SUMMARY
Nitric oxide (NO) signaling in vertebrates is well characterized and involves the Heme-Nitric oxide/OXygen binding (H-NOX) domain of soluble guanylate cyclase as a selective NO sensor. In contrast, little is known about the biological role or signaling output of bacterial H-NOX proteins. Here, we describe a molecular pathway for H-NOX signaling in Shewanella oneidensis. NO stimulates biofilm formation by controlling the levels of the bacterial secondary messenger cyclic diguanosine monophosphate (c-di-GMP). Phosphotransfer profiling was used to map the connectivity of a multi-component signaling network that involves integration from two histidine kinases and branching to three response regulators. A feed-forward loop between response regulators with phosphodiesterase domains and phosphorylation-mediated activation intricately regulated c-di-GMP levels. Phenotypic characterization established a link between NO signaling and biofilm formation. Cellular adhesion may provide a protection mechanism for bacteria against reactive and damaging NO. These results are broadly applicable to H-NOX-mediated NO signaling in bacteria.
INTRODUCTION
Nitric oxide (NO) is a ubiquitous signaling molecule in nature despite its inherent toxicity and reactivity. NO signaling in vertebrates is a well-understood process in which NO activates soluble guanylate cyclase (sGC), leading to the formation of cyclic GMP, which controls diverse downstream processes such as vasodilation and neurotransmission (Derbyshire and Marletta, 2009). sGC contains a unique heme domain, which selectively binds NO and has no measurable affinity for O2. This domain is a member of a larger family of hemoprotein sensors for diatomic gases termed Heme-Nitric oxide/OXygen binding (H-NOX) proteins (Boon et al., 2006; Iyer et al., 2003; Karow et al., 2004). H-NOX domains are also present in many bacteria including a number of pathogens. Bacteria encounter NO as an intermediate of denitrification or as a product of bacterial or host nitric oxide synthases. NO plays a particularly important role in host-pathogen interactions and macrophages produce NO as an antimicrobial agent (MacMicking et al., 1997). A variety of bacterial iron and heme-containing NO sensors confer protection by regulating expression of enzymes that convert NO to less toxic species (Poole, 2005; Spiro, 2007). The majority of these NO sensors are direct transcriptional regulators. In contrast, bacterial NO-selective H-NOX proteins are often found in the same operon as signaling proteins such as histidine kinases or diguanylate cyclases, suggesting a role as sensors in prokaryotic NO signaling pathways.
The biological function of H-NOX proteins has been studied in few organisms. In Legionella pneumophila and Shewanella woodyi, the H-NOX protein was shown to be involved in biofilm repression by directly influencing the activity of a diguanylate cyclase/phosphodiesterase protein, which controls the level of the bacterial secondary messenger cyclic diguanosine monophosphate (c-di-GMP) (Carlson et al., 2010; Liu et al., 2012). In Vibrio fischeri, the H-NOX protein plays a role in symbiosis with the squid Euprymna scolopes by regulating the expression of iron uptake genes during NO exposure in the course of colonization (Wang et al., 2010). In the majority of bacteria with H-NOX proteins, including V. fischeri, the hnoX gene is located adjacent to that of a histidine kinase (HK). Direct interaction between the H-NOX protein and the HK has been demonstrated in Shewanella oneidensis MR-1, a metabolically versatile bacterium with applications in bioremediation. HK autophosphorylation is controlled by the ligation state of the H-NOX protein and NO binding suppresses HK activity (Price et al., 2007). The present study addresses signal transduction from the HK to the physiological output of the signaling pathway.
HKs are part of two-component signal transduction systems, the predominant means by which bacteria sense and adapt to diverse stimuli (Stock et al., 2000). In the simplest form, two-component signaling involves a sensor histidine kinase (HK) and a cognate response regulator (RR). Stimulus recognition by the HK sensory domain controls autophosphorylation of a conserved His residue. The H-NOX-associated HKs are unusual in that the H-NOX sensory protein is separately expressed and not fused to the HK. The second step in two-component signaling involves phosphoryl transfer from the His residue on the HK to a conserved Asp residue on the RR receiver domain. Receiver domain phosphorylation controls the activity of an attached effector domain, which governs the output of the signaling pathway (Galperin, 2010). The effector domains are often DNA binding domains controlling a transcriptional response but can also be enzymes (e.g. diguanylate cyclases or c-di-GMP phosphodiesterases). Specificity in the pathway is achieved by tuning the interactions between HK and RR (Skerker et al., 2008). Mapping the connectivity between particular HKs and RRs is challenging due to the large number of two-component signaling systems in most bacterial genomes and the presence of orphan HK and RRs (i.e. HK or RR encoded without a nearby partner gene) (Laub and Goulian, 2007). The majority of H-NOX-associated HKs, including the one in S. oneidensis, are orphans. Thus, the cognate RR(s) have to be identified to determine the output and the biological role of the H-NOX signaling pathway.
In this study, a complex signaling network was mapped in Shewanella oneidensis, which involves integrated and branched phosphotransfer from the H-NOX-associated HK, as well as an additional HK to three common RR targets. Phosphodiesterase assays demonstrated how two RR jointly regulated the intracellular c-di-GMP pool. Biofilm assays in S. oneidensis provided a direct link between NO sensing and the control of cellular attachment. Conservation of the pathway in other gammaproteobacteria, such as Vibrio cholerae, suggests a broad role for bacterial H-NOX proteins in controlling c-di-GMP levels and biofilm formation as a behavioral response to NO.
RESULTS
Identification of three cognate response regulators to the H-NOX-associated histidine kinase
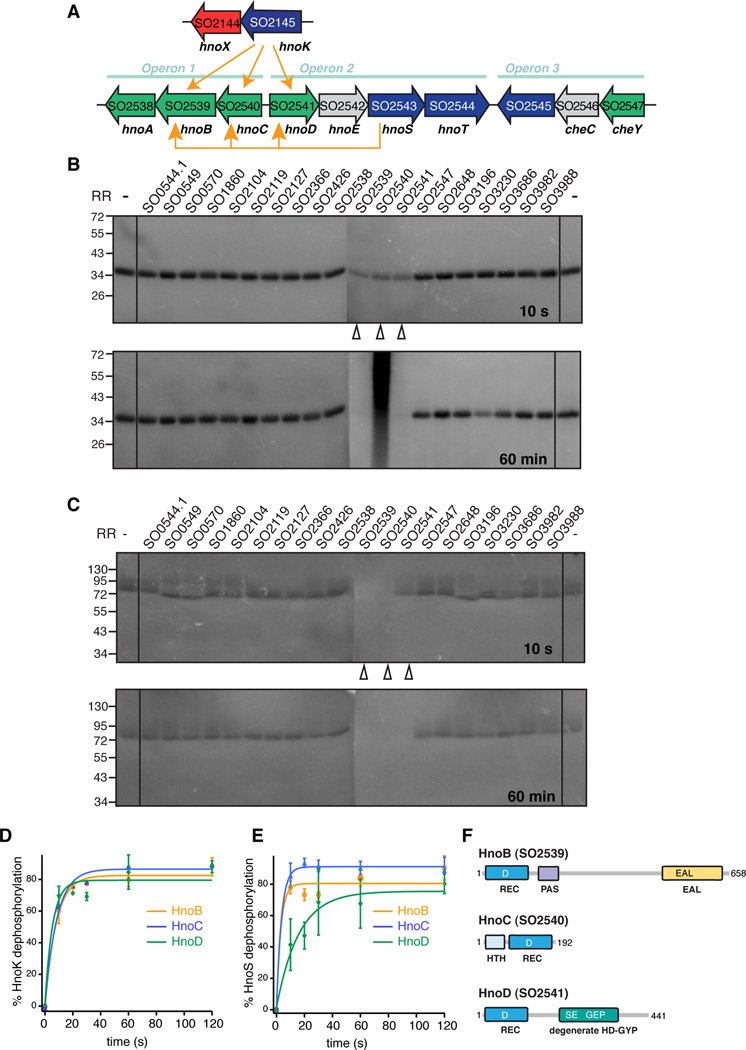
To map the two-component signaling pathway and identify the cognate RR(s) to the H-NOX-associated HK in Shewanella oneidensis, we used phosphotransfer profiling. This method probes the in vitro phosphotransfer between a purified HK and a panel of RR candidates and relies on the observation that in vivo cognate interactions display fast phosphotransfer kinetics in vitro (Laub and Goulian, 2007; Skerker et al., 2005). The H-NOX-associated HK (SO2145, HnoK) was phosphorylated with [γ-32P]-ATP prior to RR addition. Rapid loss of phosphorylation from the HK was observed within the first 10 sec for three RRs: SO2539, SO2540 and SO2541 (Figure 1B). This loss of signal occurred within a kinetic timeframe typical for cognate interactions and is indicative of specific phosphotransfer. Formation of phosphoaspartate esters on SO2539 and SO2541 was not observed, due to either instability of the esters or dephosphorylation by HnoK. In contrast, phosphorylated SO2540 was observed as an intense, unresolved signal after 60 min, possibly due to protein aggregation. Because of involvement in H-NOX signaling, the H-NOX-associated HK gene was named H-NOX/NO-regulated histidine kinase (hnoK) and the genes in the RR operons (Figure 1A) were named hnoA-E.
Figure 1. Identification of cognate response regulators to the H-NOX-associated histidine kinase in Shewanella oneidensis.
(A) The H-NOX gene (hnoX, SO2144, red) and the H-NOX-associated HK gene (hnoK, SO2145, purple) form an isolated operon in Shewanella oneidensis. The three cognate RR genes of the H-NOX-associated HK hnoB (SO2539), hnoC (SO2540) and hnoD (SO2541) (green) are located in three operons that are rich in two-component signaling proteins. The adjacent HK hnoS (SO2543, purple) had the same three response regulators targets (yellow arrows). (B) Phosphotransfer profiling of the H-NOX-associated HK (HnoK, SO2145) to 20 orphan RRs identified three cognate phosphorylation targets (SO2539, SO2540, SO2541). The HK was pre-phosphorylated with [γ-32P]-ATP and subsequently incubated with an equimolar amount of the respective RR for either 10 s or 60 min. Phosphorylation was detected by visualizing 32P radioactivity after SDS-PAGE (C) HnoS (SO2543) displayed the same phosphotransfer specificity for SO2539, SO2540, SO2541. (D–E) Comparison of phophotransfer kinetics of the two HKs. All values represent the mean (n = 2 or 3) +/− SEM (F) Domain architecture of cognate response regulators, displaying the receiver domain with the conserved aspartic acid (REC, dark blue) and the varying effector domains. HnoB contains a PAS domain and an EAL PDE domain (yellow). HnoC exhibits a helix-turn-helix (HTH) DNA binding domain (light blue) and HnoD contains a degenerate HD-GYP domain (green). See also Figure S1.
The vast majority of characterized two-component signaling systems display specific one-to-one connectivity between a HK and a single cognate RR. Phosphotransfer to multiple RRs in vitro has often been attributed to non-specific crosstalk (Laub and Goulian, 2007). To distinguish between a branched pathway and crosstalk, the phosphotransfer kinetics between HnoK and the three RRs (HnoB, HnoC and HnoD) were examined in detail (Figure 1D). Transfer to all three RRs occurred with similar rates and was essentially complete after 10 seconds, suggesting that all three represent biologically relevant cognate interactions. Consistent with the results of the phosphotransfer profiling, only HnoC phosphorylation was detected (Figure S1C).
The three RRs genes are located in a region that is particularly enriched in two-component signaling genes in three adjacent operons (Figure 1A). In addition to the RRs, three HK genes (hnoT, hnoS and SO2545) are present. HnoS and SO2545 are more similar to the H-NOX-associated HK than any other HKs in the genome. The co-localization and similarity prompted us to investigate whether these HKs could be involved in the signaling pathway. To identify the cognate RRs for each HK, phosphotransfer profiling was repeated with the panel of orphan RRs. HnoS exhibited transfer to the same three RRs as HnoK (HnoB-D) (Figure 1C). In contrast, HnoT and SO2545 led to the phosphorylation of a CheY (SO2547) (Figure S1A and S1B). HnoS demonstrated a slightly faster transfer to HnoB and HnoC over HnoD (Figure 1E). Again, HnoC was the only RR with phosphoaspartate modification that was detected (Figure S1D).
HnoT contains predicted transmembrane regions and a sensor domain, while cytoplasmic HnoS and SO2545, along with HnoK, lack these regions. To investigate whether the H-NOX protein also serves as a sensor domain for either HnoS or SO2545, like it does for HnoK (Price et al., 2007), pull-down assays were carried out between the H-NOX protein and MBP-tagged HKs (Figure S1E). Consistent with previous reports, the H-NOX protein interacts with HnoK. In contrast, HnoS and SO2545 did not pull down H-NOX above background, suggesting that distinct sensor domains control activity of these HKs or that the H-NOX interaction is weak. One candidate sensor domain is the adjacent HnoE. This protein shares similarity with FIST proteins, which are predicted to bind and sense small molecules such as amino acids (Borziak and Zhulin, 2007) but have not been characterized.
RR signaling output is governed by effector domains within the RRs (Galperin, 2010). HnoB contains a PAS domain and an EAL domain, predicted to hydrolyze the bacterial secondary messenger c-di-GMP (Figure 1F). HnoC contains a MerR superfamily helix-turn-helix DNA binding domain at the N-terminus. HnoD includes an HD-GYP domain, which is also a predicted phosphodiesterase (PDE) for c-di-GMP, but the HD-GYP consensus residues crucial for activity are altered to SE-GEP in HnoD.
Phosphotransfer to response regulator orthologs in other gammaproteobacteria
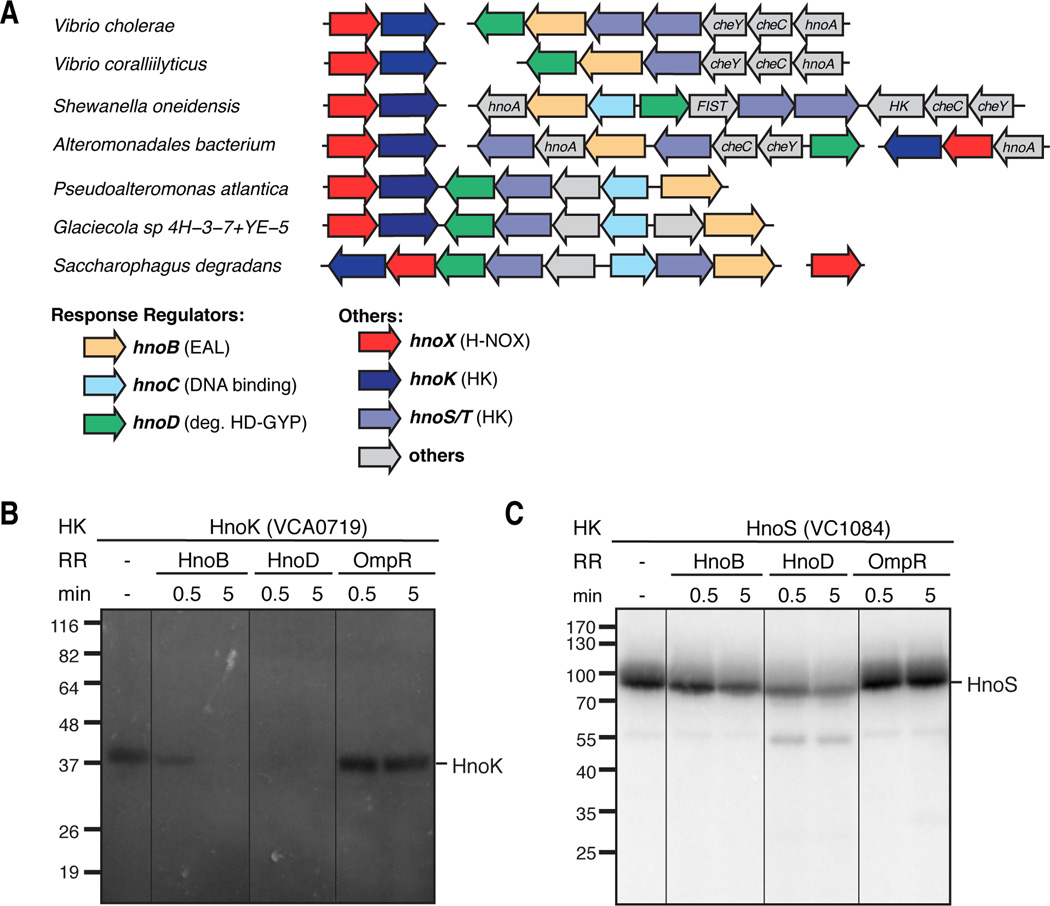
The lack of RR genes in the majority of operons with hnoXK pairs prompted the question if other bacteria contained orthologs of the three S. oneidensis cognate RRs. Discerning the ortholog of a particular RR is challenging because a single organism often contains many RRs with identical architecture and effector domains (Galperin, 2010). In particular, DNA binding domains, GGDEF diguanylate cyclase (DGC) domains, as well as EAL and HD-GYP PDE domains are very prevalent. To circumvent this multiplicity problem, we narrowed our search by taking advantage of unique sequence features in HnoD. Proteins with high sequence similarity to HnoD were discovered in 16 other gammaproteobacteria (Figure S2A). Surprisingly, genes with high similarity to hnoB and hnoC were often co-localized in operons near hnoD (Figure 2A, and Figure S2B). Indeed, four species (Saccharophagus degradans, Pseudoalteromonas atlantica, Glaciecola sp., and Neptuniibacter caesariensis) exhibited operons with the RR genes directly adjacent to the hnoXK operon. This organization and co-localization with the hnoXK genes supports a conserved functional relationship between the H-NOX and signaling proteins in other gammaproteobacteria.
Figure 2. Phosphotransfer to HnoB and HnoD orthologs.
(A) Organization of the hnoX genes (red) and associated hnoK HK genes (purple) as well as operons containing orthologs of the hnoB (yellow), hnoC (light blue) and hnoD (green) in select gammaproteobacteria. For a complete list of organisms see supplemental Figure S2B. (B) Phosphotransfer assay of HnoK from Vibrio cholerae (VCA0719) to HnoB (VC1086) and HnoD (VC1087). Assays were carried out as in Figure 1. (C) Phosphotransfer assay of Vibrio cholerae HnoS (VC1084) to HnoB and HnoD.
To determine whether the RRs are specific cognates of HnoK in other organisms, we investigated the phosphotransfer in Vibrio cholerae. This pathogen would need to avoid NO produced by the host in response to infection (Davies et al., 2011; MacMicking et al., 1997). V. cholerae harbors the hnoXK genes (VCA0720/VCA0719) in an isolated operon with hnoB (VC1086) and hnoD (VC1087) grouped into a separate operon containing two additional HKs. V. cholerae does not contain an hnoC ortholog. Indeed, HnoK rapidly transferred its phosphoryl group to HnoB and HnoD (Figure 2B) consistent with a cognate interaction. HnoS, one of the adjacent HKs, also displayed rapid phosphotransfer to the two RRs (Figure 2C). This suggests that signaling integration at the level of the RRs from two sensory inputs is a conserved feature between organisms.
The effect of phosphorylation on the c-di-GMP phosphodiesterase activity of HnoB
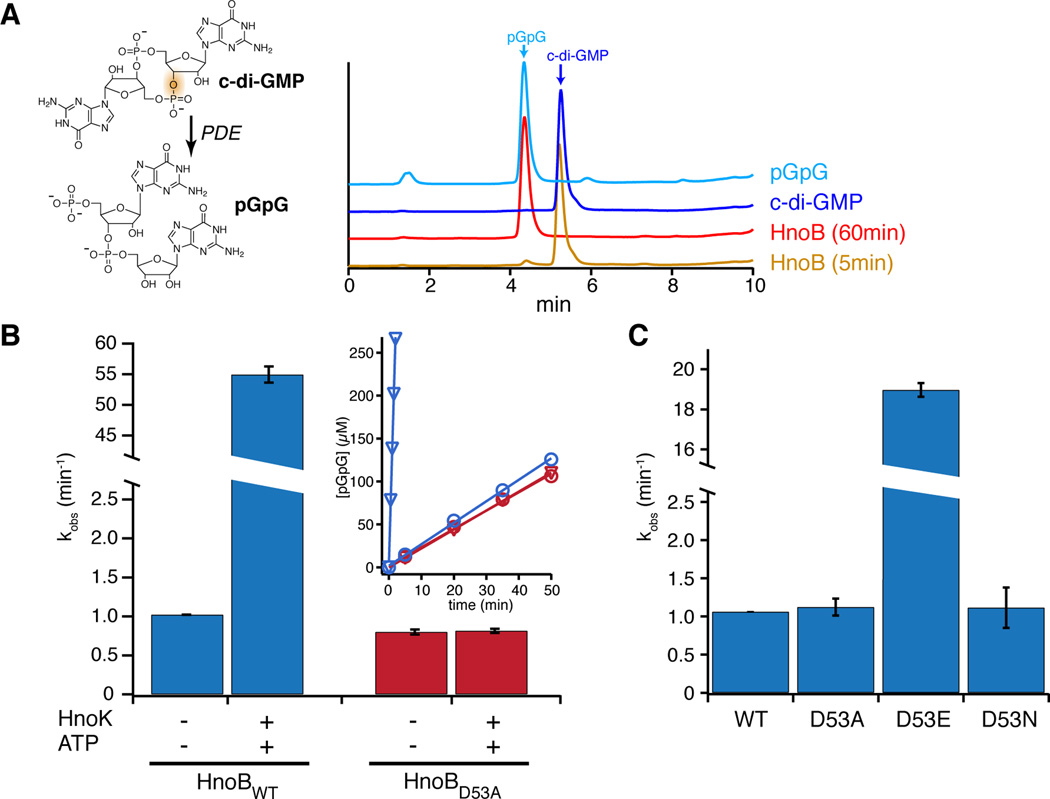
HnoB contains a C-terminal EAL domain (Figure 1F), which is predicted to hydrolyze c-di-GMP to 5'-phosphoguanylyl-(3'→5')-guanosine (pGpG) (Christen et al., 2005; Tamayo et al., 2005). Sequence alignments confirmed that the conserved residues essential for PDE activity were present in HnoB (Schirmer and Jenal, 2009). When HnoB was incubated with c-di-GMP, formation of pGpG was observed within the first 5 min and complete turnover occurred after one hour (Figure 3A), confirming that HnoB was a functional PDE for c-di-GMP.
Figure 3. C-di-GMP phosphodiesterase activity of HnoB.
(A) The structure of c-di-GMP and the hydrolysis product pGpG are shown and the bond that is broken is highlighted. HPLC chromatograms (λ = 253 nm) of a c-di-GMP standard (dark blue) and pGpG (light blue) are shown. Incubation of purified HnoB with c-di-GMP in the presence of Mg2+ led to partial hydrolysis to pGpG after 5 min (yellow) and complete hydrolysis after 60 min (red). (B) PDE activity of HnoB in the presence or absence of HK (HnoK) and ATP. Purified HnoB (2.5 µM) was incubated with 20x excess HnoK, 0.5 mM ATP and 10 mM MgCl2. Reactions were initiated with 0.5 mM c-di-GMP, time points were drawn, acid-quenched and then analyzed by HPLC. The insert shows the kinetics of the PDE assay monitored by following pGpG formation. Phosphorylation of WT HnoB by HnoK led to 50-fold enzyme activation (blue, triangles in the insert correspond to HnoK/ATP conditions). Addition of the HK to the phosphorylation-deficient D53A HnoB mutant had no effect on activity (red). (C) The phosphorylation mimic D53E mutant showed 19-fold activation of PDE activity while the phosphorylation-deficient D53A and D53N mutants displayed no change in activity compared to WT HnoB. All values represent the mean (n = 2) +/− SEM. See also Figure S3.
Next, we investigated whether RR phosphorylation regulates PDE activity. To date, regulation of EAL domain activity by the phosphosphorylation state of the receiver domain has yet to be observed. To assess the effect of HnoB phosphorylation on PDE activity, assays were carried out in the absence or presence of the H-NOX-associated HK (HnoK) and ATP. HK and ATP caused a 50-fold stimulation of HnoB PDE activity (Figure 3B). In the phosphoreceiver D53A mutant, addition of HK and ATP had no effect on activity, confirming that the stimulation was specific to phosphorylation.
To further verify stimulation of PDE activity, two different receiver domain phosphorylation mimics were tested. Mutation of the phosphoreceiver aspartate to glutamate has been shown, in certain cases, to structurally and functionally mimic the phosphorylated RR state (Davies et al., 2011; Lauriano et al., 2004). The HnoB D53E mutation resulted in a 19-fold increase in the rate of pGpG formation while the rates of the D53A and D53N mutants remained unchanged compared to WT (Figure 3C). PDE activity was also measured in the presence of the RR phosphorylation mimic beryllium fluoride (BeFx), which can serve as a structural analog for a phosphoryl group (Lee et al., 2001; Yan et al., 1999). Titration of increasing BeFx concentrations resulted in a 7-fold stimulation of HnoB PDE activity (Figure S3). Taken together, the activity of the phosphorylation mimics confirmed that the HnoB EAL domain is activated through phosphorylation of the HnoB receiver domain. Thus, HnoK and HnoS decrease c-di-GMP levels by phosphotransfer-mediated activation of the HnoB EAL domain.
The degenerate HD-GYP domain of HnoD lacks phosphodiesterase activity
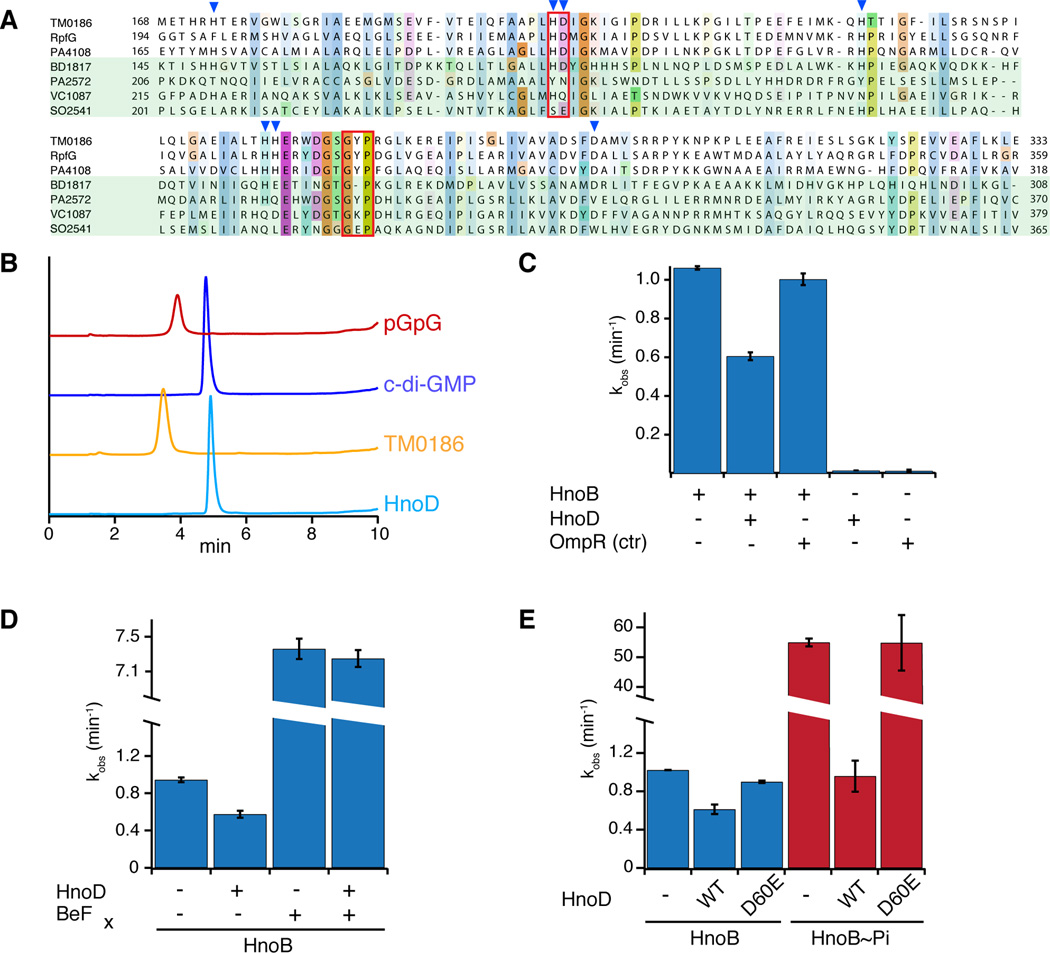
The second cognate RR (HnoD) of the H-NOX-associated HK contains a HD-GYP domain, which represents another family of c-di-GMP PDEs (Galperin et al., 1999). However, sequence alignment to other HD-GYP domains showed that the consensus residues vital for PDE activity were not conserved in HnoD (Figure 4A, Figure S2A). Therefore, we first tested if the degenerate HD-GYP domain of HnoD could hydrolyze c-di-GMP. As a positive control, TM0186, a HD-GYP RR from Thermotoga maritima containing the consensus HD-GYP motifs (Figure 4A) was expressed and purified. TM0186 completely hydrolyzed c-di-GMP, whereas HnoD displayed no detectable pGpG formation, even after 48 hours (Figure 4B).
Figure 4. Fine-tuning of HnoB c-di-GMP hydrolysis by the degenerate HD-GYP domain of HnoD.
(A) Sequence alignment of HD-GYP domains from three RRs that contain the conserved sequence motif (T. maritima TM0186, X. campestris RpfG, P. aeruginosa PA4108) and four RR that contain some degeneracy of the conserved sequence (Bdellovibrio BD1817, P. aeruginosa PA2572, V. cholerae HnoD (VC1087), S. oneidensis HnoD (SO2541)). The HD and GYP motifs are boxed in red and residues that are ligands for binuclear iron are indicated by blue triangles. (B) HPLC chromatograms of a PDE assay with a “true” HD-GYP RR (TM0186, yellow) and the degenerate HD-GYP RR (HnoD, light blue) showed that only TM0186 hydrolyzed c-di-GMP to pGpG. Standards of pGpG (red) and cyclic-di-GMP (blue) are shown at the top. (C) HnoD inhibited the PDE activity of the HnoB. Purified HnoB (2.5µM) was pre-incubated with 20x excess HnoD prior to addition of c-di-GMP (0.5mM). (D–E) Phosphorylation of HnoD abolished the inhibition of HnoB PDE activity. (D) Loss of inhibition in the presence of the phosphorylation mimic beryllium fluoride (BeFx). HnoD and HnoB were preincubated with BeFx before the reactions were initiated with c-di-GMP. (E) Inhibition of HnoB by HnoD phosphoacceptor mutants. HnoB was either unphosphorylated (blue) or was pre-phosphorylated with HK (HnoK) and ATP (red). WT HnoD or the phosphorylation mimic D60E mutant was then added prior to initiation of the PDE reaction. All values represent the mean (n = 2) +/− SEM. See also Figure S4.
HD-GYP domains belong to a larger superfamily of HD metal-dependent phosphohydrolases (Galperin et al., 1999). The HD motif forms part of the metal coordination site, along with a number of conserved histidines and acidic residues (blue triangles in Figure 4A) (Lovering et al., 2011). Purified TM0186 contained iron, but no metals were detected bound to HnoD by by inductively coupled plasma atomic emission spectroscopy (Figure S4D) or UV/vis spectroscopy (Figure S4E). Attempts to reconstitute metal binding were unsuccessful (data not shown). The inability to coordinate metal is likely one reason for the lack of HnoD PDE activity.
Degenerate EAL or GGDEF domains often retain the ability to bind nucleotides, and thereby serve a regulatory role as a receptor for c-di-GMP or GTP (Christen et al., 2005; Navarro et al., 2009). No binding of either molecule was observed by equilibrium dialysis or isothermal titration calorimetry (data not shown), suggesting that the degenerate HD-GYP domain of HnoD does not act as a nucleotide receptor.
The effect of HnoD on HnoB EAL phosphodiesterase activity
Degenerate GGDEF and EAL domains can carry out alternate regulatory roles through macromolecular interactions, especially in cases when degeneracy eliminates ligand binding (Hengge, 2009). Because of the co-localization of the hnoD and the EAL domain-containing hnoB genes in a number of species (Figure 2A, Figure S2B), we speculated that a functional relationship between these two proteins could exist. Therefore, PDE assays of HnoB were carried out in the presence of excess HnoD. Addition of HnoD decreased the rate of HnoB catalyzed c-di-GMP hydrolysis by 40% (Figure 4C). In contrast, addition of a control RR (E. coli OmpR) had no effect on HnoB activity. HnoD demonstrated detectable inhibition of HnoB activity at 10 µM HnoD (4-fold excess), and HnoB activity was further inhibited as HnoD concentration was increased (Figure S4A). Pull-down assays between MBP-tagged HnoB and HnoD suggested that the interaction between the two RRs was relatively weak (Figure S4B).
The results above demonstrated that phosphorylation of HnoB by HnoK stimulated HnoB PDE activity while the non-phosphorylated HnoD inhibited HnoB. HnoD is also a phosphotransfer target of the two HKs, creating a possibility for a feed-forward loop to control PDE activity. Therefore, we examined the effect of HnoD phosphorylation on the inhibition of HnoB activity. BeFx was employed to allow independent, simultaneous mimicking of the phosphophorylated state for both response regulators (Figure 4D). BeFx caused an activation of HnoB PDE activity as seen before, but in the presence of HnoD and BeFx, the reaction was no longer inhibited. This suggested that when phosphophorylated, HnoD was no longer inhibitory. To further validate this mechanism, conditions were devised under which HnoB and HnoD could be placed in the phosphorylated or phoshophorylation-mimicked state independently (Figure 4E). Phosphorylation of HnoB was controlled by addition of HnoK and ATP whereas phosphorylation of HnoD was mimicked by a D60E mutation in the phosphoreceiver domain. In the absence of HnoB phosphorylation (blue bars), the phosphorylation-mimic HnoD variant (D60E) was no longer inhibitory. When HnoB was phosphorylated by HnoK and ATP and the PDE was stimulated as expected (red bars), the D60E mutant had no inhibitory effect on this activated form of HnoB. PDE activity of HnoB was decreased in the presence of WT HnoD and HnoK/ATP because the large excess of HnoD out-competed HnoB as a substrate of the HK, leaving HnoB non-phosphorylated and unactivated. Overall, these results indicate that only the unphosphorylated state of HnoD is the inhibitory, suggesting that the inhibition is abolished by a conformational change upon phosphorylation. This was further confirmed by analyzing the inhibition of HnoD truncations (Figure S4C). Inhibition of HnoB activity only occurred in the presence of the HnoD receiver domain while removal of the receiver domain relieved inhibition.
Nitric oxide induces biofilm formation
C-di-GMP controls the motility behavior of bacteria and increased levels of c-di-GMP signal a transition from planktonic growth to a sessile state with biofilm formation (Hengge, 2009; Jenal and Malone, 2006). We therefore hypothesized that NO may serve as a signal for biofilm formation, as the NO-bound H-NOX inhibits autophosphorylation of HnoK, leading to inactivation of HnoB and an increase in c-di-GMP levels (Figure 5E). Therefore, the effect of NO on Shewanella oneidensis biofilm formation under static anaerobic growth was investigated. NO was introduced to the medium by adding DETA NONOate. This slow-release NO donor (t1/2 of 56 hours at 25 °C) led to a relatively steady NO concentration ranging between 0.4 and 2.7 µM (Figure S5A). Fluorescent staining of the biofilms confirmed that these were comprised of viable cells under all conditions tested (Figure S5E). In the WT strain, NO caused a significant increase in the amount of biofilm (p = 0.020 in 96-well plates in Figure 5A at and p = 0.039 in 12-well plates in Figure 5B) consistent with the signaling hypothesis outlined above.
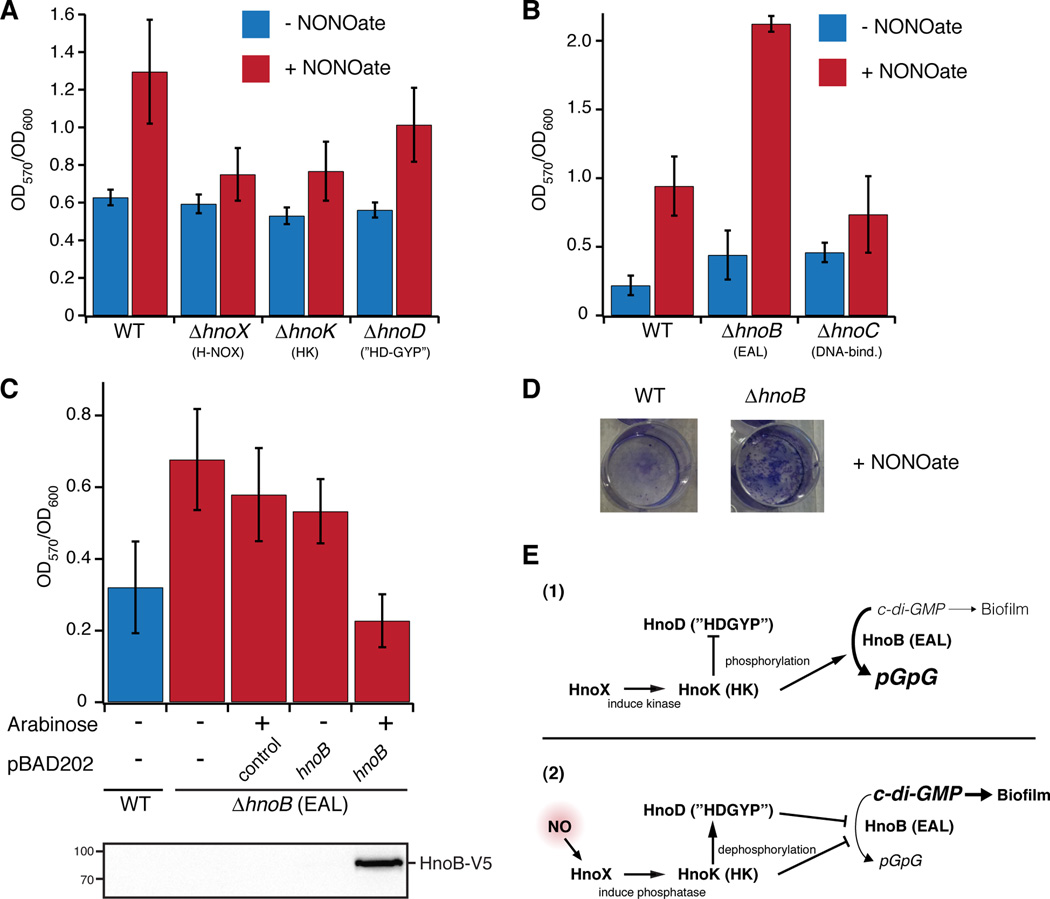
Figure 5. Phenotypic analysis of two-component signaling knockouts affecting NO induced biofilm formation.
(A–C) Static biofilm measurements of Shewanella oneidensis under anaerobic growth. NO stimulated the formation of biofilms, while cell attachment in hnoX and hnoK knockouts was insensitive to NO. Knockout of the EAL RR (hnoB) led to a hyperbiofilm phenotype. WT S. oneidensis or the respective knockouts were grown anaerobically in MM medium in 96-well plates (A) or 12-well plates (B) with fumarate as electron acceptor. DETA NONOate (200 µM), a slow release NO donor, was added for NO production. Biofilm levels were quantified by measuring the absorbance at 570 nm (OD570nm) after staining with crystal violet and normalized to cell growth (OD600nm). (C) Complementation of the hyperbiofilm phenotype of the hnoB knockout. WT S. oneidensis and the hnoB knockout were transformed with pBAD202 containing the hnoB gene or empty vector as a control. The strains were grown as described above and expression of HnoB was induced with 0.02% arabinose. Western-blot analysis (below) confirmed expression of HnoB. All values represent the mean from independent experiments (n = 12 (A), n = 3 (B–C)) +/− SEM. (D) Images of crystal violet stained biofilms display the enhanced cell attachment of the hnoB knockouts compared to WT S. oneidensis. (E) Model for signaling interactions between components of the network in the absence (1) or presence (2) of NO. See also Figure S5.
Next, we assessed the contributions of the individual two-component signaling proteins to biofilm formation. Reverse transcription PCR confirmed that the hnoX, hnoK and hnoB-D genes were transcribed in the presence and absence of NO (Figure S5C). In-frame deletion strains of hnoX, hnoK, hnoB, hnoC, and hnoD were constructed and assayed for static biofilm formation. Deletion of hnoX and hnoK resulted in biofilm formation that was comparable to WT levels in the absence of NO and had a significantly decreased response to NO (Figure 5A, Figure S5D for images). This confirmed that the H-NOX had a central role in the signaling pathway as an activator cellular attachment in response to NO (Figure 5E). HnoK also appears to stimulate NO-induced biofilm formation, based on the loss-of-function phenotype. The observed activator function of HnoK can be rationalized by invoking a dual kinase and phosphatase activity towards HnoB and HnoD (Figure 5E). Thus far, only inhibition of HnoK autophosphorylation by NO-bound H-NOX has been observed (Price et al., 2007), but HnoK, in the presence of NO-bound H-NOX, was indeed capable of dephosphorylating its RR targets (unpublished data). This NO-induced shift of HnoK kinase towards phosphatase activity and dephosphorylation of HnoB and HnoD would lead to a decrease in HnoB PDE activity.
Deletion of the DNA-binding RR (ΔhnoC) resulted in a very moderate increase in biofilm amounts both in the absence and presence of NO (Figure 5B), suggesting HnoC may control some gene targets involved in biofilm formation but likely has other cellular roles. The knockout of hnoD displayed stimulation of biofilm formation in response to NO (Figure 5A, p = 0.027), which is consistent with a more subtle role in fine-tuning c-di-GMP levels. In contrast, deletion of the PDE gene hnoB led to hyperproduction of biofilm (Figure 5B–D). Compared to WT, biofilm formation was slightly increased in the absence of NO and was significantly elevated when grown under NO (p = 0.0001 compared to WT+ NONOate). This increase in cellular attachment validates HnoB as a major contributor to NO-induced biofilm formation. At the same time, an additional regulator must suppress the overproduction of biofilms in the absence of NO. For instance, HnoD could control other DGC or PDE proteins besides HnoB in the organism. The hnoB deletion also conferred a growth advantage in the presence of NO, and the lag phase was reduced from 18 to 12 hours. (Figure S5B). To further confirm that the hyperbiofilm phenotype of the ΔhnoB strain was due to loss of EAL activity, we complemented the knockout with WT hnoB from an arabinose inducible plasmid (pBAD202). Biofilm formation was only reset to WT levels upon expression of HnoB.
DISCUSSION
H-NOX signaling proceeds through a conserved multi-component signaling system
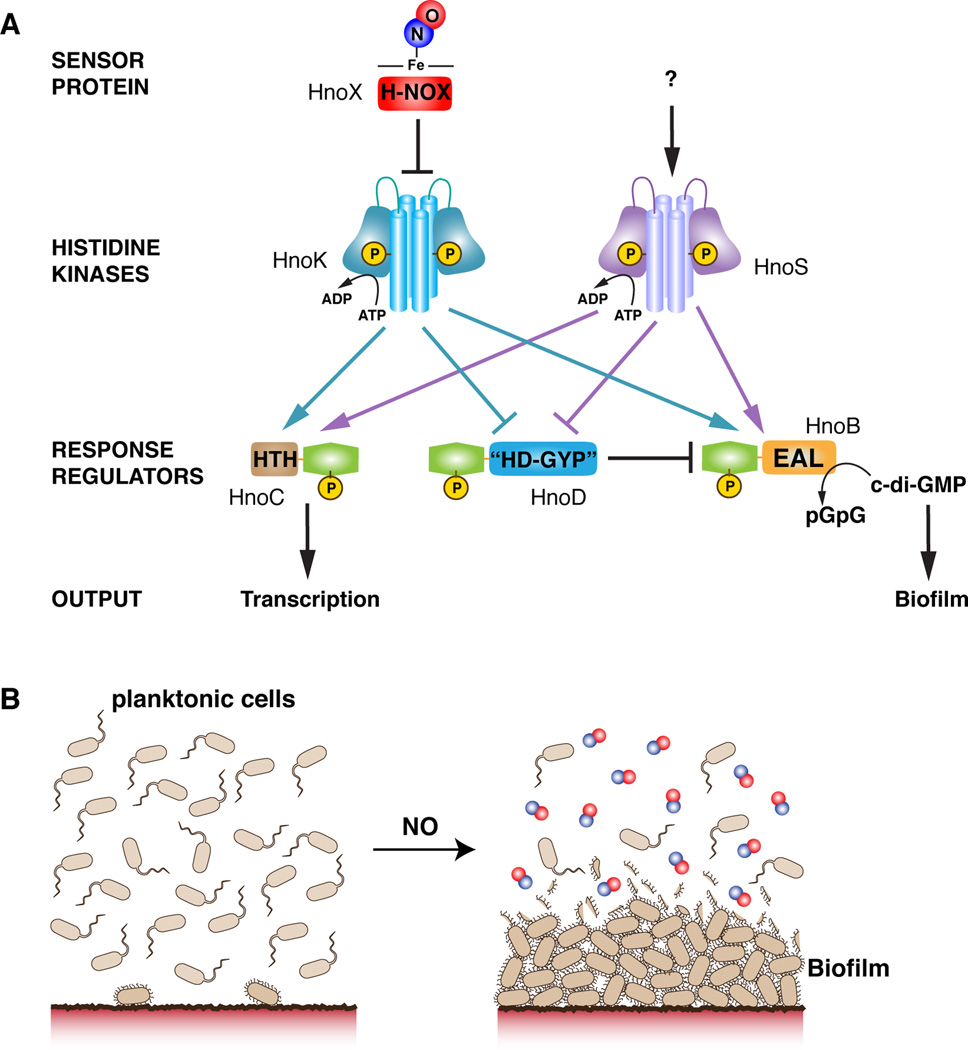
The results from phosphotransfer profiling showed the connectivity between proteins of the bacterial H-NOX signaling pathway and unexpectedly identified three RR targets of the H-NOX-associated HK HnoK (Figure 6A): HnoB containing an EAL effector domain, HnoC, containing a DNA-binding domain, and HnoD, containing a degenerate HD-GYP domain. The RRs targets of the HnoK clustered in a genome region enriched in two-component signaling proteins, containing three additional HKs, two other RRs, a CheC phosphatase and a putative FIST sensor protein. Operons containing similar proteins have been identified in a number of H-NOX-containing gammaproteobacteria, and this conserved gene organization suggests that the proteins are functionally related members of the same signaling network (Wolf et al., 2001). Based on phosphotransfer to three common RRs, HnoS and HnoK are clearly part of the same signaling network and the RRs act as a signal integration point in the network (Figure 6A). The independently expressed H-NOX protein serves as the sensor for HnoK, whereas the lack of interaction between H-NOX protein and HnoS implies that this HK must respond to an alternate stimulus.
Figure 6. Model of multi-component signaling network for NO-induced biofilm formation as a protection mechanism against NO.
(A) The complex multi-component signaling pathway is initiated by NO binding to the sensory H-NOX protein (HnoX), which then inhibits HnoK autophosphorylation. Phosphotransfer establishes a branching of the network to three response regulators. HnoB and HnoD form a feed-forward loop. Phosphorylation controls PDE activity of HnoB, which can be fine-tuned by allosteric control from HnoD. NO-controlled repression of the PDE activity leads to an increase in c-di-GMP levels, which serves as a signaling cue for cellular attachment into biofilms. (B) The NO signal switches the bacterial motility pattern from planktonic growth to increased attachment onto surfaces. The thick layers of cells provide a protective barrier against diffusion of reactive and damaging NO and may protect cells in the lower layers.
Evidence for conservation of the integrated H-NOX signaling network involving two HKs and three RRs was obtained by confirming phosphotransfer in Vibrio cholerae. V.c. HnoK and HnoS phosphorylated both V.c. HnoB and HnoD. Mutation of V.c. HnoB (VC1086) has been shown to result in an increase in biofilm formation (Waters et al., 2008). This mirrors the phenotype of the HnoB knockout in S. oneidensis and provides further evidence towards functionally conserved pathways. Interestingly, no homologs to HnoC could be identified in V. cholerae and in several of the organisms containing HnoB and HnoD (Figure S2B), suggesting that some bacteria do not depend on the HnoC-mediated transcriptional output.
A regulatory role for degenerate HD-GYP proteins in controlling EAL activity
The primary signaling response mediated by the H-NOX signaling network is to control c-di-GMP levels. The effectors of this response are the PDE RRs HnoB and HnoD, containing EAL and HD-GYP domains, respectively. Regulation of PDE activity has only been demonstrated for EAL domains that are directly fused or interact with sensor domains (Barends et al., 2009; Christen et al., 2005; Liu et al., 2012). Phosphorylation is a hallmark of RR control (Gao and Stock, 2010), and here we demonstrated that the PDE activity of the HnoB EAL RR was stimulated by phosphorylation.
HnoD, the second cognate RR linked to c-di-GMP metabolism, is identified as a HD-GYP domain, but active site residues responsible for coordination of crucial catalytic metals are missing in HnoD (Lovering et al., 2011) rendering the protein inactive for c-di-GMP hydrolysis. Compared to EAL domains, HD-GYP domains remain poorly characterized with only few reports of direct PDE activity (Hammer and Bassler, 2009; Ryan et al., 2006). Interestingly, despite the lack of PDE activity, an HnoD ortholog from P. aeruginosa is known to modulate swarming motility, a function typically controlled by c-di-GMP levels (Ryan et al., 2009). This prompted us to investigate whether HnoD regulates c-di-GMP levels indirectly by controlling HnoB activity. HnoD significantly inhibited the PDE activity of HnoB and this inhibition was dependent on the phosphorylation state of HnoD. Phosphorylated HnoD was no longer inhibitory, and this loss of inhibition was independent of HnoB phosphorylation. HnoD and HnoB thus form a coherent feed-forward loop to control c-di-GMP levels (Mangan and Alon, 2003). Unphosphorylated HnoB exhibited low activity that was further suppressed by HnoD. Phosphotransfer from HnoK or HnoS activated the PDE activity of HnoB while simultaneously preventing the inhibition by HnoD. This example of cross-regulation between two RRs to form a feed-forward loop in the same signaling network is in contrast to typical cross-regulation, which connects distinct signaling pathways, often by use of separate connector proteins (Mitrophanov and Groisman, 2008).
This feed-forward loop in the H-NOX signaling network may allow for an extra level of tenability over the intracellular pool of c-di-GMP. Precise control is required because the affinities of c-di-GMP receptors span several orders of magnitude (Hengge, 2009), thus the cellular processes controlled by c-di-GMP are likely sensitive to a wide concentration range. The dual control provides the opportunity for fine-tuning the signal integration from HnoK and HnoS. The in vivo phosphorylation levels of the two RRs will be influenced by the dueling kinase and phosphatase activities from each HK. Gradual attenuation of PDE activity of HnoB could be achieved by biasing phosphotransfer activity from the kinases slightly towards either RR. Lastly, HnoD may regulate the activity of additional unknown downstream signaling effectors, and indeed serve as a global regulator for other EAL domain proteins, and possibly, GGDEF diguanylate cyclases. It has been shown that the functional HD-GYP RR RpfG from X. campestris interacts physically and functionally with multiple GGDEF proteins (Ryan et al., 2010). C-di-GMP modulators are particularly enriched in the genomes of gammaproteobacteria and S. oneidensis contains 51 proteins with GGDEF domains, 27 with EAL domains and 20 GGDEF-EAL hybrids (Thormann et al., 2006). Thus to effectively modulate the intracellular c-di-GMP concentration, it may be necessary to exert a global effect on the activities of multiple EAL and GGDEF proteins.
Biofilm formation as a defense mechanism against NO
A variety of cellular processes are mediated by receptor proteins and riboswitches specific to c-di-GMP (Schirmer and Jenal, 2009). C-di-GMP controls flagellar motor speed (Boehm et al., 2010), and in Vibrio cholerae and Shewanella oneidensis, it has been shown to upregulate gene clusters for biosynthesis of extracellular polysaccharides, which facilitate the adhesion between cells (Beyhan et al., 2006; Krasteva et al., 2010; Thormann et al., 2006; Thormann et al., 2004). We focused the phenotypic characterization on biofilm formation in S. oneidensis because biofilms represent an important aspect of the life cycle for S. oneidensis, allowing the colonization of metal surfaces for dissimilatory metal reduction. Biofilms are equally important in the life cycle of pathogens, enhancing survival and protection from predators and antimicrobial agents (Donlan and Costerton, 2002). Static biofilm assays demonstrated a clear increase in cellular adhesion with NO treatment, which is consistent with the biochemical interactions in the signaling network (Figure 6A). H-NOX protein and the associated HK, HnoK, were positive regulators of biofilm formation because deletions of the genes rendered these cells insensitive to NO. In contrast, HnoB was a negative regulator of biofilm formation and the hnoB knockout resulted in elevated biofilm formation. Interestingly, in the absence of NO, the large biofilm increase of the ΔhnoB strain was attenuated, suggesting that other regulators were able to suppress biofilm formation. HnoD could act as suppressor and control the activity of additional c-di-GMP modulators, supporting a function for HnoD as a global regulator.
To date, the only links between NO/H-NOX signaling and c-di-GMP dependent changes in biofilm formation have been established in Legionella pneumophila and in Shewanella woodyi. However, in both organisms, the H-NOX controlled the activity of an adjacent diguanylate cyclase/phosphodiesterase protein directly and repressed biofilm formation (Carlson et al., 2010; Liu et al., 2012). Despite a paucity of studies addressing the biological function of bacterial NO-specific H-NOX proteins, a common role for controlling motility via c-di-GMP levels is emerging. Genes encoding GGDEF and EAL proteins are occasionally found adjacent to H-NOX genes and their direct regulation by the H-NOX protein represents a simplified one-component signaling mode with a role in biofilm dispersal in response to NO. Alternatively, other gammaproteobacteria appear to have evolved a more complex signaling network, homologous to the system characterized here. This network structure can meet additional demands for fine-tuned regulation and integration from multiple signal inputs, and it has the opposite role of signaling increased biofilm formation in response to NO.
Encapsulation of bacteria into a biofilm provides a significant protective mechanism against environmental toxins and antimicrobial agents (Donlan and Costerton, 2002). At high concentration, the free radical NO and related reactive nitrogen species (RNS), such as peroxynitrite, are active antimicrobials and are detrimental to cellular growth. Thick biofilm layers and extracellular matrix encapsulation provide a protective diffusional barrier, buffering nitrosative stress at the top layers and protecting the underlying cells (Figure 6B). This mechanical protection could be especially important for S. oneidensis as the organism lacks any genes encoding flavohemoglobins or nitric oxide reductases, which usually remove NO enzymatically. Indeed, the ΔhnoB strain, which produced the highest biofilm levels, displayed a significant growth advantages in the presence of NO. Although exact NO concentrations in the natural soil or water environment of S. oneidensis are not known, the organism is certainly exposed to NO. S. oneidensis itself generates low micromolar NO as a byproduct of denitrification (Price et al., 2007). The organism is also well known for its capacity to utilize a diverse range of metals as electron acceptors. For instance, it can grow on Fe(III) mineral surfaces releasing Fe(II) into solution (Myers and Nealson, 1988). When concomitant with denitrification, reaction of nitrite with Fe(II) will lead to production of NO (Brons et al., 1991). Signaling for increased biofilm formation may serve as protection against the NO byproduct. Alternatively, it could serve as a signaling cue to increase surface adhesion onto the metal mineral for increased respiration efficiency.
H-NOX proteins are also present in pathogens, and an orthologous H-NOX/NO signaling network was discovered in Vibrio cholerae. Pathogens are exposed to high NO concentrations and nitrosative stress during the host innate immune response (MacMicking et al., 1997). V. cholerae colonizes the small intestine causing the severe diarrhea associated with cholera disease. V. cholerae encounters NO formed by acidified nitrite while passing through the stomach and NO generated by macrophages in the intestine (Davies et al., 2011; Duncan et al., 1995). It was recently demonstrated that V. cholerae strains defective in RNS defense exhibited a colonization defect in mouse intestines (Davies et al., 2011). Furthermore, biofilm formation plays an important role during V. cholerae infection and colonization (Zhu and Mekalanos, 2003). For instance, biofilm-like multicellular clumps are more infectious than planktonic cells (Faruque et al., 2006). NO sensing by the H-NOX protein and the subsequent control over biofilm formation may therefore play a virulence role in V. cholerae.
Emergence of complex bacterial signal transduction networks
The complex architecture of the H-NOX signaling network characterized in this study is highly unusual and stands in contrast with the mostly linear two-component signaling pathways. Branched and integrated networks still represent a minority of the characterized pathways. However, there are emerging examples of more complex signaling structures, such as chemotaxis, differentiation in Caulobacter crescentus (Paul et al., 2008; Skerker et al., 2005), quorum sensing in Vibrio harveyi (Ng and Bassler, 2009), and fimbriae production in Pseudomonas aeruginosa (Mikkelsen et al., 2011; Sivaneson et al., 2011). The results reported here describe a further example of a highly complex bacterial signaling network with an intricate relationship between two HKs and three RRs. It involves signal integration, branching of outputs, and an unprecedented allosteric feed-forward loop between RR components, hallmarks of signaling cascades in higher organisms. The complexity exhibited by the NO-sensing pathway reported here suggests that sophisticated prokaryotic signaling mechanisms may be more prevalent than previously imagined.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Genes were PCR amplified from S. oneidensis MR-1 or Vibrio cholerae genomic DNA and cloned into pHMGWA appending His6-MBP at the N-terminus using TOPO and Gateway cloning (Invitrogen). Proteins were expressed in E. coli BL21(DE3)pLysS and affinity-purified on amylose resin (NEB) and Ni-NTA resin (Qiagen). A more detailed purification protocol can be found in the supplemental experimental procedures.
Phosphotransfer profiling and phosphotransfer kinetics
Phosphotransfer profiling was performed as described (Laub et al., 2007). Briefly, purified HK was pre-incubated with 1 mM ATP, 10 µCi [γ-32P]-ATP and 5mM MgCl2 prior to addition of the respective purified RR. Reactions were quenched after 10 sec or 60 min by addition of 6x SDS loading dye, products were separated by SDS-PAGE and the gels were imaged for 32P radioactivity. Phosphotransfer kinetics were measured in the same way and the phosphorylation signal was quantified using ImageQuant. Two independent experiments were averaged. A more detailed protocol can be found in the supplemental experimental procedures.
Phosphodiesterase assays
Phosphodiesterase reactions of purified EAL protein (or HD-GYP protein) were initiated by addition of 0.5 mM c-di-GMP. If necessary, tryptophan or cXMP internal standards were included. Aliquots were quenched at different time points by addition of trifluoroacetic acid (2%) and CaCl2 and analyzed by reverse phase HPLC. Detailed HPLC conditions are described in the supplemental experimental procedures. HnoK was pre-phosphorylated with ATP for 30 min prior to addition of HnoB and incubated for 15 min before initiating reactions with c-di-GMP. Beryllium fluoride was prepared freshly from BeCl2 and NaF was added to the assay 15 min prior to initiation. HnoD was also added 15 min before initiation. All experiments were conducted in duplicates or triplicates.
Static biofilm assays
Biofilm assays were performed in an anaerobic glovebag (Coy Laboratory Products) in 12-well or 96-well polystyrene microtiter plates. S. oneidensis was grown aerobically in LB medium at 30°C overnight. Anaerobic MM mineral medium (Thormann et al., 2006) supplemented with 50 mM lactate, 50 mM fumarate and 0.1% casamino acids was inoculated with 100-fold diluted overnight cultures. DETA NONOate (Cayman Chemical), prepared freshly in 10 mM NaOH, was added to 200 µM if necessary. Cells were grown statically at 29°C and a 3 mL reference culture was grown for OD measurements and normalization. Addition of NO to S. oneidensis caused an 18-hour lag in growth but cultures reached the same optical density after 25 hrs (Figure S5B). Therefore, biofilm formation was quantified after 20 hours during log phase (Figure 5A) and at 44 hours during stationary phase (Figure 5B,D) and normalized to growth. Crystal violet staining after 20 hrs or 44 hrs was performed as described (Lassak et al., 2010). Measurements from individual wells were combined and normalized to growth (OD600nm) and twelve (96-well) or three (12-well) experiments from separate days were averaged.
HIGHLIGHTS.
A complex multi-component bacterial H-NOX signaling network was mapped
EAL response regulator phosphorylation stimulated c-di-GMP hydrolysis activity
A degenerate HD-GYP response regulator inhibited the EAL response regulator
NO induced biofilm formation through c-di-GMP modulation by the signaling network
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Alfred Spormann (Stanford University) for providing strains and materials to generate S. oneidensis knockouts, and we are grateful to Lily Chao and members of the Spormann lab for advice. We thank Hans Carlson and members of the Marletta lab for helpful discussions and critical review of the manuscript. This work was supported in part by NIH grant GM070671 and a Chang-Lin Tien Graduate Fellowship in the Environmental Sciences to L. P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, Marletta MA. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase beta1 H-NOX domain. J. Biol. Chem. 2006;281:21892–21902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- Borziak K, Zhulin IB. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics. 2007;23:2518–2521. doi: 10.1093/bioinformatics/btm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons HJ, Hagen WR, Zehnder AJ. Ferrous iron dependent nitric oxide production in nitrate reducing cultures of Escherichia coli. Arch. Microbiol. 1991;155:341–347. doi: 10.1007/BF00243453. [DOI] [PubMed] [Google Scholar]
- Carlson HK, Vance RE, Marletta MA. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 2010;77:930–942. doi: 10.1111/j.1365-2958.2010.07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Dupes NM, Gerstenfeld TAI, Simmons LA, Mekalanos JJ. DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 2011;7:e1001295. doi: 10.1371/journal.ppat.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb. Exp. Pharmacol. 2009:17–31. doi: 10.1007/978-3-540-68964-5_2. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat. Med. 1995;1:546–551. doi: 10.1038/nm0695-546. [DOI] [PubMed] [Google Scholar]
- Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 2010;13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J. Bacteriol. 2009;191:169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochem. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak J, Henche A-L, Binnenkade L, Thormann KM. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2010;76:3263–3274. doi: 10.1128/AEM.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Biondi EG, Skerker JM. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Meth. Enzymol. 2007;423:531–548. doi: 10.1016/S0076-6879(07)23026-5. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 2004;186:4864–4874. doi: 10.1128/JB.186.15.4864-4874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick J, Boon EM. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochem. 2012 doi: 10.1021/bi201753f. (ahead of print) [DOI] [PubMed] [Google Scholar]
- Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. The Structure of an Unconventional HD-GYP Protein from Bdellovibrio Reveals the Roles of Conserved Residues in this Class of Cyclic-di-GMP Phosphodiesterases. MBio. 2011:2. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CR, Nealson KH. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. T. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochem. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Lucey J, O'Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 2009;11:1126–1136. doi: 10.1111/j.1462-2920.2008.01842.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. USA. 2010:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol. Microbiol. 2011;79:1353–1366. doi: 10.1111/j.1365-2958.2010.07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the Specificity of Two-Component Signal Transduction Systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. Initial Phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 2004;186:8096–8104. doi: 10.1128/JB.186.23.8096-8104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc. Natl. Acad. Sci. USA. 2010;107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- Yan D, Cho HS, Hastings CA, Igo MM, Lee SY, Pelton JG, Stewart V, Wemmer DE, Kustu S. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.