Abstract
REST/NRSF (the RE-1 silencing transcription factor or neuron-restrictive silencer factor) was originally identified as a transcriptional repressor of a number of neuronal-specific genes in neural stem cells and non-neuronal cells. REST functions as a master regulator in the maintenance of neural stem cells. During tumorigenesis, REST shows opposing roles in different type of cells. In human epithelial cancers such as colon cancer, REST act as a tumor suppressor. In contrast, REST plays an oncogenic role in the development of brain tumors and other cancers. Abnormal upregulation of REST has been found in medulloblastoma, neuroblastoma and glioblastoma (GBM). Recent studies in GBMs suggest that REST exert its oncogenic function by maintaining self-renewal potential of glioma stem cells (GSCs).
Keywords: REST, Ubiquitination, Deubiquitination, Neural Stem Cell, Tumorigenesis
Introduction
The RE-1 silencing transcription factor (REST) or neuron-restrictive silencer factor (NRSF) is a master transcriptional repressor with a critical role in suppressing expression of neuronal-specific genes. Since its original identification in 1995 [1; 2], REST has been shown to play critical roles during embryonic development and neurogenesis. In addition, abnormal expression or dysfunction of REST has been associated with diverse diseases, including Down’s syndrome, Huntington’s disease and various types of cancer. This review will focus on the roles of REST to act as a tumor suppressor or oncogene in diverse types of cancers.
1. The Repressor Complex Formed by REST
REST is a Krüppel type zinc finger transcription factor containing eight zinc fingers and two repressor domains (RD1 and RD2) located at N-terminal and C-terminal respectively. REST protein contains a DNA binding domain that is localized within the cluster of zinc fingers and usually binds to the 21-bp repressor element 1 (RE-1) in the regulatory regions of target genes. The DNA binding domain can recognize two types of RE-1 motifs, including the canonical RE-1 motif and non-canonical RE1 motif. The canonical RE1 motif consists of two 10 bp conserved sequences separated by a single non-conserved nucleotide, whereas the non-canonical RE1 motif has variable length of insertion between these two conserved sequences [3; 4]. The canonical RE-1 motif has a higher affinity for REST than the non-canonical motif [5], suggesting that tissue-specific function of REST may depend on binding to different RE-1 motifs. Once REST binds to the RE-1 motifs in target genes, it acts as a molecular scaffold to recruit several cofactors to its amino-terminal RD1 and carboxy-terminal RD2 repression domains. The macromolecular complexes thus generated epigenetically modulate target gene expression. The most important transcriptional co-regulators that REST recruits are CoREST and mSin3A [6–13]. The N-terminal RD1 domain of REST interacts with mSin3A, subsequently recruits the histone deacetylase complex (HDAC1, HDAC2, HDAC4 and HDAC5) and a nuclear hormone receptor co-repressor N-CoR [12; 14]. Meanwhile, the C-terminal RD2 domain of REST associates with CoREST, which also interacts with methyl-CpG binding protein 2 (Mecp2), lysine-specific demethylase-1 (LSD1) and histone H3-lysine 9 methyltransferases G9a [15–17]. In addition, several studies have identified new members of the REST complex, such as DNA methyltransferases (DNMTs), the SWI/SNF complex and the chromodomain on Y-like (CDYL) [17; 18]. These studies demonstrated that REST regulates expression of target genes through a dynamic macromolecular complex. The composition of the repressor complex may affect specificity of REST regulation on target genes.
2. Regulation of REST by Ubiquitination and Deubiquitination
It has been well known that REST protein level is critical for cell fate determination of neural stem or progenitor cells (NSCs or NPCs). Reduced REST could trigger cell differentiation of NSCs or NPCs. Although several pathways have been implicated in the translational regulation of REST [19; 20], the post-translational control mediated by ubiquitination and deubiquitination also plays important roles in regulating protein levels of REST. During neuronal differentiation, REST undergoes proteasomal degradation through β-TrCP-mediated ubiquitination. For example, during neuronal differentiation induced by retinoic acid (RA), REST protein was reduced by the β-TrCP-mediated ubiquitination that leads to degradation of REST [21]. However, during astrocytic differentiation from neural progenitor cells, REST expression is maintained by bone morphogenetic proteins (BMPs) to repress neuronal gene expression [19]. The canonical Wnt pathway has also been shown to regulate the expression of REST [20]. Interestingly, REST promoter also contains an RE-1 sequence, suggesting that there may be a negative feedback loop in regulating REST expression at the transcriptional level. At the post-translational level, REST protein is tightly controlled by β-TrCP through ubiquitin-proteasomal degradation [22] and the HAUSP-mediated deubiquitination [23]. Two β-TrCP binding motifs have been identified near the RD2 domain of REST. A conserved phosphodegron near the carboxy-terminal of REST is required for its interaction with β-TrCP [22]. Dephosphorylation of REST by treatment with λ-phosphatase or phosphodegron-mutant REST reduces its interaction with β-TrCP, resulting in increased stability of REST protein. Other protein such as a telomere binding protein TRF2 has been shown to stabilize REST in pluripotent human stem cells (NTera2) by direct interaction with REST [24]. Recently, we identified a critical deubiquitinase HAUSP (the herpesvirus-associated ubiquitin-specific protease, also known as USP7) that can counterbalance REST ubiquitination and prevents NPC differentiation through deubiquitination [23]. REST protein level declines concordantly with HAUSP upon neuronal differentiation and reciprocally with β-TrCP levels. Functional disruption of HAUSP in NPCs reduces REST protein and induces neuronal differentiation. In contrast, HAUSP overexpression stabilizes REST protein by overriding β-TrCP-mediated ubiquitination. We also showed that a consensus site (310-PYSS-313) of human REST protein is required for the HAUSP-mediated REST deubiquitination. Our studies demonstrated that HAUSP stabilizes REST through deubiquitination and antagonizes β-TrCP in regulating REST at the post-translational level [23]. Thus, the HAUSP-mediated deubiquitination and the β-TrCP-mediated ubiquitination represent critical regulatory mechanisms involved in the post-translational control of REST protein and its biological functions. We proposed that deubiquitination and ubiquitination system functions as “Ying-Yang” control system to regulate REST protein levels. The net balance between the β-TrCP-mediated ubiquitination and HAUSP-mediated deubiquitination controls REST protein levels to direct cell fate determination (Figure 1). The relative activity of deubiquitination and ubiquitination for REST may define the maintenance of “stemness” or initiation of neuronal differentiation. It is likely that the abnormal regulation of REST by deubiquitination and ubiquitination may be associated with tumorigenesis.
Figure 1.
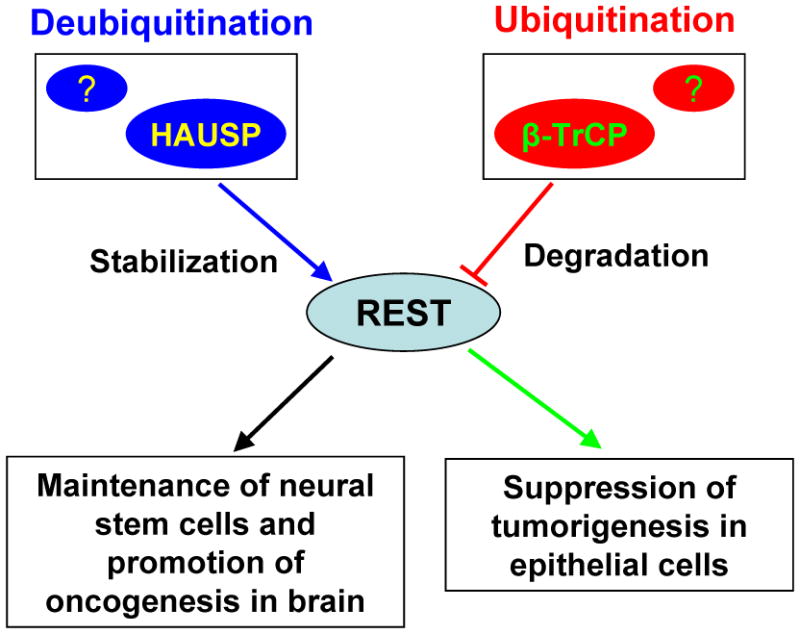
Post-translational control of REST by β-TrCP-mediated ubiquitination and HAUSP-mediated deubiquitination to regulate REST protein level and its biological roles. REST functions as a tumor suppressor in epithelial cells. But abnormal expression or dysfunction of REST may exert oncogenic activity during brain development.
3. The Tumor Suppressor Role of REST in Non-nervous Tumors
As REST play crucial roles in the regulation of gene expression, abnormal function or genetic mutation of REST has been linked to several cancer types. Interestingly, REST appears to have opposite roles in non-nervous cancers and tumors in central nervous system (CNS). REST was identified as a tumor suppressor in epithelial cells during an RNAi-based genetic screening [25]. Functional inhibition of REST in epithelial cells increased cellular capability for oncogenic transformation. Consistently, genetic deletion of REST was found to be a frequent event in colon cancer. A premature termination mutant REST-FS, which lacks the carboxyl terminus and acts as dominant negative isoform, also promotes anchorage-independent growth of colon cancer cells. Normal bronchial epithelial cells express high level of REST to block the neuroendocrine gene expression, while some small cell lung cancers (SCLCs) express both epithelial and neuronal markers [26; 27]. In these SCLC primary tumor samples, a new REST splice variant with a 50-bp insertion resulting in a truncated REST mutant was identified. Similar to the REST-FS mutant, the truncated REST isoform also functions as a dominant-negative inhibitor of wild type REST, causing upregulation of neuronal markers in those cancer cells. In addition, the reduced REST expression or activity and abnormal expression of the REST target gene glycine receptor α1 subunit were detected in other SCLC cancer cell lines [28; 29]. Based on the potential link between REST activity and human SCLCs, a recent study suggested that the transcriptional profile of SCG3 (a REST-dependent gene) can be used as a sensitive prognostic biomarker for neuroendocrine lung cancer [30]. In contrast to SCLC cells, most non-SCLC cells display normal levels of REST mRNA and protein. However, approximately 10% of non-SCLC cancers show loss of function of the SWI/SNF complex, a cofactor required for REST activity, resulting in increased expression of some neuronal genes in these cancers. In addition to colon cancer and lung cancer, prostate cancer LNCaP cells and breast cancer cells have been shown to have the loss of REST function and the subsequent increase in neuroendocrine genes expression [31–33]. Thus, in those non-nervous cancers, loss of REST activity leads to neuroendocrine carcinoma-like tumor, a highly aggressive type of carcinoma [34], suggesting that REST dysfunction could contribute to tumor development and malignant progression.
The tumor suppressor role of REST in human epithelial cancers has been further validated by other important studies [22; 35]. Overexpression of β-TrCP, an E3 ubiquitin ligase responsible for REST degradation, has been found to cause oncogenic transformation of human mammary epithelial cells (HMECs). This transformation depends on REST degradation induced by β-TrCP. Furthermore, REST has been shown to have the ability to repress transcription of oncogenes. The REST co-repressor CDYL (chromodomain on Y-like) can bridge REST and the histone methylase G9a to inhibit the expression of proto-oncogene TrkC, which can induce oncogenic transformation [17]. In addition to suppression of oncogene expression, REST acts as a tumor repressor in these non-neuronal cancers by regulating cell apoptosis and proliferation. Knockdown of REST enhances the EGF-induced Akt phosphorylation, and inhibition of PI(3)K signaling reduces the REST-shRNA-induced transformation in HMECs [25]. Loss of REST function also enhances cell proliferation and survival by promoting Akt phosphorylation and upregulation of BCL-2 expression in lung cancers and breast cancers [32; 36]. Collectively, these studies have demonstrated that the critical role of REST in human non-neuronal cells is tumor suppression and growth inhibition. However, one most recent study using conditional deletion of REST gene in the colonic crypts indicated that REST might have a different role in mouse colon cancer development [37]. Conditional deletion of REST gene in the colonic crypts increases the expression of REST-targeted genes such as syt4 and tubb3 in these cells, but showed no significant effect on colon tumor development. These findings suggest that the ablation of REST tumor suppressor is not sufficient for the development of colon tumors in mice and other genetic or epigenetic alteration might be required for the tumorigenic effect of REST deletion in mouse colon. Further investigation on the role of REST in tumor suppression in epithelial cells is clearly necessary.
4. The Oncogenic Function of REST in Brain Tumors
As REST plays a crucial role in the maintenance of neural stem cells in brain, the functional significance of REST abnormality has been examined in several types of brain tumors, including medulloblastoma and glioblastoma. It has been demonstrated that REST mainly acts as an oncogenic promoter in the development of these brain tumors. In medulloblastoma, the most common malignant pediatric brain tumor that was thought to arise from neural stem cells or neural progenitor cells, REST has been shown to promote medulloblastoma development [38; 39]. REST is highly expressed in medulloblastomas relative to fully differentiated human neuronal teratocarcinomas and neural progenitor cells. Ectopic expression of REST-VP16 mutant in medulloblastoma cells significantly reduced tumorigenic potential of these cancer cells in mice. In addition, inhibition of REST activity in medulloblastoma cells leads to apoptosis. But the cellular and molecular mechanisms associated with the apoptosis induced by REST inhibition remain unclear. A recent study showed that REST can regulate Hedgehog signaling in a context-dependent manner during embryonic development [40]. The Hedgehog signaling also plays an important role in regulating proliferation and differentiation of neural stem cells [41]. During medulloblastoma development, the Hedgehog signaling was found to be highly activated [42–44]. In addition, the Wnt signaling pathway that controls the property of the neural stem cells has been shown to regulate REST expression [20]. Thus, the cross talk between REST and the hedgehog signaling or Wnt signaling may play important roles during oncogenesis of medulloblastoma. Notably, the effect of REST inhibition on cell survival in medulloblastoma cells and human epithelial cancer cells appears totally opposite. Inhibition of REST activity promotes survival of human epithelial cell but induces apoptosis of medulloblastoma cells. This may partially explain the different role of REST in epithelial cancers and brain tumors. It has shown that REST needs cooperation with other oncogenes to promote tumorigenesis of medulloblastomas. Overexpression of REST alone in neural stem cells is not sufficient to induce medulloblastoma. But co-expression of REST and oncoprotein c-Myc in neural stem cells potently induce medulloblastoma [45]. Thus, despite different roles of REST in varied types of human tumors, genetic mutation or abnormal expression of REST may contribute to only one of the initial steps of tumorigenesis. It seems that REST has to work with other oncogenes or oncogenic pathways induced by genetic and/or epigenetic alterations to push tumorigenesis.
Glioblastoma multiforme (GBM) is the most common and lethal type of primary brain tumor in adults. GBM displays remarkable cellular heterogeneity and hierarchy with tumor-propagating cells or glioma stem cells (GSCs) at the apex of the hierarchy. GSCs are functionally defined by self-renewal, multi-lineage differentiation and in vivo propagation of tumor that recapitulates the cellular hierarchy and tissue architecture of the parental GBM. Several studies including ours demonstrated that GSCs promote tumor angiogenesis, cancer invasion and therapeutic resistance [46–48]. It has been suggested that targeting REST for proteasomal degradation might disrupt GSCs [49]. GSCs derived from GBM primary tumors express varied levels of REST [50]. However, the GSCs that highly express REST (HR-GSCs) also have high levels of the key GSC transcription factor SOX2 and show greater ability of tumorsphere formation relative to the GSCs with lower REST (LR-GSCs) [50]. Furthermore, tumor formation assay in the GBM xenograft model demonstrated that HR-GSCs have greater tumorigenic potential in vivo. Knockdown of REST in HR-GSCs induced apoptosis of tumor cells, resulting in reduction of tumor growth and invasion. These studies suggest that the main role of REST in GSCs is to maintain the self-renewal and tumorigenic potential of GSCs. In neural stem cells, REST can bind to a number of regulatory regions of target genes that are members of the pluripotency network or Oct4/Sox2/Nanog transcriptional network [51]. In addition, REST has been found to control the expression of several important miRNAs (miR-124a, miR-9 and miR-132) that regulate the functions of neural stem cells and cancer stem cells in brain tumors. Further studies are required to determine whether REST controls the similar signaling pathway to regulate the self-renewal potential of GSCs and NSCs.
5. Concluding Remarks
Recent studies clearly showed that REST plays opposite roles in epithelial cancers and brain tumors. REST acts as a tumor suppressor to inhibit tumorigenesis in epithelial cells, while REST plays an oncogenic role during the development of brain tumors. It is not clear why REST play differential roles in different cell types. The precise molecular mechanisms associated with this cell type-specific role of REST need to be further investigated. It is likely that REST forms different dynamic repressor complex with other co-activators in different cell types to exert its varied biological roles. In order to better understand the functional significance of REST in regulating tumor development, more characterizations on transcriptional and post-translational regulations of REST as well as REST-mediated signaling pathway in different type of cells are certainly necessary. As REST plays a critical role in the development of several malignant brain tumors, REST may be a potential therapeutic target for brain tumors. However, as REST also plays crucial roles in the maintenance of neural stem cells, it is important to design a novel therapeutic approach specifically targeting REST in cancer cells.
Acknowledgments
We thank other members in the Bao’s laboratory for helpful scientific discussions. This work was supported by the Cleveland Clinic Foundation and a NIH R01 grant (NS070315) to S.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chong JA, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 2.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 3.Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, Wood IC, Jackson RM, Buckley NJ. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic acids research. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto SJ, et al. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce AW, et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome research. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Current opinion in neurobiology. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Current biology : CB. 2005;15:R665–668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 9.Lunyak VV, Rosenfeld MG. No rest for REST: REST/NRSF regulation of neurogenesis. Cell. 2005;121:499–501. doi: 10.1016/j.cell.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Current opinion in cell biology. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annual review of neuroscience. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nature neuroscience. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 13.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 14.Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. Journal of molecular biology. 2005;354:903–915. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome biology. 2009;10:R9. doi: 10.1186/gb-2009-10-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Mulligan P, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Molecular cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battaglioli E, Andres ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G. REST repression of neuronal genes requires components of the hSWI.SNF complex. The Journal of biological chemistry. 2002;277:41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- 19.Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, Nakashima K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. The Journal of cell biology. 2010;189:159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishihara S, Tsuda L, Ogura T. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochemical and biophysical research communications. 2003;311:55–63. doi: 10.1016/j.bbrc.2003.09.158. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, Stewart J, Zage P, Gopalakrishnan V. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TRCP) in neuroblastoma cells. Cancer. 2011;117:5189–5202. doi: 10.1002/cncr.26145. [DOI] [PubMed] [Google Scholar]
- 22.Westbrook TF, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, Bao S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nature cell biology. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, Pazin MJ, Schwartz CM, Becker KG, Wersto RP, Dilley CM, Mattson MP. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Current biology : CB. 2008;18:1489–1494. doi: 10.1016/j.cub.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westbrook TF, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Coulson JM, Fiskerstrand CE, Woll PJ, Quinn JP. Arginine vasopressin promoter regulation is mediated by a neuron-restrictive silencer element in small cell lung cancer. Cancer research. 1999;59:5123–5127. [PubMed] [Google Scholar]
- 27.Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer research. 2000;60:1840–1844. [PubMed] [Google Scholar]
- 28.Gurrola-Diaz C, Lacroix J, Dihlmann S, Becker CM, von Knebel Doeberitz M. Reduced expression of the neuron restrictive silencer factor permits transcription of glycine receptor alpha1 subunit in small-cell lung cancer cells. Oncogene. 2003;22:5636–5645. doi: 10.1038/sj.onc.1206790. [DOI] [PubMed] [Google Scholar]
- 29.Neumann SB, Seitz R, Gorzella A, Heister A, Doeberitz MK, Becker CM. Relaxation of glycine receptor and onconeural gene transcription control in NRSF deficient small cell lung cancer cell lines. Brain research Molecular brain research. 2004;120:173–181. doi: 10.1016/j.molbrainres.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Moss AC, Jacobson GM, Walker LE, Blake NW, Marshall E, Coulson JM. SCG3 transcript in peripheral blood is a prognostic biomarker for REST-deficient small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:274–283. doi: 10.1158/1078-0432.CCR-08-1163. [DOI] [PubMed] [Google Scholar]
- 31.Tawadros T, Martin D, Abderrahmani A, Leisinger HJ, Waeber G, Haefliger JA. IB1/JIP-1 controls JNK activation and increased during prostatic LNCaP cells neuroendocrine differentiation. Cellular signalling. 2005;17:929–939. doi: 10.1016/j.cellsig.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Lv H, Pan G, Zheng G, Wu X, Ren H, Liu Y, Wen J. Expression and functions of the repressor element 1 (RE-1)-silencing transcription factor (REST) in breast cancer. Journal of cellular biochemistry. 2010;110:968–974. doi: 10.1002/jcb.22610. [DOI] [PubMed] [Google Scholar]
- 33.Wagoner MP, Gunsalus KT, Schoenike B, Richardson AL, Friedl A, Roopra A. The transcription factor REST is lost in aggressive breast cancer. PLoS genetics. 2010;6:e1000979. doi: 10.1371/journal.pgen.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker GE, Antoniono RJ, Ross HJ, Paisley TE, Oh Y. Neuroendocrine-like differentiation of non-small cell lung carcinoma cells: regulation by cAMP and the interaction of mac25/IGFBP-rP1 and 25.1. Oncogene. 2006;25:1943–1954. doi: 10.1038/sj.onc.1209213. [DOI] [PubMed] [Google Scholar]
- 35.Guardavaccaro D, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene. 2010;29:5828–5838. doi: 10.1038/onc.2010.321. [DOI] [PubMed] [Google Scholar]
- 37.Hatano Y, Yamada Y, Hata K, Phutthaphadoong S, Aoki H, Hara A. Genetic ablation of a candidate tumor suppressor gene, Rest, does not promote mouse colon carcinogenesis. Cancer science. 2011;102:1659–1664. doi: 10.1111/j.1349-7006.2011.02006.x. [DOI] [PubMed] [Google Scholar]
- 38.Lawinger P, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nature medicine. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 39.Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, Majumder S. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Molecular cancer therapeutics. 2005;4:343–349. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- 40.Gates KP, Mentzer L, Karlstrom RO, Sirotkin HI. The transcriptional repressor REST/NRSF modulates hedgehog signaling. Developmental biology. 2010;340:293–305. doi: 10.1016/j.ydbio.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, Alvarez-Buylla A. Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron. 2011;71:250–262. doi: 10.1016/j.neuron.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatton BA, et al. Notch signaling is not essential in sonic hedgehog-activated medulloblastoma. Oncogene. 2010;29:3865–3872. doi: 10.1038/onc.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Molecular and cellular biology. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guryanova OA, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma--molecular signaling and therapeutic targeting. Protein & cell. 2010;1:638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 49.Zhang P, Lathia JD, Flavahan WA, Rich JN, Mattson MP. Squelching glioblastoma stem cells by targeting REST for proteasomal degradation. Trends in neurosciences. 2009;32:559–565. doi: 10.1016/j.tins.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamal MM, et al. REST Regulates Oncogenic Properties of Glioblastoma Stem Cells. Stem Cells. 2012 doi: 10.1002/stem.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]


