifs (motifs identified in clusters plus CpG - CG/CG and the AID target motif – WRC/GYW). Numbers above bars indicate the fold enrichment of mutations occurring at a motif over the frequency of the motif in a fraction of sequence representative of the each genome. The enrichments of TC/GA and TCW/WGA also were compared by Wilcoxon signed rank test. (B) Fraction of total mutations occurring at the motifs described in (A) for individual sequenced tumors (circles). The horizontal bar indicates the median value (see also Figure S4).
Discussion
Clusters of simultaneous mutations are a general phenomenon
Our results revealed the presence of mutation clusters apparently associated with long ssDNA in two very different biological systems: yeast cells proliferating under chronic DNA damaging conditions and in human malignant tumors, whose only similarity is the high density of mutations accumulated over generations (0.1–2 per Mb). Once a mutagen is present (MMS in yeast or APOBEC in cancers), the limiting factor in cluster formation appears to be the formation of ssDNA, where the length of ssDNA region and the time it persists are the key parameters determining the cluster’s mutation density and length. Importantly, many DNA damaging agents, in addition to being mutagenic, can increase formation of ssDNA by causing single and double strand breaks or fork uncoupling (Friedberg, 2006 and refs within). Therefore, formation of ssDNA in MMS-treated yeast and in cancers could well be associated with the same DNA damage that led to mutations. Both MMS base damage and APOBEC cytosine deamination are handled by base excision repair (BER) systems. DSBs could occur either through creation of an unstable backbone at AP sites or through BER-associated breakage (Ma et al., 2008), thereby increasing the likelihood of cluster formation. Alternatively, as generation of ssDNA is a part of many cellular processes including transcription, DNA repair, and even the DNA damage checkpoint, it may be formed independently of mutagenic damage. Mammals employ safeguards against the production of ssDNA potentially to combat hyper-mutability. Specifically for the case of end-resection, the amount of ssDNA produced in mammalian cells is limited due to the preferential use of non-homologous end joining for repair of DSBs (Beucher et al., 2009). Also, BRCA1 and 53BP1 control the length of ssDNA created once a DNA end has committed to resection (Bunting et al., 2010). Since we found that human tumors contain mutation clusters co-localizing with rearrangement breakpoints, we suggest that the mammalian end resection machinery is capable of generating long ssDNA stretches which are prone to mutation cluster formation. Based on our yeast results, dysfunctional (potentially uncoupled) replication forks should also be considered as a source of ssDNA, either continuous or in patches, that initiates mutation clusters in human cancers. Thus, it is important to uncover the genetic defects and conditions that could lead to formation of persistent long ssDNA during break resection and/or fork uncoupling.
A permanent record of hyper-mutation in human cancers
Our analysis revealed that mutations in C- or G-coordinated clusters in sequenced human cancers often fell into motifs characteristic of a subgroup of APOBEC family cytosine deaminases. These enzymes have been implicated in carcinogenesis based on their mutagenic capacity and presence of mutations in TCW motifs among oncogenic mutations (Beale et al., 2004). TC or TCW base substitutions are also highlighted in the lists of mutations from several cancer samples (Stephens et al., 2005). Our findings of many clusters composed almost entirely of strand coordinated mutations in TC(W)/(W)GA motifs and the co-localization of these clusters with rearrangement breakpoints support a speculation that TC(W)-specific APOBECs could have acted as potent ssDNA mutagens during the history of some cancers. Consistent with this view, over-expression of APOBEC1 is tumorigenic in mice (Yamanaka et al., 1995). Thus, a detailed understanding of the regulation of the APOBECs and mechanisms limiting their access to chromosomal DNA is important, especially in light of recent work implicating specific APOBEC family members in active demethylation of chromosomal DNA in stem cells (Chahwan et al., 2010 and refs within). Based on a combination of the biochemical capacity of the APOBEC enzymes to induce simultaneous mutations in ssDNA and the mutation signatures observed in cancers, we speculate that genome-wide mutations may occur within one cell cycle through APOBEC activity at multiple, concurrently formed ssDNA regions such as replication forks or at abnormally persistent transcription bubbles (Aguilera, 2002). This provides a specific means by which the increase in mutability often ascribed to carcinogenesis (Loeb, 1991), could operate via a transient hyper-mutability that generates multiple mutations and results in selective growth advantage over just one or a very few cell generations.
Multiple paths to mutation clusters
We show here that the combination of strong selective pressure and chronic non-lethal DNA damage can result in clusters of up to 30 simultaneous mutations in yeast suggesting that localized, transient hyper-mutation is the most efficient means to produce change in this situation. Furthermore, we discovered that several sequenced human cancers harbor clusters with features similar to those found in yeast. While damaged ssDNA appears to be the primary cause of mutation clusters in yeast and even in a certain significant fraction of clusters in cancer mutation datasets, several other mechanisms and mutagenic factors likely contribute to cluster formation depending on the biological system in which they occur. In addition to C- or G-coordinated clusters that could be mediated by TC-specific APOBECs, our analysis of cancer mutation data sets also uncovered A- or T-coordinated clusters which strongly associated with the mutation motif WA/TW. This is consistent with a role of Pol eta in the formation of these clusters; however, the high likelihood of random incidence of WA/TW sequence does not allow discrimination among several potential mechanisms based upon the WA/TW motif alone. Regardless of the details of A- or T-coordinated cluster formation, the mechanism of formation is clearly distinct from those mediating C- or G-coordinated clusters in human cancers and the MMS-induced clusters in yeast. Beyond the systems we investigated here, we also expect mutation clusters to be prevalent in tumors that have a known exogenous damage component as they are likely to experience the lesions required to generate mutation clusters. DNA repair inhibition due to heterochromatin (Livingstone-Zatchej et al., 2003), telomeric sequences (Rochette and Brash, 2010), or high transcription (Datta and Jinks-Robertson, 1995), could also be candidate mechanisms. In fact, future studies may show that some of these, or other unknown mechanisms, could be responsible for clustered mutations that compose the additional portion of the MMS-induced spectra beyond the coordinated mutations in C or G that highlight the ssDNA pathways in the yeast clusters.
Our studies with yeast and cancer mutations suggest that simultaneous multiple changes may actively drive disease and evolution. The production of several simultaneous mutations within biologically functioning sequences appears to be ubiquitous and is potentially an efficient means to produce rapid change in the face of strong selection. Additionally, the analysis of mutation clusters within cancers as well as various other biological systems is an effective tool for unraveling a complex “archeological record bearing the imprint of mutagenic and DNA repair processes” (Stratton, 2011).
Experimental Procedures
Mutagenesis and selection of yeast
Yeast were grown at 30°C in 5 mL of liquid YPDA media for 2 days to stationary phase (~2x108 cells/mL for ade5-1 strains and ~9x108 cells/mL for ADE+ strains), diluted to 1x107 cells/mL (for mutagenesis) and 2x103 cells/mL (for survival), and plated using a multi-pronging device (Jin et al., 2003 and refs within) as 120 independent 1uL exposure cultures on each plate of fresh YPDA containing either 0, 2 or 4 mM MMS. Densely plated yeast was then grown for 2 days, and replica-plated to YPDA or synthetic complete media containing either: 5-FOA, canavanine, or both drugs. Replicas on 5-FOA, canavanine, and YPDA media were incubated for 6 days; plates with both 5-FOA and canavanine were incubated for 7 days. Resistant colonies were counted and independent isolates (1 per pronged culture) were sequenced. Unselected independent isolates were obtained from cultures replica plated to YPDA media.
Estimation of diploid FOA-CAN resistance independent of Loss of Heterozygosity (LOH)
Since FOA-CAN resistance is a recessive phenotype, a cross-over and LOH event is required to select diploid cells containing mutation clusters with the URA3-CAN1 reporter. We determined the frequency of LOH during growth on 4 mM MMS in wild type diploid yeast heterozygous for the URA3-CAN1 reporter and carrying a/alpha, alpha/alpha, and a/a mating type alleles. The median LOH frequency was determined as the number of FOA-CAN resistant cells/survivor among 6 replicates of the experiment (Table S2). To adjust FOA-CAN resistance for LOH frequency, the mean number of FOA-CAN resistant colonies/plate was divided by the median frequency of LOH calculated for the same genotype.
Whole genome sequencing
Yeast containing 3 or more mutations in the LYS2::URA3-CAN1 reporter, or strand-coordination of two single mutations in URA3 and CAN1 were whole-genome sequenced. Genome sequencing and analysis was conducted similar to (Burch et al., 2011 and refs within) using S288C (http://www.yeastgenome.org/) as a starting DNA sequence for reference-based mapping and mutation calling. Yeast genomic DNA was sheared and 300 to 400 bp fragments were extracted after gel electrophoresis. Libraries were constructed and sequenced by Illumina protocols on either an Illumina GAII or HiSeq2000 sequencer. Reads were mapped to a reference genome (>20x average coverage) and mutations identified with CLC Bio Genomics Workbench 4.7.2. For haploid genomes, we required all mutations to be covered by at least 4 paired reads and exist in at least 90% of these reads. For diploid genomes, we required coverage of 9 paired reads and presence in at least 90% of reads for homozygous mutations and in between 45 and 55% of reads for heterozygous mutations. We further required mutations to be unique among all samples sequenced except for those mutations occurring in URA3 and CAN1 where bottlenecks of URA3+ and CAN1+ single cells separated the sequenced isolates. For spectra analysis, mutations in repeat regions (i.e., telomeric regions, retro-transposons, and LTRs) were ignored. False positive and negative rates of 2.2% and 2.2% respectively were estimated by comparing the mutations found in the LYS2 reporter region, including URA3 and CAN1, by Illumina sequencing to corresponding Sanger sequencing.
Identifying clusters
We searched for clusters among lists of genomic locations (chromosome and nucleotide position) of mutations defined by sequencing of individual yeast or human genomes. We first identified complex mutations defined as groups of mutations where consecutive mutations are less than 10 nucleotides apart, based on the estimated capacity of DNA polymerases to make consecutive errors in a single round of synthesis (Sakamoto et al., 2007). Each complex mutation was counted as a single event and only accounted for a small fraction of all events. They were practically absent in clusters (Tables S4 and S6).
The following categories of mutations in human cancer datasets were excluded (filtered) before the cluster search:
. All mutations that were listed in the original analyses as identical to known dbSNPs (6.3 – 8.9% of all mutations).
. All mutations that fell into the simple Repeat Track (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=204690969&c=chr8&g=simpleRepeat), a list of short and long tandem repeats identified in the corresponding release of human genome sequence. These mutations (5.3 – 8.5% of all mutations) were excluded because of the high chance of false positives within low complexity sequences.
When calculating Cluster P-values, mutations excluded while assessing clustering in the human cancer data sets were included in the total number of changes that could be randomly distributed in the genome. This is a conservative approach that can lead only to an increase in the calculated P-value, never to a decrease.
After any filtering, the groups of consecutive mutations, within which any pair of adjacent mutations was separated by less than 100,000 nucleotides (for yeast) or 10,000 nucleotides (for cancer data sets), were identified and the probability for each mutation cluster occurring by chance (P-value) was calculated (see Supplemental Experimental Procedures).
Other Experimental Procedures
Complete experimental procedure information is provided in the Supplemental Experimental Procedures.
Accession Numbers
Yeast S288C chromosomal DNA sequences used for Illumina read mapping and mutation calling were taken from the Saccharomyces Genome Database (http://www.yeastgenome.org/). Sequencing data containing human cancer mutations analyzed in this paper were submitted at the time of original publication to the dbGaP repository (http://www.ncbi.nlm.nih.gov/gap) under the following accession numbers: multiple myelomas (Chapman et al., 2011) - phs000348.v1.p1; prostate carcinomas (Berger et al., 2011) - phs000330.v1.p1; head and neck squamous cell carcinomas (Stransky et al., 2011) - phs000370.v1.p1.
Supplementary Material
Highlights.
Clusters of simultaneous multiple mutations occur in yeast and human genomes
Mutation clusters can occur in damaged ssDNA during DSB repair or replication
Clusters of coordinated C or G mutations in cancers colocalize with rearrangements
Clustered mutations in cancers occur at motifs of cytosine deamination by APOBECs
Acknowledgments
We would like to thank Drs. Tom Kunkel, Paul Wade, Kin Chan, and Scott Lujan for critical reading of the manuscript and advice. This work was supported by the Intramural Research Program of the NIEHS (NIH, DHHS) project ES065073 to M.A.R., by NIH Grant RC1 ES018091-02 and UNC University Cancer Research Fund to P.A.M, and by NIEHS/NIH Contract GS-23F-9806H and Order: HHSN273201000086U to R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A. The connection between transcription and genomic instability. Embo J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. Embo J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch LH, Yang Y, Sterling JF, Roberts SA, Chao FG, Xu H, Zhang L, Walsh J, Resnick MA, Mieczkowski PA, et al. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–1085. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–326. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahwan R, Wontakal SN, Roa S. Crosstalk between genetic and epigenetic information through cytosine deamination. Trends Genet. 2010;26:443–448. doi: 10.1016/j.tig.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Drake JW. Too many mutants with multiple mutations. Crit Rev Biochem Mol Biol. 2007;42:247–258. doi: 10.1080/10409230701495631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC. DNA repair and mutagenesis. 2. Washington, D.C: ASM Press; 2006. [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Henry M, Guetard D, Suspene R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Clark AB, Slebos RJ, Al-Refai H, Taylor JA, Kunkel TA, Resnick MA, Gordenin DA. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat Genet. 2003;34:326–329. doi: 10.1038/ng1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Livingstone-Zatchej M, Marcionelli R, Moller K, de Pril R, Thoma F. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J Biol Chem. 2003;278:37471–37479. doi: 10.1074/jbc.M306335200. [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS Genet. 2010;6:e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair (Amst) 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005;33:4404–4411. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- Swartz MN, Trautner TA, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962;237:1961–1967. [PubMed] [Google Scholar]
- Thielen BK, McNevin JP, McElrath MJ, Hunt BV, Klein KC, Lingappa JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Burton M, Oki M, Johnson J, Seier TA, Kamakaka R, Haber JE. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics. 2006;174:41–55. doi: 10.1534/genetics.106.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gonzalez KD, Scaringe WA, Tsai K, Liu N, Gu D, Li W, Hill KA, Sommer SS. Evidence for mutation showers. Proc Natl Acad Sci U S A. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland J, Ma W, Yan Y, Van Hulle K, Malkova A, Resnick MA. RAD50 is required for efficient initiation of resection and recombinational repair at random, gamma-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA repair. 2010;9:914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.