SUMMARY
The coordination of nutrient and energy availability with cell growth and division is essential for proper immune cell development and function. Using a chemical mutagenesis strategy in mice, we identified a pedigree that has a complete block in B cell development at the pre-B cell stage due to a deletion in the Fnip1 gene. Enforced expression of an immunoglobulin transgene failed to rescue B cell development. Whereas essential pre-B cell signaling molecules were activated normally in Fnip1-null pre-B cells, the metabolic regulators AMPK and mTOR were dysregulated resulting in excessive cell growth and enhanced sensitivity to apoptosis in response to metabolic stress (pre-B cell receptor cross-linking, oncogene activation). These results indicate that Folliculin-interacting protein 1 (Fnip1) is vital for B cell development and metabolic homeostasis, and reveal a metabolic checkpoint which may ensure that pre-B cells have sufficient metabolic capacity to support division, while limiting lymphomagenesis caused by deregulated growth.
INTRODUCTION
The development of all metazoan organisms involves the strict coordination of cell growth, apoptosis, division, and differentiation. During B lymphocyte development, these processes are carefully linked to an orchestrated series of “checkpoints” which sensor autoreactivity and ensure that mature B cells can mount an immunogenic response against foreign antigens, without reacting against self. The earliest committed B cell progenitor (pro-B cell) is defined by having its immunoglobulin (Ig) genes in germline (unrearranged) configuration. The Ig heavy (IgH) chain gene segments rearrange first and following successful in-frame VH- to DHJH juxtaposition, the IgH chain “μ” pairs with surrogate light chain chaperones λ5 and Vpre-B, and signal transducing Igα and Igβ chains, as a component of the pre-B cell receptor (pre-BCR) complex on the surface of pre-B cells. The pre-BCR complex then signals pre-B cells to stop VH-to-DHJH rearrangement of the other IgH allele (a process called “allelic exclusion”), proliferate (synergistically with IL-7 receptor stimulation), start IgL chain (κ and λ) gene transcription and rearrangement (VL –JL), and mature. Following in-frame IgL chain rearrangement, the IgH and IgL chains pair to form IgM on the surface of immature B cells, at which point B cells with low self-reactivity exit the BM and migrate to peripheral lymphoid tissues, where development continues (see (Goodnow et al., 2005) for review)).
The importance of particular signaling proteins in pre-B cell development has been demonstrated through a series of biochemical and gene-targeting studies in mice (see (Hendriks and Middendorp, 2004; Herzog et al., 2009) for review). For example, formation of the pre-BCR results in activation of the Src-family protein tyrosine kinases (Ptks) Fyn, Lyn, and Blk, which phosphorylate membrane bound Igα and Igβ on their immunoreceptor tyrosine-based activation motifs (ITAMs). This results in the recruitment and activation of the Syk Ptk, which activates the Ras-Raf signaling pathway and phosphorylates the adaptor protein SLP-65 (also known as BLNK). Phosphorylated SLP-65 then forms a signaling module which recruits and activates multiple pathways including phosphatidylinositol 3- kinase (PI3K), and the Tec-family Ptks Btk and Tec, which activate PLCγ1/2 and ultimately Ca2+ flux. These events altogether lead to activation of essential transcriptional mediators including Erk, Myc (Habib et al., 2007), IRF-4/8, Foxo1, and NFκB (see (Herzog et al., 2009; Kurosaki et al., 2010) for review).
In this study, we utilized a phenotype-driven “reverse-genetic”, recessive ethylnitrosourea (ENU) mutagenesis strategy in mice to discover genes involved in immune cell development and function (see (Appleby and Ramsdell, 2003; Cook et al., 2006) for review). In the course of this program, we identified a pedigree (denoted LPAB.1) that selectively lacks B cells in peripheral blood. By genome scanning, the B cell immunodeficiency phenotype was mapped to a non-coding deletion in the Fnip1 gene, which encodes for the 160 kDa protein Folliculin interacting protein-1 (Fnip1)(Baba et al., 2006). Although the biological functions of Fnip1 and the related Fnip-family member Fnip2 (Hasumi et al., 2008; Takagi et al., 2008) are unknown, both molecules interact with Folliculin and AMP Kinase (Baba et al., 2006; Hasumi et al., 2008), a highly-conserved, master sensor and regulator of cellular metabolism (see (Towler and Hardie, 2007) for review). In response to low energy (low ATP, high AMP), AMPK is activated by phosphorylation at threonine 172 by LKB1 kinase. Activated AMPK then stimulates ATP production by increasing glucose uptake, stimulating mitochondrial biogenesis, and increasing glycolysis and oxidative phosphorylation (by inducing expression of the PGC1α and PPAR-γ transcription factors). AMPK also decreases ATP consumption by inhibiting mammalian target of rapamycin (mTOR)-driven cell growth, in part by phosphorylating and activating the mTOR inhibitor tuberous sclerosis protein 2 (TSC2) (Inoki et al., 2003), and by phosphorylating and inactivating the mTOR positive regulatory protein Raptor (Gwinn et al., 2008). Our studies indicate that Fnip1 maintains metabolic homeostasis in developing B cells, and reveal a metabolic “checkpoint” during B cell development, which we hypothesize may ensure that mature B cells are adequately equipped to fuel clonal expansion and antibody production, while protecting the host against excessive growth and transformation.
RESULTS
Generation of Fnip1-null mice using ENU mutagenesis
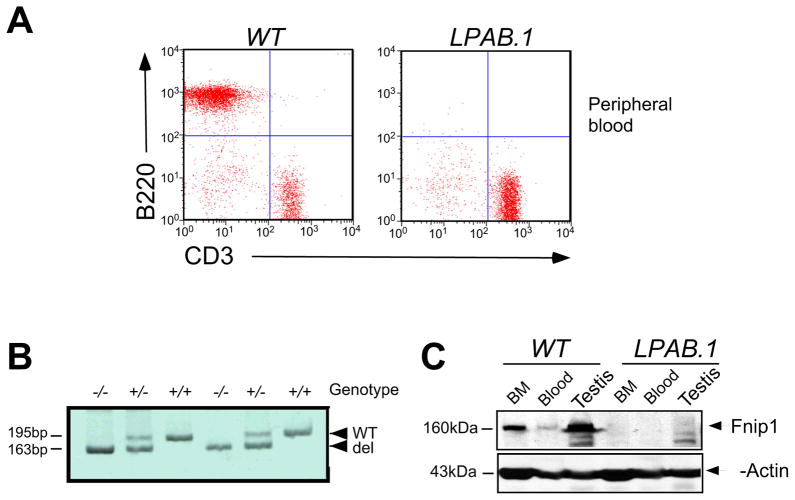
We screened G3 mice from a large-scale ENU mutagenesis project for recessive mutations leading to specific immunodeficiencies. Utilizing flow cytometry to assess the representation of immune cells in peripheral blood, we identified the LPAB.1 pedigree based on an absence of B lymphocytes (Figure 1A), while myeloid and T cells were represented normally. By mapping affected G3 animals using positional cloning strategies, the LPAB.1 mutation was localized to a 1.7 Mb interval on chromosome 11. We sequenced candidate genes and identified a 32-bp deletion in exon 9 of a putative gene (A730024A03Rik), which resulted in the generation of a premature stop codon at residue 293 (Figure S1A). The product of A730024A03Rik was subsequently identified as the murine homologue of Fnip1 (Baba et al., 2006). PCR analysis confirmed that the deletion (Figure 1B) tracked with the B cell immunodeficiency, and immunoblotting with α-Fnip1 revealed the absence of Fnip1 protein in tissues from LPAB.1 (Fnip1−/−) mice (Figure 1C). Fnip1−/− mice were viable and fertile, but exhibited several additional phenotypes relative to wildtype (WT) littermates, including alterations in skeletal muscle (which appeared deep-red due to high mitochondria content), increased liver glycogen content, and hypertrophic cardiomyopathy (data not shown). Taken together, these results indicate that the lack of B cells in LPAB.1 mice maps to a deletion in the Fnip1 gene, which results in the absence of Fnip1 protein.
Figure 1. LPAB.1 mice lack peripheral B cells and have a deletion in the Fnip1 gene.
(A) Flow cytometric analyses of peripheral blood samples from LPAB.1 and WT mice revealed the absence of B220+ B cells, while CD3ε+ T cells were represented normally. (B) A PCR genotyping strategy was designed using oligonucleotides spanning the 32-bp deleted region in the Fnip1 gene. Shown is an ethidium-stained acrylamide gel containing PCR products amplified from WT, Fnip1+/−, and Fnip1−/− genomic DNA. (C) Representative immunoblot showing the absence of Fnip1 protein in testis, bone marrow cells (BM), and blood from Fnip1−/− mice relative to WT littermates.
Fnip1 is expressed in multiple tissues
We examined the expression of Fnip1 in 25 different normal mouse tissues (Park et al., 2008), using real-time PCR. We found that Fnip1 was highly expressed in testes, kidney, skeletal muscle, liver, heart, and embryo; in addition to thymus, spleen, and bone marrow (BM) (Figure S1B). Fnip1 was equally expressed in FACs-sorted B lineage cells throughout B cell development, whereas Fnip2 expression sharply increased during B cell development, reaching maximal levels in immature B cells (Figure S1C). Whereas both Fnip1 and Fnip2 were expressed in thymocytes, neither showed regulated expression during T cell development (Figure S1D). Transfection of Flag-tagged Fnip1 into the WEHI B cell line indicated that Fnip1 protein resides in the cytoplasm (Figure S1E), as was previously shown in a kidney cell line (Baba et al., 2006). These results collectively suggest that Fnip1 is normally expressed in multiple tissues including hematopoietic cells, and encodes for a cytoplasmic protein in B cells.
Fnip1 deficiency blocks B cell development at the large, pre-B cell stage
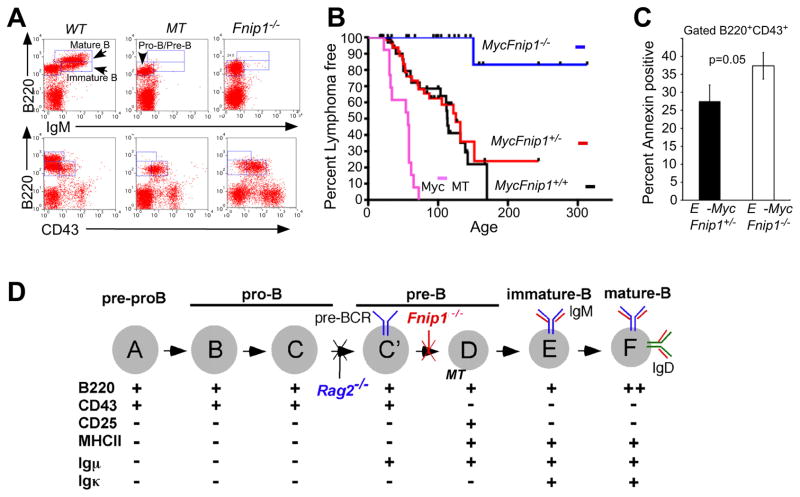
To examine where loss of Fnip1 blocks B cell development, total BM cells and splenocytes were stained with antibodies against proteins that are differentially expressed during B cell development (Figure 7D). Analysis of Fnip1−/− BM revealed a complete block at the B220+CD43+ CD25− MHCII− large pre-B cell stage (Figure 2A), which resulted in the absence of mature B cells bearing IgM, CD21, and CD23 in the BM and spleen (Figures 2A and 2B). Fnip1−/− mice also lacked “B1” B lymphocytes, which represent a subset of B cells that are found in the peritoneal and pleural cavities (Hardy and Hayakawa, 2001) (Figure 2C). Fnip1−/− mice failed to produce serum Igs of all isotypes following immunization with keyhole limpet hemocyanin (KLH) (Figure 2D). Intracellular (IC) flow cytometric analyses indicate that Fnip1−/− BM B cells contain IC IgH “μ” protein, but lack IC IgL “κ” (Figure 2E). These results suggest that loss of Fnip1 results in a complete block in B cell development at the large pre-B-to-small pre-B cell transition mediated by formation of the pre-BCR ((Figure 7D and (Hardy and Hayakawa, 2001)).
Figure 7. Loss of Fnip1 inhibits pre-B cell lymphomagenesis induced by the Eμ-c-Myc transgene.
(A) Disruption of the transmembrane portion of Igμ (μMT) or Fnip1 (Fnip1−/−) in mice blocks B cell development at the pre-B cell stage. BM cells from B cell deficient Fnip1−/− or μMT (control) mice were stained with fluorescent-conjugated antibodies and analyzed by flow cytometry. (B) Fnip1−/− or μMT mice were bred to Eμ-Myc transgenic mice. Mice were euthanized when lymphomas were detected. Kaplan-Meier lymphoma-free estimates are shown for MycFnip1+/+ (n=33), MycFnip1+/− (n=66), MycFnip1−/− (n=24), and MycμMT (n=13). Lymphoma-free curves are significantly different between MycFnip1+/− and MycFnip1−/− (p<0.0001), MycFnip1+/+ and MycFnip1−/− (p<0.0004), MycμMT and MycFnip1−/− (<0.0001), and MycμMT and MycFnip1+/− (<0.0001) using Log Rank Mantel-Cox Test. (C) Disruption of Fnip1 results in increased apoptosis of Myc-expressing pre-B cells. MycFnip1−/− and MycFnip1+/− BM were stained with α-B220, α-CD43, and Annexin V. Shown is a bar graph (mean ± SEM, 3 mice per group) depicting the percent Annexin V positive cells within a B220+CD43+ gate. (D) A model of B cell development (modified from (Hardy and Hayakawa, 2001), characterized by the differential expression of surface and intracellular molecules. Shown are the stages whereby loss of Fnip1, Rag2, or Igμ (μMT) result in arrested B cell development.
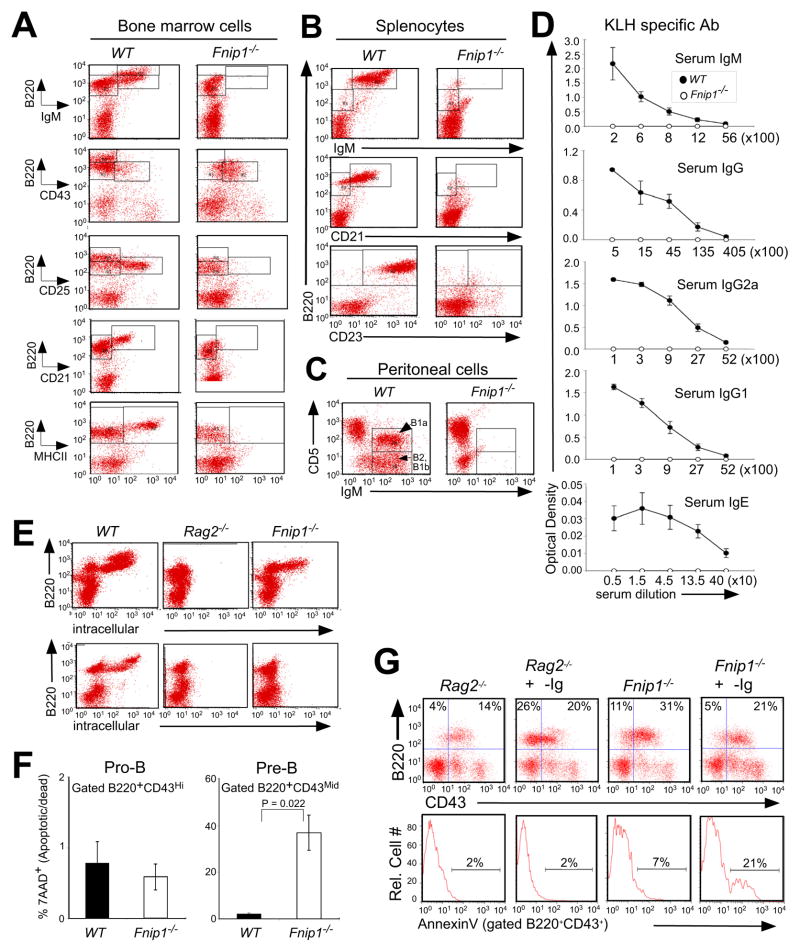
Figure 2. Loss of Fnip1 results in a complete block in B cell development at the pre-B cell stage.
BM cells (A), splenocytes (B), and peritoneal cells (C) derived from Fnip1−/− and WT mice were stained with fluorescent-conjugated antibodies and analyzed by flow cytometry. Shown are representative histograms (of 8 mice/group). B1a, B1b, and B2 refer to types of B cells (Hardy and Hayakawa, 2001). (D) Fnip1−/− and WT mice (5 mice/group) were immunized with KLH, and sera were tested for KLH-specific antibodies by ELISA. (E) BM cells from Fnip1−/−, Rag2−/− and WT mice were intracellularly stained with fluorescent-conjugated α-Igμ (upper) and α-Igκ (lower) after α-B220 surface staining. Shown are representative flow cytometric histograms obtained from five mice/group. (F) Decreased cell viability of B220lowCD43mid BM cells from Fnip1−/− mice. BM cells were stained with α-B220, α-CD43 and 7AAD. Cell viability was determined by flow cytometry. Bar graphs depict percent apoptotic/dead (7AAD+) cells (mean ± SEM) on gated B220+CD43hi cells, or B220+CD43mid cells. p-value is shown from 3 mice/group. (G) Rag2−/− and Fnip1−/− mice were injected intraperitoneally with 250μg of α-Igβ MAb HM79. On day 7 post-injection, bone marrow cells were stained with antibodies specific to B220, CD43, and Annexin V. Anti-Igβ stimulates the development of Rag2−/− pro-B cells but induces apoptosis of Fnip1−/− pro-B cells. (See also Figure S2)
We next examined the ability of Fnip1−/− pre-B cells to divide and survive in the presence or absence of the growth factor IL-7. CFSE analyses indicated that Fnip1−/− pre-B cells divide equally well as WT pre-B cells in vitro in response to IL-7 (Figure S2A), whereas the proliferation of Rag2−/− pro-B cells is blunted due to the absence of pre-BCR formation, which normally synergizes with IL-7 for maximal proliferation (Fleming and Paige, 2002). Fnip1−/− pre-B cells also divided equally well as WT pre-B cells in vivo, based on equivalent incorporation of BrdU after 48 hrs (Figure S2B). Fnip1−/− B220+CD43mid pre-B cells, but not B220+CD43hi pro-B cells, exhibited significantly decreased viability relative to WT pre-B cells (Figure 2F). Antibody-mediated crosslinking of Igβ, which mimics pre-BCR crosslinking, induced apoptosis of Fnip1−/− pre-B cells and failed to stimulate pre-B cell development, whereas Rag2−/− pro-B cells were induced to mature based on downregulation of CD43 (Figure 2G) and upregulation of CD25 (Kouro et al., 2001). Relative to WT pre-B cells, Fnip1−/− pre-B cells expressed normal mRNA levels of the anti-apoptotic molecule Bcl2, slightly decreased levels of the pro-apoptotic molecule Bim (Figure S2C), and were equally sensitive to apoptosis in response to the protein kinase inhibitor staurosporine (Figure S2D) or Fas crosslinking (Figures S2E), suggesting that apoptosis is relatively specific to activation. These results suggest that Fnip1−/− pre-B cells fail to mature in response to pre-BCR signals, whereas proliferative signals emanating from the pre-BCR and the IL-7 receptor are transmitted relatively normally.
Enforced expression of IgH, IgL, or IgM fails to rescue B cell development in Fnip1-null mice
We next tested whether the experimental provision of rearranged IgH and/or IgL chains in Fnip1−/− mice could overcome the arrest in B cell development. Fnip1−/− and Rag2−/− (control) mice were crossed with IgMHEL mice, which carry a rearranged IgH chain “knock-in” (denoted IgHHEL) and a separate IgL “κ transgene (denoted IgLHEL), which together encode surface IgM specific against hen egg lysozyme (HEL) protein (Allen et al., 2007). We assessed whether IgHHEL, IgLHEL, or IgHHEL plus IgLHEL (IgMHEL) could stimulate B cell development in Fnip1−/− and Rag2−/− control mice. Expression of a rearranged IgH chain (IgHHEL), which mimics pre-BCR expression, efficiently drove B cell maturation in Rag2−/− mice based on downregulation of CD43, and upregulation of CD25 and MHC II (Figure S3A), and a reduction in cell size (data not shown). In contrast, IgHHEL was unable to stimulate B cell maturation in Fnip1−/− mice, suggesting that pre-BCR signaling is impaired. Similarly, expression of a rearranged IgL chain (IgLHEL Tg), which pairs with endogenous Fnip1−/− IgH chains to form surface IgM, also failed to stimulate Fnip1−/− pre-B cell progenitors to mature (Figures S3B, S3C, S3D). Remarkably, live-gated Fnip1−/− B cell progenitors that expressed both IgHHEL and IgLHEL (IgMHEL) still failed to mature (Figure 3A) and migrate to the periphery (Figure 3B) despite high-level IgM expression, which potently drove B cell development and migration to the periphery in Rag2−/− mice. Fnip1−/− IgMHEL BM B cells exhibited a “tolerant-like” phenotype consisting of (CD62Lneg, B220lo, IgMlo), and were more sensitive to apoptosis in response to anti-IgM (data not shown). These results demonstrate that Fnip1 is absolutely required for pre-BCR-mediated B cell differentiation, and suggest a potential role for Fnip1 in BCR-mediated maturation and survival.
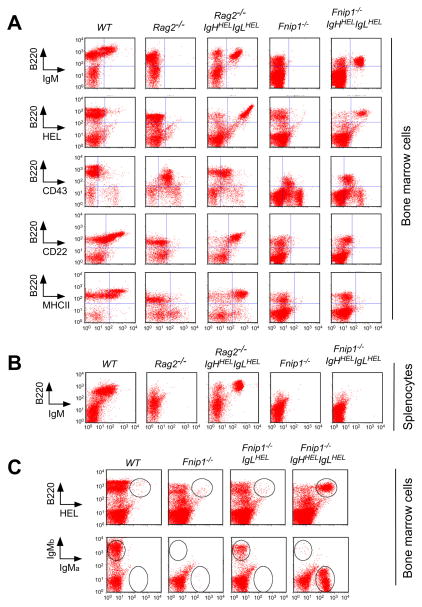
Figure 3. Enforced expression of Ig heavy and Ig light chains fail to drive B cell development in Fnip1-null mice, whereas allelic exclusion occurs normally.
Total BM cells (A) and splenocytes (B) from WT, Rag2−/−, Rag2−/−IgHHELIgLHEL, Fnip1−/−, Fnip1−/−IgHHELIgLHEL mice were analyzed by flow cytometry utilizing the indicated fluorescent-conjugated antibodies. IgMHEL stimulates the development of Rag2−/− pro-B cells but fails to stimulate the maturation of Fnip1−/− pre-B cells. Representative histograms of 5 mice/group are shown ((A)(B)). (C) Allelic exclusion occurs normally in Fnip1−/− mice. Total BM cells from the indicated mice were stained with fluorescent-conjugated α-B220 and α-HEL (top), or α-IgMa and α-IgMb (bottom). Shown are representative flow cytometric histograms from 4 mice/group.
Allelic exclusion of the Ig heavy chain occurs normally in the absence of Fnip1
It has previously been shown that expression of a rearranged IgH chain in pro-B cells mimics pre-BCR formation, resulting in suppression of endogenous IgH chain gene rearrangement (allelic exclusion) (Nussenzweig et al., 1987). Hence, we utilized IgMHEL mice to ask whether expression of IgHHEL and IgLHEL, which together form IgM of the “a” isotype (IgMa), would extinguish expression of endogenous μ chains (IgMb) in Fnip1−/− B cells. As shown in Figure 3C, WT C57BL6 mice express surface IgMb, whereas Fnip1−/− mice lack IgM expression due to the absence of IgL chains. Expression of IgLHEL Tg in Fnip1−/− mice restored surface expression of IgMb. However, when Fnip1−/− mice were bred to IgHHEL/+IgLHEL/+ mice (IgMa), all B cells expressed IgMa and not endogenous IgMb, indicating that signals delivered from the IgMHEL efficiently extinguished rearrangement and expression of the endogenous Igμ in Fnip1−/− mice.
Impaired B cell development in Fnip1-null mice is cell autonomous
To determine whether the defects in B cell development are autonomous to Fnip1−/− pre-B cells, we transferred total BM cells from WT (CD45.1+), Fnip1−/− (CD45.2+) or mixed WT and Fnip1−/− cells into lethally irradiated Rag2−/−Il2rg−/− (recombinase activating gene 2/common γ chain double-null) hosts (CD45.2+)(Garcia et al., 1999). The development of donor hematopoietic cells were analyzed ~15 weeks after transplantation. As shown in Figure S6, Rag2−/−Il2rg−/− completely lacked all lymphocytes due to complete blocks in B and T cell development at the pro-B and the CD4−CD8− cell stages respectively. Transfer of WT BM cells rescued the development of B220+IgM+ B cells, GR1+ myeloid, and CD4 and CD8 T cells. In contrast, transfer of Fnip1−/− BM rescued the development of CD4+ and CD8+T cells, but failed to restore the development of Fnip1−/− B cells. Transfer of a 1:1 mixture of CD45.1+ WT: CD45.2+ Fnip1−/− BM cells rescued the development of both WT and Fnip1−/− T cells and GR1+ myeloid cells, but only WT CD45.1+ (CD45.2−) IgM+ B cells. These results indicate that the defect in the development of Fnip1−/− B cells is cell autonomous and is not due to a defective BM environment or secretion of a B cell inhibitory factor.
Retroviral expression of Fnip1 rescues B cell development in LPAB.1 mice
To confirm that the absence of Fnip1 is directly responsible for the LPAB.1 phenotype, we infected lineage-negative (Lin−) BM cells from Fnip1−/− mice with empty MSCV retrovirus, or MSCV containing full-length Fnip1 cDNA, and transplanted the cells into Rag2−/−/Il2rg−/− mice. Empty MSCV-infected Fnip1−/− BM cells failed to restore B cell development following transplant into Rag2−/−/Il2rg−/− hosts, while CD4+ and CD8+ T cells are efficiently rescued to normal levels, confirming successful engraftment of Fnip1−/− donor HSCs (Figure S7A). In contrast, transfer of MSCV-Fnip1-infected Fnip1−/− BM also rescued the development of conventional B220+IgM+ B cells and CD5+IgM+ “B1” B cells. These results demonstrate that loss of Fnip1 is directly responsible for the B cell immunodeficiency phenotype in LPAB.1 mice.
Fnip1 null pre-B cells exhibit changes in gene expression indicative of altered cellular metabolism
To examine why loss of Fnip1 might result in impaired pre-B cell development, we FACS-sorted B220+CD43+ cells from Fnip1−/− and WT littermate mice, and examined the global differences in gene expression using cDNA microarray technology. We found 293 genes significantly increased and 246 genes decreased in Fnip1−/− pre-B cells relative to WT pre-B cells, based on p-values <0.05 (Figure S4). Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed a large percentage of the changes involved genes controlling cell metabolism and mitochondrial biogenesis and function. In particular, we found substantial increases in the expression of AMPK-regulated “catabolic” genes including the transcriptional co-activators peroxisome proliferator-activated receptor gamma coactivator-1α and β (Ppargc1a (Pgc1α) and Ppargc1b (Pgc1β)), which are “master” regulators of fatty acid oxidation (FAO) and mitochondrial biogenesis (Figure S4). This correlated with a significant increase in the expression of genes involved in fatty acid metabolism and mitochondrial function. Surprisingly, we also found increases in “anabolic” genes involved in cellular growth such as ribosomal S6 kinase-1 (Rps6kb1), ribosome protein L3 (Rpl3), and Ras-related GTP binding proteins B (Rragb) and D (Rragd), which are positive regulators of mTOR activation and/or function (Figure S4).
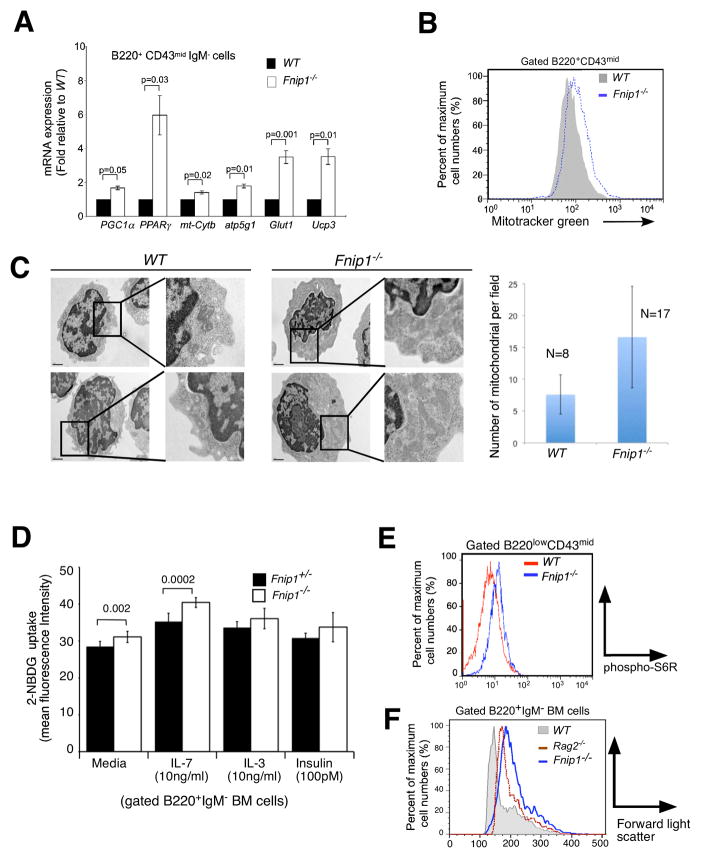
We next determined whether these global changes in gene expression were reflective of the biology of Fnip1−/− pre-B cells. Firstly, utilizing real-time PCR we confirmed significantly increased expression of essential AMPK-regulated catabolic genes including Pgc1α (Pparγ coactivator 1α), Pparγ(peroxisome proliferator-activated receptor γ) (Narkar et al., 2008), and 4f2hc (solute carrier family 3, member 2)(Towler and Hardie, 2007); and the glucose transporter Glut1(Rathmell et al., 2000), which is the main glucose transporter in lymphocytes, in Fnip1−/− versus WT pre-B cells (Figure 4A). In addition, essential mitochondrial genes including Ucp3 (uncoupling protein 3), Atp5g1 (mitochondrial ATPase, C1 subunit), and mt-Cytb (Cytochrome b) were also significantly increased in pre-B cells from Fnip1−/− mice relative to WT mice (Figure 4A). Flow cytometry and transmission electron microscopy (EM) analyses further revealed that Fnip1−/− B cell progenitors exhibited increased labeling with the mitochondrial dye MitoTracker® green relative to WT B cell progenitors (Figure 4B), and contained significantly increased numbers of mitochondria (Figure 4C). Fnip1−/− B cell progenitors took up the fluorescent deoxyglucose analog 2-NBDG more effectively than WT pre-B cells in response to IL-7, IL-3, and insulin (Figure 4D). These results confirmed that the upregulated AMPK and PGC-1α gene signature correlates with increased AMPK and PGC1α function in Fnip1-null pre-B cells. To determine whether the upregulated mTOR gene signature correlated with increased cell growth, we examined mTOR activation and cell size by flow cytometry. We found that relative to WT pre-B cells, Fnip1−/− pre-B cells exhibited increased intracellular phospho-S6 ribosomal protein (pS6R) (Figure 4E), which is a downstream mediator of mTOR signaling that stimulates ribosome biogenesis and translation, and are significantly larger in size relative to WT pre-B cells and Rag2−/− pro-B cells (Figure 4F). These results collectively suggest that loss of Fnip1 results in parallel increases in catabolic metabolism, and mTOR-mediated anabolic metabolism; two normally antagonistic energy regulating processes which typically occur separately.
Figure 4. Loss of Fnip1 results in metabolic dysregulation in pre-B cells defined by increased expression of AMPK and mTOR regulated genes, increased mitochondrial biogenesis, and increased mTOR-mediated cell growth.
(A) Purified pre-B cell cDNA from Fnip1−/− and WT mice were subjected to real-time PCR specific for the indicated genes. Bars = means ± SEM of 3 mice/group. P-values are shown. (B) Increased mitochondria in Fnip1−/− pre-B cells. Fnip1−/− and WT BM cells were stained with α-B220, α-CD43 plus the mitochondrial dye MitoTracker® green. Shown is a representative flow cytometric histogram (n=3 mice/group) of gated B220+CD43+ cells. (C) Transmission electron micrographs were obtained on FACS-sorted pre-B cells from 3 WT and 3 Fnip1−/− mice. (left) Shown are representative high-power images (20,000X). (Right) The number of mitochondria per field were enumerated in the cytoplasmic compartment for 8 WT and 17 Fnip1−/− pre-B cells. Bars = means ± SEM. p<0.00006. (D) Increased glucose uptake in Fnip1−/− pre-B cells. Total BM cells were cultured in media, IL-7, IL-3, or insulin at the indicated concentrations for 48 hrs. 2-NDBG was added for the last 3 hrs. Shown is bar graph (n=3 per group for IL-7, n=2 per group in triplicate for IL-3, insulin) showing the relative fluorescence intensity of B220+IgM− gated cells as assessed by flow cytometry. Error bars= mean+/−SEM. P-values are shown. (F) Fnip1−/− and WT BM cells were stained with α-B220, α-CD43, and intracellular pS6R. (G) Shown is a representative forward-light scatter (FSC) histogram overlay comparing the size of gated B220+IgM− pro-B and pre-B cells from WT, Rag2−/−, and Fnip1−/− mice. (see also Figure S4)
Fnip1 controls the ability of AMPK to inhibit mTOR-mediated cell growth
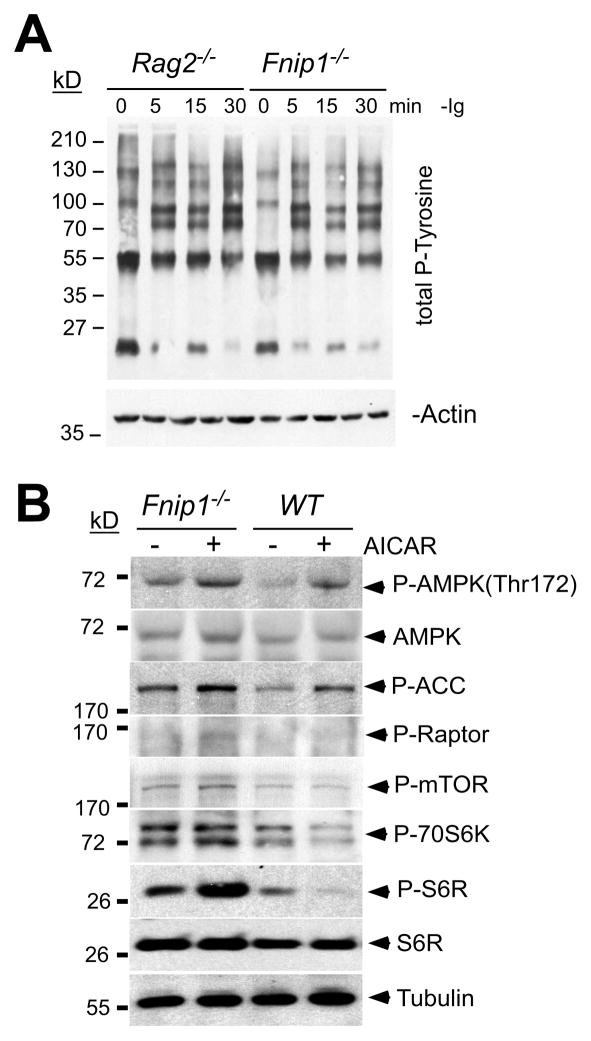
Although the cellular and molecular functions of Fnip1 are unknown, both Fnip1 (Baba et al., 2006) and Fnip2 (Hasumi et al., 2008; Takagi et al., 2008) have been shown to interact with all 3 subunits (α,β,γ) of AMPK. To examine how Fnip1 might interact with pre-BCR-signaling and metabolism, we assessed the presence and activation of known pre-BCR signaling molecules following stimulation with α-Igβ-coated beads (Nagata et al., 1997). Crosslinking of Igβ on purified B cell progenitors from Fnip1−/− and Rag2−/− control mice resulted in equivalent increases in total tyrosine or serine/threonine phosphorylated proteins (Figure 5A) including Syk, Blnk (Slp65), Btk, Akt, Erk and IKKα/β (Figure S5A), suggesting that pre-BCR signaling is activated normally in the absence of Fnip1. Fnip1−/− mice also expressed normal levels of Cd79a (Igβ), Tcf3 (E2a), Irf4 and Irf8, Pax5, and other important pre-BCR signaling molecules relative to WT mice (Figure S5B). Activation of Fnip1−/− B cell progenitors with the AMPK activator 5-aminoimidazole-4-carboxyamide-1-beta-D-ribofuranoside (AICAR) resulted in equivalent activation of AMPK, based on phosphorylation levels of AMPK (Thr172) and the AMPK-target acetyl-CoA carboxylase (ACC) (Figure 5B). However, whereas activation of AMPK with AICAR inhibited S6 kinase-induced phosphorylation of S6 ribosomal protein (S6R) in WT pre-B cells (indicative of decreased mTOR function), activation of AMPK failed to inhibit phosphorylation of S6R in Fnip1−/− pre-B cells (Figure 5B), which paralleled our findings using flow cytometry (Figure 4E). AMPK appeared to phosphorylate (and presumably inhibit) the mTOR partner Raptor normally in the absence of Fnip1 (Gwinn et al., 2008). Akt (a positive regulator of mTOR) was phosphorylated normally at the Thr-308 and Ser-473 activation sites (Sengupta et al., 2010) in the absence of Fnip1 (Figure S5A and S5C). These results collectively suggest that Fnip1 is not required for activation of AMPK or AMPK-mediated catabolism, but is important for AMPK to inhibit mTOR.
Figure 5. Dysregulated mTOR signaling in Fnip1−/− pre-B cells.
(A) B cell progenitors from Rag2−/− and Fnip1−/− mice were activated with α-Igβ at the indicated time points. Immunoblots were probed with antibodies against total phosphotyrosine. (B) Fnip1−/− and WT mice were injected with the AMPK-activator AICAR. Total BM cells from untreated or AICAR treated mice were isolated 18 hrs post-injection and B220+CD43+ cells were FACs-sorted. Immunoblots (representative of 3–4 experiments) were performed using the indicated antibodies. α-Tubulin was utilized as a loading control. (see also Figure S5)
Loss of Fnip1 increases sensitivity to nutrient deprivation-induced apoptosis and developmental arrest
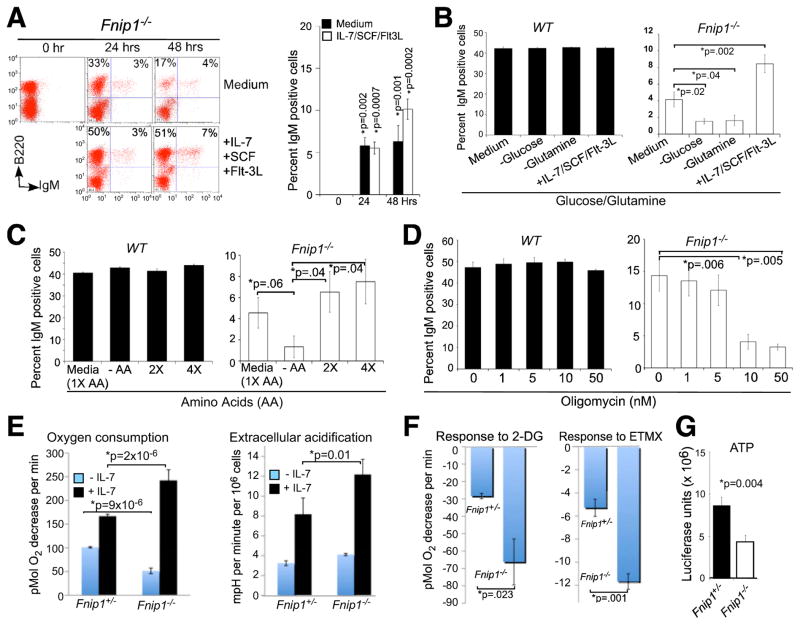
Since Fnip1−/− pre-B cells exhibited increased mTOR activation and cell growth (which consumes nutrients), even in presence of activated AMPK (low energy), we reasoned that perhaps Fnip1 loss results in “nutrient and energy deficit” which may trigger apoptosis or anergy in Fnip1−/− pre-B and Fnip1−/− IgMHEL B cells following pre-BCR or BCR crosslinking, when energy and nutrient demands increase sharply to accommodate cell division. Firstly, we tested whether a “nutrient-rich” environment would increase survival of Fnip1−/− B cells, by culturing BM cells or sorted pro-B cells from Fnip1−/− or WT mice in nutrient rich media. Surprisingly, culture of total BM (Figure 6A) or sorted B220+CD43+ pre-B cells (Figures 6B and data not shown) in B cell growth media (10% fetal calf serum, 1% non-essential amino acids, 1% L-glutamine, 1% sodium pyruvate, β-mercaptoethanol) rescued a small population of IgM+ Fnip1−/− B cell progenitors, which increased by 48 hrs, but decreased thereafter due to increased apoptosis. The total number of IgM+ Fnip1−/− BM cells emerging in vitro increased up to two-fold by adding a cocktail of IL-7/SCF/Flt3 ligand (Figures 6A and 6B), and was associated with increased intracellular Igκ protein expression (data not shown).
Figure 6. Loss of Fnip1 alters metabolic homeostasis and sensitizes pre-B cells to nutrient and IL-7 restriction.
(A) In vitro nutrient supplementation partially rescues IgM expression in Fnip1−/− B cell progenitors. Total BM derived from WT and Fnip1−/− mice were grown in complete B cell growth media for 0, 24, or 48 hrs in the presence or absence of growth factors (IL-7, SCF and Flt-3L). Cells were analyzed by a flow cytometry using fluorescent conjugated α-B220 and α-IgM. Bars represent the means ± SEM from 6 mice per group. (B) Depletion of glucose or glutamine inhibits the development of Fnip1−/− pre-B cells in vitro. FACS-sorted B220low IgM− cells were grown in complete media in the presence or absence of growth factors, glucose, or glutamine for 48 hrs. Cells were analyzed by a flow cytometry using fluorescent conjugated α-B220 and α-IgM. Bars represent the means ± SEM from WT (n=3) and Fnip1−/− (n=6) mice. (C–D) Total BM were cultured in complete media for 48 hrs in the presence of (C) different amounts of essential amino acids (no AA, or 1X, 2X, or 4X AA), or (D) oligomycin (0, 1, 5, 10, 50 nM). Cells were analyzed by a flow cytometry using fluorescent conjugated α-B220 and α-IgM. Bars represent the means ± SEM from WT (n=4) and Fnip1−/− mice (n=4) on gated B220low populations. (E, F) Oxygen consumption rate (OCR) and Extracellular acidification (ECAR) were measured using a Seahorse Bioscience® extracellular flux analyzer over the course of 150 minutes. FACs-sorted pro/pre-B cells from Fnip1+/− and Fnip1−/− BM were cultured in the absence or presence of growth factors (10 ng/ml of IL-7 and SCF) for 24 hrs prior to analyses. (E) OCR and ECAR were then measured within 5 minutes of culture. Shown is the mean ± SEM from 5 mice per group. P-values are shown. (F) OCR were measured within 5 minutes of culture in the presence of 2-deoxyglucose (2-DG) or etomoxir (ETMX). Shown is the decrease in OCR following treatment relative to pre-treatment baseline. Mean ± SEM (3 mice per group). (G) ATP levels from FACs-sorted WT (n=3) and Fnip1−/− (n=3) pre-B cells were determined using a luciferase assay. Mean ± SEM (3 mice per group). P-values are shown.
We next determined which nutrients are most critical for the development of IgM+ Fnip1−/− B cells. We found that culturing pre-B cells in glucose- or glutamine-free media prevented the emergence of Fnip1−/− IgM+ B cells at concentrations where the emergence of WT IgM+ B cells were relatively unaffected (Figure 6B and data not shown). Furthermore, removal of essential amino acids (EAA) significantly inhibited the development of Fnip1−/− IgM+ B cells, which could be rescued in a dose-dependent manner by the addition of EAAs (Figure 6C). Administration of oligomycin, which blocks ATP synthase and oxidative phosphorylation, also inhibited the emergence of Fnip1−/− IgM+ B cells at concentrations that do not affect the development of WT IgM+ B cells (Figure 6D).
To examine why Fnip1−/− B cells might be in metabolic deficit, we analyzed pre-B cell metabolism on FACS-sorted pre-B cells using the Seahorse® Extracellular Flux Analyzer. Fnip1−/− pre-B cells have decreased basal oxygen consumption rate (OCR), which is a measure of oxidative phosphorylation (OXPHOS)(Figure 6E). OCR and extracellular acidification rate (ECAR), which is a measure of glycolysis, could be restored to a hypermetabolic state in Fnip1−/− pre-B cells by the addition of IL-7, which stimulates glucose uptake (Figure 4D) and restores energy balance (Figures 6E) (Rathmell et al., 2001). Fnip1−/− pre-B cells also exhibited increased sensitivity to 2-deoxyglucose (glycolysis inhibitor) and etoxomir (fatty acid oxidation inhibitor) (Figure 6F), which correlated with decreased basal ATP levels relative to WT pre-B cells (Figure 6G).
To examine whether increased PPARγ or dysregulated mTOR (Figure S5D) are responsible for impaired B cell development in Fnip1−/− mice, we first cultured BM cells from Fnip1−/− mice in B cell growth media with or without different concentrations of the specific PPARγ-inhibitor GW9662 (Bendixen et al., 2001). We found that inhibiting PPAR-γ did not rescue nutrient-mediated IgM expression in Fnip1−/− pre-B cells (Figure S7B), suggesting that increased expression of PPARγ (Figure 4A) may reflect an attempt to maintain energy balance in response to dysregulated mTOR. Likewise, activation of PPARγ in wildtype pre-B cells with the agonist troglitazone did not inhibit pre-B to immature-B cell development, although the cells exhibited increased non-specific apoptosis (Figure S7C). Injection of Fnip1−/− mice with the mTORC1-inhibitor rapamycin also failed to rescue B cell development in vivo. However, delivery of a combination of rapamcyin and IL-7 (which increases glucose uptake) were sufficient to rescue the emergence of IgM+ cells in Fnip1−/− mice after 2 weeks (Figure S7D). These results collectively suggest that loss of Fnip1 results in excessive mTOR-mediated metabolite consumption; increased nutrient demand; impaired OXPHOS; and IL-7 “addiction”, which prevent the development of pre-B cells in response to nutrient stress and pre-BCR crosslinking.
Fnip1 deficiency inhibits pre-B cell transformation by c-Myc
Since cancer cells have high metabolic demands, we hypothesized that disruption of Fnip1 might sensitize pre-B cells to oncogene-induced apoptosis. To test this hypothesis, we bred Fnip1−/− mice to mice that overexpress the c-Myc oncogene under control of the Ig heavy chain enhancer Eμ (Eμ-Myc). Eμ-Myc transgenic mice typically develop pre-B cell lymphomas at around 8–30 weeks of age (Adams et al., 1985). As a control, we also bred Fnip1−/− mice to Ighmtm1Cgn (also known as μMT) mice, which are also arrested in B cell development at the pre-B cell stage due to targeted disruption of the transmembrane portion of Igμ (Kitamura and Rajewsky, 1992) (Figure 7A). Eμ-MycFnip1−/+ or +/+ mice exhibited nearly identical Kaplan-Meier lymphoma free curves (Figure 7B), indicating that loss of one allele of Fnip1 has no effect on pre-B cell transformation by c-Myc. Targeted-disruption of Igμ resulted in accelerated B cell transformation by c-Myc (Eμ-MycμMT), likely because B cell development is arrested at an IL-7-responsive pre-B cell stage (Herzog et al., 2009). In contrast, despite a similar block in B cell development (Figure 7A) disruption of Fnip1 in Eμ-MycFnip1−/− mice nearly completely inhibited pre-B cell lymphomagenesis induced by c-Myc. Inhibition of pre-B cell transformation was associated with increased apoptosis of B220+CD43+ Eμ-Myc Fnip1−/− pre-B cells relative to Eμ-Myc Fnip1−/+ pre-B cells (Figure 7C), whereas cell division was unaffected (data not shown). These results collectively demonstrate that Fnip1 is important for the generation of pre-B cell lymphomas induced by c-Myc.
DISCUSSION
Resting B lymphocytes respond to foreign pathogens by rapidly shifting from a quiescent state to a metabolically-activated phenotype, characterized by massive increases in macromolecular synthesis necessary for cell growth, division, and eventually Ig synthesis (see (Rathmell, 2004) for review). The inability to accommodate these demanding bioenergetic processes would result in dysfunctional B cells and an immunodeficient host. Conversely, deregulated cell growth could result in lymphomagenesis and the demise of the host. In this study, we utilized an ENU mutagenesis screen in mice to identify a new B cell immunodeficient strain, caused by a non-coding deletion in the Fnip1 gene. Our studies suggest that Fnip1 normally acts as a molecular switch to permit pre-B cell differentiation and survival in response to adequate metabolic balance, while turning off cellular growth. Disruption of Fnip1 results in metabolic imbalance, which triggers apoptosis in response to “metabolic stress” such as pre-BCR stimulation, nutrient deprivation, or oncogene activation. We hypothesize that this “metabolic checkpoint” may ensure that maturing B cells have adequate metabolic capacity to fuel clonal expansion and to produce Igs, while also attenuating deregulated cell growth and transformation.
Our analysis of Fnip1−/− B cell progenitors reveals that Fnip1 is very unique in how it intersects with known pre-BCR signaling pathways. Firstly, enforced expression of potent IgMHEL signaling complexes failed to rescue B cell development in Fnip1−/− mice, indicating that Fnip1 is required for B cell development downstream of the pre-BCR (and perhaps the BCR as well). Whereas mice deficient in other signaling molecules including Src-, Syk-, and Tec-family PTKs; the adapter protein SLP-65; and the Ca2+ signaling molecules PLCγ1/2 and the p85 subunit of PI3K, also exhibit impaired B cell development at the large pre-B cell stage (see (Herzog et al., 2009) for review), we find that these pathways (as well as important transcriptional mediators) are expressed or activated normally in Fnip1−/− pre-B cells following α-Igβ crosslinking. A second unique feature of Fnip1 is that loss of Fnip1 blocks pre-BCR-induced differentiation, whereas pre-BCR-induced proliferation (synergistic with IL-7) and allelic exclusion of the IgH chain loci remain intact. This is in contrast to disruption of the membrane portion of IgH, Igα and Igβ, or Syk and Zap70, which result in blocked proliferation, differentiation, and allelic exclusion (Kurosaki et al., 2010). A third unique feature is that loss of Fnip1 potently inhibits transformation and/or survival by Myc, whereas disruption of another pre-BCR signaling molecule Igμ (which also blocks B cell development at the pre-B cell stage), results in significantly accelerated Myc-induced transformation. These results collectively highlight that Fnip1 functions in a distinct manner from most of the known pre-BCR signaling molecules.
Fnip1 was cloned based on its interactions with Folliculin and AMPK in kidney cells (Baba et al., 2006), and was subsequently shown to interact with Fnip2. Here, we find that AMPK is activated normally in pre-B cells from Fnip1−/− mice relative to WT mice. In addition, we find increased expression of essential AMPK-induced mitochondrial metabolic genes, and increased glucose uptake and mitochondrial biogenesis in Fnip1−/− pre-B cells. However, whereas AMPK activation in WT pre-B cells shuts down mTOR-mediated anabolic (ATP and bioprecursor-consuming) growth (Gwinn et al., 2008), AMPK fails to inhibit mTOR-mediated growth in Fnip1−/− pre-B cells, which correlates with decreased energy balance (decreased ATP) and increased sensitivity to apoptosis following pre-BCR activation, nutrient or IL-7 deprivation, or oncogene activation. These results, combined with previous studies by Baba et al. in HEK cells, suggest that Fnip1 acts downstream of AMPK to help maintain “metabolic homeostasis” in B lineage cells during metabolic stress (see Figure S5D for Model). Recent studies indicate that disruption of AMPK-activator LKB also inhibits pre-TCR-mediated T cell development (Tamas et al., 2010), and disruption of LKB1 in hematopoietic stem cells results in dysregulated mTOR, increased cell growth, and decreased survival of differentiated hematopoietic cells (Gan et al., 2010; Gurumurthy et al., 2010; Nakada et al., 2010). Similar to Fnip1 deficiency, hematopoietic cell development is not rescued by rapamycin alone, indicating that Fnip1 and LKB1 may function in both mTOR-dependent and -independent manners. Indeed, B cell development in Fnip1−/− mice can only be partially rescued by a combination of rapamycin (which inhibits mTORC1) and IL-7 (which stimulates glucose uptake and OXPHOS).
Whereas both Fnip1 and Fnip2 are expressed during B cell development in WT mice, only Fnip2 is significantly regulated with expression peaking in immature B cells. In addition, Fnip2 mRNA levels increase in Fnip1-null pre-B cells (data not shown). These results suggest that Fnip1 and Fnip2 may only be active as heterodimers, which would explain why Fnip2 does not compensate for the absence of Fnip1. Targeted disruption of the Fnip2 gene in mice should shed light on this issue.
An important distinction between Fnip1-null pre-B cells, which exhibit increased mTOR activity, and other human diseases whereby mTOR is dysregulated, is the absence of benign or malignant overgrowth. For example, mutations in LKB1, PTEN, FLCN, and TSC1/2 result in deregulated mTOR and overgrowth (hamartoma) syndromes or cancer in humans (see (Shackelford and Shaw, 2009) for review). In addition, loss-of-function mutations in the PPKAG2 gene encoding the α subunit of AMPK result in Familial Cardiac Hypertrophy and Wolf-Parkinson’s-White Syndrome, overgrowth diseases characterized by cardiac hypertrophy and arrhythmias due to dysregulated mTOR (Arad et al., 2007). We find that loss of Fnip1 also results in cardiac hypertrophy and excessive growth of pre-B cells characterized by dysregulated mTOR. However, we have not observed transformation of any cell types including pre-B cells, and we find that loss of Fnip1 inhibits pre-B cell lymphomagenesis by the c-Myc oncogene, which potently drives transformation in part by increasing the metabolic capacity of cancer cells (see (Dang et al., 2009) for review)). Thus, our results suggest the existence of a “metabolic checkpoint” during B cell development, which inhibits the maturation and survival of pre-B cells that are “metabolically insufficient”, either due to mutations that disrupt energy or bioprecursor generating pathways, and/or due to excessive energy or bioprecursor consumption. Inhibition of Fnip1 could provide a novel mechanism to limit the fuel necessary to support cancer cell metabolism (Vander Heiden et al., 2009), thus selectively sensitizing highly metabolic cancer cells for apoptosis in response to chemotherapy or metabolic stress.
EXPERIMENTAL PROCEDURES
Mice
VDJ9HyHEL10 heavy chain knockin mice and HyHEL10 light chain transgenic mice were provided by Jason Cyster (Allen et al., 2007). Ighmtm1Cgn, Eμ-Myc, Rag2−/− mice and Rag2−/−Il2rg−/− mice were purchased from Jackson Laboratories and Taconic Farms Inc. Fnip1−/− mice were screened and maintained by genomic PCR analysis, following amplification with Fnip1 forward (5′-ACTTGTCGCCATAGTTCTTCACTG-3′) and reverse (5′-CCAACTTACCATCTAGGAAACACCC-3′) oligonucleotides. Mice were housed under Specific Pathogen-Free conditions. Mouse procedures were approved by the Institutional Animal Care and Use Committees of Celltech R&D, Inc. and the University of Washington.
Immunoblot analysis
Immunoblot analyses were performed as previously described (Iritani et al., 1997) using rabbit polyclonal antibodies specific for Fnip1 (anti-hFNIP1; Abcam, MA); βActin, Syk, Btk (Santa Cruz Biotech); Blnk, P-Tyr96 Blnk, Akt, P-Ser473 Akt, P-Thr308 Akt, P-Ser176/180IKKα/β, P-Ser240/244 S6 Ribosomal Protein, S6 Ribosomal Protein, P-Thr172AMPK, P-Erk, total Erk (Cell Signaling), donkey α-goat IgG HRP (Santa Cruz), goat α-rabbit IgG HRP and goat α-mouse IgG HRP (Bio-Rad Laboratories), 4G10 antibody (Upstate Biotech).
Flow cytometry and cell sorting
BM, thymus, and spleen were stained with fluorescent-conjugated antibodies specific for CD5, B220, CD21, CD25, CD22.2, CD62L, IAb, CD23, CD43, IgM, IgMa, IgMb, λ5, Igμ, Igκ, pS6R, BrdU, Annexin V, BP1, HSA, CD5, CD4, CD8, Lysozyme, and analyzed by flow cytometry as previously described (Habib et al., 2007). Mitochondrial staining was performed using MitoTracker® green dye (Molecular Probes).
Cell proliferation and viability
BM cells were labeled with CFSE and then cultured in the presence or absence of IL-7 (10ng/ml) for 4 days. Cell division was determined by flow cytometry on gated B220low cells (Habib et al., 2007). BM cells were stained with α-B220, α-CD43, and 7AAD (Invitrogen) or α-annexinV and cell viability was analyzed by flow cytometry.
Statistical analysis
Data were analyzed using the Student’s one or two-tailed t-test. Kaplan-Meier curve statistics were performed using Log-rank Mantel-Cox test. P<0.05 were considered as significant values.
Supplementary Material
HIGHLIGHTS.
A deletion in the Fnip1 gene blocks B cell development at the pre-B cell stage
Fnip1 controls metabolic homeostasis and survival during pre-B cell activation
Fnip1 is required for AMPK to inhibit mTOR-mediated pre-B cell growth
Fnip1−/− pre-B cells are resistant to lymphoma formation induced by c-Myc
Acknowledgments
This study was supported by the University of Washington Royalty Research Fund #4162, NIH K26RR024462, MMPC 09MCG96, ITHS UL1RR05014, P30 CA15704 grants to B.M.I., and funding from Celltech R&D, Inc. We thank Jason Cyster for kindly providing IgMHEL mice.
Footnotes
The authors have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- Appleby MW, Ramsdell F. A forward-genetic approach for analysis of the immune system. Nat Rev Immunol. 2003;3:463–471. doi: 10.1038/nri1109. [DOI] [PubMed] [Google Scholar]
- Arad M, Seidman CE, Seidman JG. AMP-activated protein kinase in the heart: role during health and disease. Circ Res. 2007;100:474–488. doi: 10.1161/01.RES.0000258446.23525.37. [DOI] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendixen AC, Shevde NK, Dienger KM, Willson TM, Funk CD, Pike JW. IL-4 inhibits osteoclast formation through a direct action on osteoclast precursors via peroxisome proliferator-activated receptor gamma 1. Proc Natl Acad Sci U S A. 2001;98:2443–2448. doi: 10.1073/pnas.041493198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MC, Vinuesa CG, Goodnow CC. ENU-mutagenesis: insight into immune function and pathology. Curr Opin Immunol. 2006;18:627–633. doi: 10.1016/j.coi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming HE, Paige CJ. Cooperation between IL-7 and the pre-B cell receptor: a key to B cell selection. Semin Immunol. 2002;14:423–430. doi: 10.1016/s1044532302000775. [DOI] [PubMed] [Google Scholar]
- Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang YA, Chin L, Depinho RA. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, DiSanto J, Stockinger B. Following the development of a CD4 T cell response in vivo: from activation to memory formation. Immunity. 1999;11:163–171. doi: 10.1016/s1074-7613(00)80091-6. [DOI] [PubMed] [Google Scholar]
- Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435:590–597. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, Knoepfler PS, Andrews S, Rawlings DJ, Eisenman RN, Iritani BM. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415:60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks RW, Middendorp S. The pre-BCR checkpoint as a cell-autonomous proliferation switch. Trends Immunol. 2004;25:249–256. doi: 10.1016/j.it.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Forbush KA, Farrar MA, Perlmutter RM. Control of B cell development by Ras-mediated activation of Raf. Embo J. 1997;16:7019–7031. doi: 10.1093/emboj/16.23.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Rajewsky K. Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- Kouro T, Nagata K, Takaki S, Nisitani S, Hirano M, Wahl MI, Witte ON, Karasuyama H, Takatsu K. Bruton’s tyrosine kinase is required for signaling the CD79b-mediated pro-B to pre-B cell transition. Int Immunol. 2001;13:485–493. doi: 10.1093/intimm/13.4.485. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Shinohara H, Baba Y. B cell signaling and fate decision. Annu Rev Immunol. 2010;28:21–55. doi: 10.1146/annurev.immunol.021908.132541. [DOI] [PubMed] [Google Scholar]
- Nagata K, Nakamura T, Kitamura F, Kuramochi S, Taki S, Campbell KS, Karasuyama H. The Ig alpha/Igbeta heterodimer on mu-negative proB cells is competent for transducing signals to induce early B cell differentiation. Immunity. 1997;7:559–570. doi: 10.1016/s1074-7613(00)80377-5. [DOI] [PubMed] [Google Scholar]
- Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Holmes KL, Morse HC, 3rd, Leder P. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin mu. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- Park H, Staehling-Hampton K, Appleby MW, Brunkow ME, Habib T, Zhang Y, Ramsdell F, Liggitt HD, Freie B, Tsang M, et al. A point mutation in the murine Hem1 gene reveals an essential role for Hematopoietic protein 1 in lymphopoiesis and innate immunity. J Exp Med. 2008;205:2899–2913. doi: 10.1084/jem.20080340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC. B-cell homeostasis: digital survival or analog growth? Immunol Rev. 2004;197:116–128. doi: 10.1111/j.0105-2896.2004.0096.x. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000;6:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Kobayashi T, Shiono M, Wang L, Piao X, Sun G, Zhang D, Abe M, Hagiwara Y, Takahashi K, Hino O. Interaction of folliculin (Birt-Hogg-Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27:5339–5347. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- Tamas P, Macintyre A, Finlay D, Clarke R, Feijoo-Carnero C, Ashworth A, Cantrell D. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur J Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.