Abstract
Antibody switching involves class switch recombination (CSR) events between switch (S) regions located upstream of heavy chain constant (C) genes. Mechanisms targeting CSR to S-regions are not clear. Deletion of Sμ tandem repeat (SμTR) sequences causes CSR to shift into downstream regions that do not undergo CSR in WT B-cells, including the Cμ-region. We now find that, in SμTR−/− B cells, Sμ chromatin histone modification patterns also shift downstream relative to WT and coincide with SμTR−/− CSR locations. Our results suggest that histone H3 acetylation and methylation are involved in accessibility of switch regions and that these modifications are not dependent on the underlying sequence, but may be controlled by the location of upstream promoter or regulatory elements. Our studies also show RNA polymerase II (RNAPII) loading increases in the Eμ/Iμ region in stimulated B cells; these increases are independent of SμTR sequences. Longer Sμ deletions have been reported to eliminate increases in RNAPII density, therefore we suggest that sequences between Iμ and Sμ (possibly the Iμ splicing region as well as G-tracts that are involved in stable RNA:DNA complex formation during transcription) might control the RNAPII density increases.
Keywords: Class Switching, Histone Modification, RNA Polymerase
1. Introduction
The process of class switch recombination (CSR) alters antibody heavy (H) – chain constant (CH) region gene segments through deletional DNA recombination events between switch (S) region sequences located upstream of each CH (except Cδ)(reviewed in (Stavnezer et al., 2008)). S-regions contain highly repeated tandem arrays of G-rich DNA. Activation-induced cytidine deaminase (AID) and transcription through donor and acceptor S-region sequences have been shown to be required for CSR (Muramatsu et al., 2000; Stavnezer et al., 1988; Stavnezer-Nordgren and Sirlin, 1986). AID deaminates dC residues in single stranded DNA and in transcribed double stranded DNA (Chaudhuri et al., 2003; Di Noia and Neuberger, 2002; Muramatsu et al., 2000; Petersen-Mahrt et al., 2002; Rada et al., 2002). In single-stranded DNA, AID preferentially deaminates within WRC sequence motifs (W = A or T and R = purine) (Beale et al., 2004; Pham et al., 2003; Yu et al., 2004) that are often enriched in S regions.
For CSR to occur, DNA breaks must be made in S-regions. Individual S-regions have different nucleotide sequences and vary from ~1 kb to 12 kb in length (Arakawa et al., 1993; Kataoka et al., 1981; Mowatt and Dunnick, 1986; Nikaido et al., 1981; Nikaido et al., 1982; Szurek et al., 1985). The basis for targeting of CSR to S regions is not understood. Transcription through S-regions can form stable R-loop structures that leave the non-template strand as a favorable single-stranded target of AID activity (Shinkura et al., 2004; Tian and Alt, 2000; Yu et al., 2003). However, substantial CSR can occur in S-regions where no R-loops have been found (Zarrin et al., 2004), and RNA transcription appears sufficient to provide AID access to double-stranded DNA (Chaudhuri et al., 2003).
Chromatin remodeling could be important for targeting CSR. Post-translational modifications alter DNA-histone interactions in the nucleosome (Kouzarides, 2007) and could regulate access of the CSR machinery to S-regions. Covalent modifications, such as acetylation (AcH3 and AcH4) and H3 K4 methylation (di-me H3K4) alter transcription by modifying chromatin structure to an active state (Kouzarides, 2007). Induction of SHM in a B-cell lymphoma correlates with increased histone acetylation of the transcribed and mutated V region but not of the equivalently transcribed, but unmutated, Cμ region (Woo et al., 2003). Reports have indicated increases in active histone modifications of downstream switch regions that coincide with CSR, however these increases, if any, are modest in the Sμ region (Beale et al., 2004; Kuang et al., 2009; Li et al., 2004; Nambu et al., 2003; Wang et al., 2006; Wang et al., 2009).
In stimulated B cells that lack Sμ tandem repeat sequences (SμTR), CSR recombination break points shift downstream into regions where switch events are not found in wild-type (WT) mice, including in the Cμ region (Min et al., 2005). This shift demonstrates that these downstream sequences are fully capable of supporting CSR, but that, in WT cells, the sequences are inaccessible to the CSR machinery. We have compared the histone acetylation and methylation patterns within the μ locus in stimulated WT and SμTR−/− B cells to see if chromatin modifications associate with shifts in CSR locations. We find differences in acetylation and methylation in the μ locus that appear to designate accessible regions and boundaries in targeting CSR. Furthermore, our results show that removal of the SμTR results in increased active histone modifications in the Cμ region; suggesting that this increase allows the Cμ region to become CSR accessible. Our data indicate that histone H3 acetylation and methylation are involved in specific accessibility to switch regions and that these modifications are not dependent on the underlying sequence, but may be controlled by the location of a nearby promoter or regulatory elements. In addition, histone modifications of the WT Sμ region do not change when resting B cells are activated, indicating that other mechanisms control the activity of AID on accessible Sμ chromatin. We find that activated B cells show increased RNA polymerase density in the Eμ/Iμ region upstream of Sμ, suggesting that polymerase loading in this region could promote the entry of AID onto the accessible Sμ DNA target.
2. Materials and Methods
2.1. Mice and cell culture
Animal experiments were approved by the Tufts University Institutional Animal Care and Use Committee. C57BL/6 mice were purchased from Jackson Laboratories. All mice were 8–12 weeks of age. Splenic B cells were purified using StemSep B Cell Enrichment kit (StemCell Technologies). Splenic B cells were cultured at 5 × 105 cells/ml in RPMI-1640/10% FCS. Cultures were supplemented with LPS (25 μg/ml; Sigma Chemical Co.) and 10 ng/ml murine IL4 (Preprotech) to induce isotype switching to IgG1.
2.2. Chromatin Immunoprecipitations (ChIPs)
Cells (~40 × 106 stimulated and ~100 × 106 resting splenic B cells per ChIP) were fixed in 1% formaldehyde for 10 min at room temperature. Glycine (125 mM final concentration) stopped the reaction. Cells were washed 2x with cold PBS, then 3x with cold lysis buffer (10 mM Tris•HCl, pH 7.5/10 mM NaCl/3 mM MgCl2/0.5% NP-40). The nuclear pellet was resuspended in 2 ml of micrococcal nuclease (MNase) Reaction buffer (10 mM Tris•HCl, pH 7.5/10 mM NaCl/3 mM MgCl2/1 mM CaCl2/4% NP-40/1 mM PMSF) and incubated with 4 units of MNase (Sigma) for 10 min at 37°C. MNase digestion was stopped with 3 mM EGTA. SDS (1% final concentration), NaCl (200 mM final concentration), and protease inhibitors were added. The samples were sonicated to lyse nuclei, and cellular debris was removed by centrifugation. Conditions were optimized to produce 100–500 bp DNA fragments. Chromatin samples were diluted with buffer (20 mM Tris•HCl, pH 8.0/2 mM EDTA/1% Triton X-100/150 mM NaCl/protease inhibitors) and precleared for 15 min at 4°C with 50 μl of Protein A Sepharose (Sigma) resin. A DNA aliquot of this precleared sample was the “input” sample. Immunoprecipitations were rocked at 4°C overnight with either 5ug of anti-Acetyl H3 (K9/K14), Upstate Biotechnology, Lake Placid, NY (U.B.) catalogue no. 06-599, 5ug of anti-di-Me H3-K4, Abcam Inc., Cambridge, Ma (Abcam) catalogue no. ab7766, 5 ug of anti-tri-Me H3-K4, (Abcam catalogue no. ab7766), 5 ug of anti-RNAPII CTD phosphorylated on serine 5 (Abcam catalogue no. ab5131), or 5 ug of anti-RNAPII CTD phosphoryated on serine 2, (Abcam catalogue no. ab5095), together with 50ul of Protein A Sepharose resin. According to the manufacturer, RNAPII antibodies are specific for single-phosphorylated forms, neither antibody will bind to RNAPII phosphorylated on both serine 5 and serine 2. The resin was washed with IP dilution buffer, twice with IP dilution buffer plus 0.1% SDS, with IP dilution buffer plus 0.1% SDS and 500 mM NaCl, with ChIP wash 3 (10 mM Tris•HCl, pH 8.0/1 mM EDTA/250 mM LiCl/1% NP-40/1% deoxycholate), and twice with TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). Chromatin was eluted and decrosslinked. DNA was phenol/chloroform extracted, ethanol precipitated, and quantified.
2.3. Real-Time PCR analysis
Differences in amounts of specific DNA sequences enriched in an IP sample relative to an input sample were determined by SYBR green-mediated real-time PCR analysis using the Bio-Rad iCycler iQ. Triplicate reactions containing 1 ng input or IP DNA and 10 pmol of primers (Table I) were amplified in SYBR Green PCR Master Mix (Applied Biosystems). Primers were tested in reactions containing α32P dCTP, ensuring a single product with appropriate size. Fold enrichment values for amplified DNA sequences were determined by cycle threshold (Ct; average of the three values, measured at a threshold level of fluorescence in the linear range of amplification) for input and IP samples and the rate of amplification (R, ranging between 1.7 and 2.3, with 2 being the theoretical optimum) of each primer pair – Fold enrichment = R^(Ctinput−CtIP).
Table I.
PCR primers
| Primer Name | Primer Sequence Reference |
|---|---|
| EμU | 5′-TGGCAGGAAGCAGGTCAT-3′ |
| EμD | 5′-GGACTTTCGGTTTGGTGG-3′ |
| 5Sμ1U | 5′-TGCTCTGTGTGAACTCCCTCTG-3′ |
| 5Sμ1D | 5′-AGCCACAACCATACATTCCCAGG-3′ |
| 5Sμ2U | 5′-GTAAATGTACTTCCTGGTTG-3′ |
| 5Sμ2D | 5′-GGTCTCTATTCTTTCTCAA-3′ |
| 3Sμ1U | 5′-ATAAGTTAGGCTGAGTAGGGC-3′ |
| 3Sμ2D | 5′-ACTGGCTGGGAGAACTATT-3′ |
| CμU | 5′-CACCATTTCCTTCACCTG-3′ |
| CμD | 5′-TGTTTTTGCCTCCGTAGT-3′ |
| 3Cμ1U | 5′-GAATGAGCAATAGGCAGTA-3′ |
| 3Cμ1D | 5′-GATGGTGAAGGTTAGGATG-3′ |
| 3Cμ2U | 5′-CCGAGAGGACCGTGGACA-3′ |
| 3Cμ2D | 5′-AGAGCAAGCAAAACACAACT-3′ |
| CδU | 5′-CCATCACTTTTTGTCCAT-3′ |
| CδD | 5′-AGCAAGAGGTGTAAGGTT-3′ |
| 5Sγ1U | 5′-TATGCCACCCACTGTCAATCCTGT-3′ |
| 5Sγ1D | 5′-TGGTCCTGCCCTTCTCTTGTCTTT-3′ |
| Sγ1U | 5′-GGTCCCAGGTTCAATCCCAGC-3′ |
| Sγ1D | 5′-TTTGCAGGTGCTCAGTCTTGTGTCCT-3′ |
| 3Sγ1U | 5′-ACAGGTCAAG GCTGAGTAGAAGCA-3′ |
| 3Sγ1D | 5′-TCCCACAACTCCCACTGGTTTAGTT-3′ |
| 3Sγ12ndU | 5′-GGAACTGCTGCAGGCACAAAGAAT-3′ |
| 3Sγ12ndD | 5′-CTCCAGCCTGTATGTTTCCACT-3′ |
| Cγ1U | 5′-AGCCAGCGGAGAACTACAAGAACA-3′ |
| Cγ1D | 5′-TGCTCTTCTGCACATTGAGCTTGC-3′ |
| Cγ12ndU | 5′-TCACACTGTCTGCTCATCTCGCTT-3′ |
| Cγ12ndD | 5′-CTTTGGTGCTGCTGTGATGGTGTT-3′ |
| β-globinU | 5′-GCCTTGCCTGTTCCTGCTC-3′ |
| β-globinD | 5′-CAGACCATAAACTGTATTTTTCTTATTGAGCCC-3′ |
| μGLTF | 5′-CTCGGTGGCTTTGAAGGAAC-3′ |
| μGLTR | 5′-TGGTGCTGGGCAGGAAGT-3′ |
| γ1GLTF | 5′-TCGAGAAGCCT-GAGGAATGTG -3′ |
| γ1GLTR | 5′-ATGGAGTTAGTTTGGGCAGCA-3′ |
| IgβF | 5′-CAGAAATGTGACAGCGCCAACCAT-3′ |
| IgβR | 5′-TGTCAAGTAGCAGGAAGATGGGCA-3′ |
| β-actinF | 5′-AGGTATCCTGACCCTGAAG-3′ |
| β-actinR | 5′-CACGCAGCTCATTGTAG-3′ |
3. Results
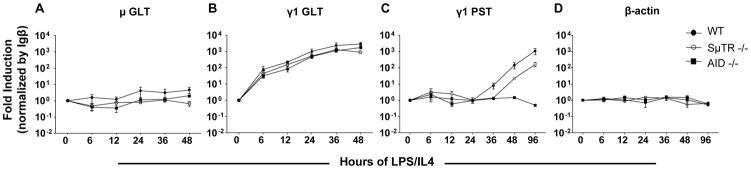
Chromatin immunoprecipitation (ChIP) assays were performed to determine the status of histone H3 acetylation (AcH3) and methylation (di-MeH3K4 and tri-MeH3K4) for different segments of the μ and γ1 loci in both resting and activated B cells. Splenic mouse B cells were activated by stimulation in culture with LPS and IL4 to induce class switching to γ1. After 48 hours of stimulation, B cells were collected and analyzed by ChIP. At this time point, germline γ1 transcripts have been induced, but little CSR has occurred (Figure 1). Extending stimulation to 96 hours leads to significant CSR as indicated in the increased levels of post-switched transcripts (Figure 1C). Significant levels of DSB can be detected in Sμ after 48 hours of B-cell stimulation (Schrader et al., 2005).
Figure 1. IgM and IgG1 transcript expression levels in resting and stimulated splenic B cells.
Expression levels for (A) μ germline transcripts (μGLT), (B) γ1 germline transcripts (γ1GLT), (C) γ1 post switched transcripts (γ1PST), and (D) β-actin were measured by quantitative real time RT- PCR using cDNAs from splenic B cells of C57BL/6 wild type (solid circles), SμTR−/− (open circles), and AID−/− (solid squares) mice. Splenic B cells were stimulated with LPS and IL4 over a 48 hour time course (0hr, 12hr, 24hr, 36, and 48hr). PST samples included a 96 hour time point. Results were normalized to Igβ expression. Real time PCR was performed in triplicate. Error bars represent standard estimated mean (SEM) from three independent experiments.
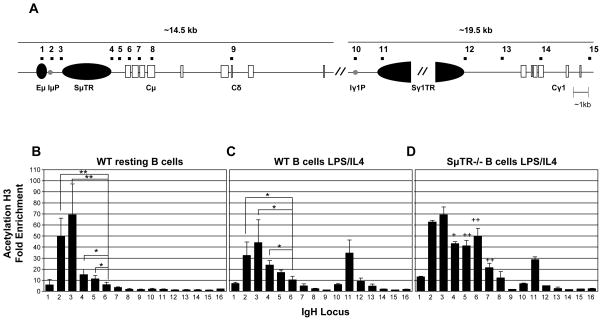
3.1. Sμ region is hyperacetylated in resting and stimulated WT B cells, while Sγ1 region is hyperacetylated only after stimulation
To determine the extent and pattern of changes in AcH3 levels in the μ and γ1 locus of resting and stimulated splenic B cells, we performed a series of ChIP assays using real time PCR with 15 primer pairs distributed across the μ and γ1 region (Table I, Figure 2A). Primers specific for β-globin were included as a negative control. Our results indicate that Sμ AcH3 is enriched in both resting and stimulated WT splenic B cells (Figure 2B & C). This finding confirms previous reports that Sμ is acetylated before and after B-cell activation (Chowdhury et al., 2008; Kuang et al., 2009; Li et al., 2004; Wang et al., 2006; Wang et al., 2009). Our analyses now demonstrate that acetylation levels are highest in regions near the SμTR segment and decrease in neighboring segments that are not accessible to CSR (Figure 2B& C). We also find that Sμ AcH3 is identical in WT and AID-deficient B cells (data not shown), consistent with the lack of Sμ AcH3 changes upon B-cell stimulation. In resting WT B cells, acetyl H3 enrichment was not observed across the Ig γ1 region. Upon activation, however, significant enrichment was induced in the γ1 region and appeared to focus primarily in the Sγ1 region (Figure 2B& C).
Figure 2. Acetyl H3 localizes to specific segments of the Ig heavy chain μ and γ1 regions in splenic B cells.
(A) Diagram of the Ig heavy chain locus. Black squares above the diagram mark locations of PCR primer pairs in the ChIP analyses; primer pair 16 is for a control gene, β-globin (Chowdhury and Sen, 2003; Garrett et al., 2005). Black vertical oval depicts the Eμ enhancer. Black horizontal ovals depict switch sequence, whereas white rectangles depict constant segments. Gray circles depict promoter regions. Two slashes depict sequence gaps. (B). Acetyl H3 at the IgH μ and γ1 loci in WT resting splenic B cells, n=4. (C). Acetyl H3 at the IgH μ and γ1 loci in WT splenic B cells stimulated with LPS and IL4 for 48 hours, n=4. (D). Acetyl H3 at the IgH μ and γ1 loci in SμTR−/− splenic B cells stimulated with LPS and IL4 for 48 hours, n=2. Chromatin Immunoprecipitation was performed with antibody specific for acetylated histone 3. Real-time PCR was performed in triplicate on input and antibody bound ChIP fractions. Fold enrichment of each DNA sequence in an IP sample relative to an Input sample is shown. Values were averaged; standard deviations are shown as error bars. Student t-test statistical analysis was performed. * or + indicates p-value < 0.05, ** or ++ indicates p-value < 0.01. Asterisks (*) indicate significant differences between different DNA segments. Crosses (+) indicate significant differences between SμTR−/− B cells LPS/IL4 and WT B cells LPS/IL4 48 within the same DNA segments.
To determine whether Sμ AcH3 enrichment is dependent on motifs within the underlying SμTR sequence, we performed ChIP analysis on stimulated B cells from mice lacking the SμTR region. These mice offer a Sμ region that is short and lacks highly repetitive sequences, but that is able to support substantial levels of CSR (Luby et al., 2001). WT and SμTR−/− B cells exhibit similar levels of AcH3 enrichment in the Sμ and Sγ1 regions (Figure 2C & D). In addition, both WT and SμTR−/− B cells exhibit AcH3 levels that increase dramatically ~ 500bp downstream of Eμ. However, SμTR−/− B cells show high levels of acetylation extending into Cμ sequences, whereas, in WT B cells, high levels of acetylation are limited to sequences more than 1.4 kb upstream of Cμ. These results indicate that Sμ acetylation levels are not dependent on the underlying sequence, and that the ~3kb SμTR deletion causes the previously inaccessible Cμ gene segment to shift into a region that is acetylated and accessible to CSR. These results correlate with previous studies showing that SμTR deletion results in CSR within regions that only rarely undergo CSR in WT mice (Min et al., 2005), both downstream of the SμTR and within the Cμ region. The removal of the ~3kb SμTR segment does not affect the location or pattern of acetylation enrichment within the γ1 region (Figure 2D).
3.2. Tri-methylation of histone 3 on lysine 4 coincides with regions targeted by the CSR machinery
In both resting and stimulated WT B cells, the Sμ region exhibits enrichment of the tri-MeH3K4 activation modification (Figure 3B &C), primarily in regions where CSR has been found to occur (Min et al., 2005). In SμTR−/− B cells the upstream portion of the JH-Cμ intron exhibits tri-MeH3K4 levels similar to those in WT B cells (Figure 3C & D). However, in SμTR−/− B cells, tri-MeH3K4 modification remains high in Cμ compared to the lower Cμ levels in WT B cells, similar to our findings with H3 acetylation. This pattern correlates with the active CSR within Cμ in SμTR−/− B cells, compared to the lack of Cμ CSR in WT.
Figure 3. Tri-methylation of H3K4 at the Ig heavy chain region in splenic B cells.
(A) Diagram of the Ig heavy chain locus as in Fig. 2. (B). Tri-Methyl H3K4 at the IgH μ and γ1 loci in WT resting splenic B cells, n=3. (C). Tri-Methyl H3K4 at the IgH μ and γ1 loci in WT splenic B cells stimulatedwith LPS and IL4 for 48 hours, n=3. (D). Tri-Methyl H3K4 at the IgH μ and γ1 loci in SμTR−/− splenic B cells stimulated with LPS and IL4 for 48 hours, n=3. Chromatin Immunoprecipitation used antibody specific for di-methylation of histone 3 on lysine 4. Analyses and data presentation are as in Fig. 2.
For the Ig γ1 region, we found no enrichment of tri-methylation in resting B cells, similar to our analyses of AcH3 in γ1. However, upon activation, significant MeH3K4 enrichment is induced in γ1, and appears to focus primarily in Sγ1 (Figure 3). As in Sμ, patterns of H3 trimethylation and H3 acetylation in γ1 correlate closely. SμTR deletion does not affect either AcH3 or MeH3K4 patterns in Sγ1, indicating that SμTR sequences do not influence chromatin modification patterns of downstream S regions when these are activated by B-cell stimulation.
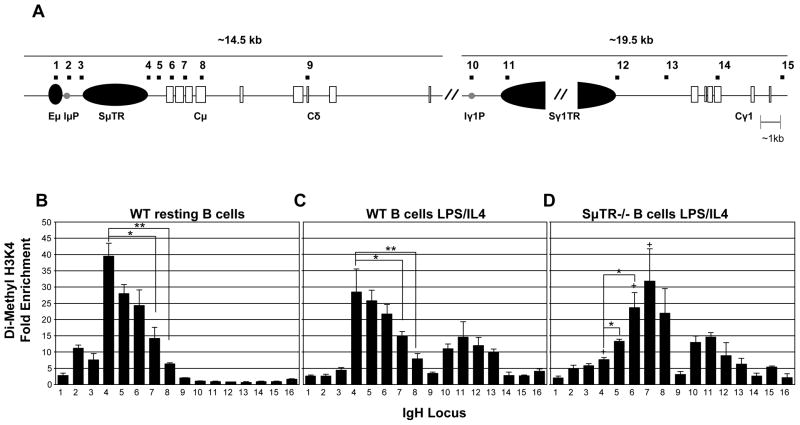
3.3. Di-methylation of histone 3 on lysine 4 marks 3′ boundary of CSR domain
In pro–B cells, peaks of di-methylation of histone 3 at lysine 4 (di-MeH3K4) appear to delineate the boundaries of active chromatin (Morshead et al., 2003). We assessed Sμ di-MeH3K4 in resting and stimulated WT B cells and found the highest enrichment immediately downstream of the Sμ tandem repeats. Di-MeH3K4 enrichment levels reach ~30–40 fold at this position and then appear to drop rapidly in the Cμ region (Figure 4B &C). The highest levels of di-MeH3K4 correspond to the end of the CSR domain in WT B cells as defined previously (Min et al., 2005). This suggests that the distribution of the di-MeH3K4 modification could designate the end of the Sμ CSR domain.
Figure 4. Di-Methyl H3K4 peak delineates 3′ border in CSR.
(A) Diagram of the Ig heavy chain locus as in Fig. 2. (B). Di-Methyl H3K4 at the IgH μ and γ1 loci in WT resting splenic B cells, n=2. (C). Di-Methyl H3K4 at the IgH μ and γ1 loci in WT splenic B cells stimulated with LPS and IL4 for 48 hours, n=5. (D). Di-Methyl H3K4 at the IgH μ and γ1 loci in SμTR−/− splenic B cells stimulated with LPS and IL4 for 48 hours, n=2. Chromatin Immunoprecipitation was performed on freshly isolated B cells or after LPS/IL4 stimulation for 48 hours with antibody specific for di-methylation of histone 3 on lysine 4. Analyses and data presentation are as in Fig. 2.
To assess whether di-MeH3K4 consistently marks the 3′ boundary of the active CSR domain in the μ region, we analyzed di-MeH3K4 in activated SμTR−/− B cells. We find that the pattern of increased H3 di-methylation shifts downstream in stimulated SμTR−/− B cells (Figure 4D). In the position where di-MeH3K4 is highest in WT B cells, we observe only ~ 8 fold enrichment in SμTR−/− B cells. The SμTR−/− B cell di-MeH3K4 increases to 30–40 fold enrichment between Cμ exons 2 and 3. CSR in SμTR−/− B cells rarely occurs beyond the third exon of Cμ (Min et al., 2005). The di-MeH3K4 patterns in WT and SμTR−/− B cells both coincide with CSR boundaries, suggesting that this modification might designate the 3′ boundary of the μ CSR domain.
The di-MeH3K4 pattern seen in activated B cells in the γ1 region (Figure 4) is different from that in the μ region. In resting WT B cells, the γ1 region does not display any di-MeH3K4 enrichment. Upon induction of CSR, di-MeH3K4 levels are enriched upstream of Sγ1. The di-MeH3K4 levels appear to continue to increase within Sγ1 and then drop. These results show that chromatin modification structures of S regions can exhibit unique, isotype-specific features.
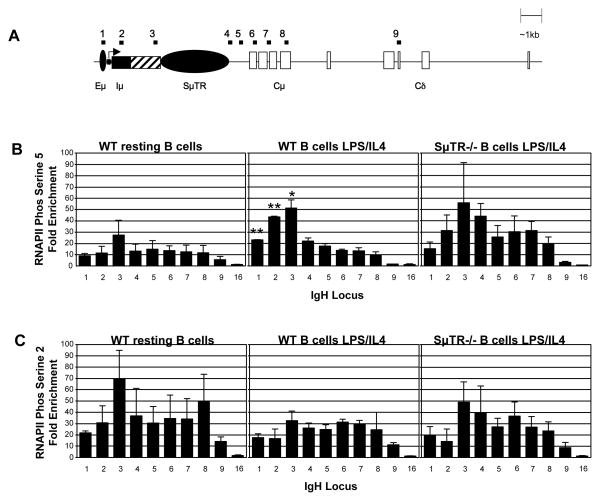
3.4. RNA polymerase II (RNAPII) enrichment is independent of the SμTR sequence
We find that AcH3, tri-MeH3K4, and di-MeH3K4 modification levels and patterns of the WT Sμ region do not appear to be altered significantly upon B-cell stimulation. Our results are similar to findings in other previous reports regarding Sμ histone modifications and chromosomal interactions during B-cell activation, even though large changes in chromatin modifications and germline transcription are observed for downstream S regions after B-cell stimulation (Chowdhury et al., 2008; Kuang et al., 2009; Li et al., 2004; Nambu et al., 2003; Wang et al., 2006; Wang et al., 2009; Wuerffel et al., 2007). Correlations between germline transcription and CSR activity (Bottaro et al., 1994; Jung et al., 1993; Yancopoulos et al., 1986) suggest that changes in RNAPII association to S regions might be important for inducing CSR. RNAPII co-immunoprecipitates with AID (Nambu et al., 2003), suggesting that increases in RNAPII loading could recruit AID to S-regions. One previous study has shown increased density of RNAPII) in the Sμ region after B-cell stimulation (Li et al., 2004). Two further studies have shown an accumulation of RNAPII close to the SμTR that is dependent on the Sμ sequence (Rajagopal et al., 2009; Wang et al., 2009). Reports differ as to whether B-cell activation increases this enrichment. To probe transcription throughout the Sμ region during B-cell stimulation, we performed ChIP analyses using two different RNAPII antibodies; (1) RNAPII C-terminal domain (CTD) phosphorylated at serine 5 (Pser5 RNAPII) and (2) RNAPII CTD phosphorylated at serine 2 (Pser2 RNAPII) (see Materials and Methods). Pser5 RNAPII is associated with transcription initiation whereas Pser2 RNAPII is associated with transcription elongation (Sims et al., 2004).
In unstimulated B cells, the distribution of Pser5 RNAPII within the JH-Cμ region displays a modestly increased level of enrichment in the region just upstream of the SμTR as compared to the region downstream of the SμTR (Figure 5B). In stimulated B cells, however, this enrichment intensifies, with the greatest increase (~ 4 fold) over unstimulated levels specifically in the region containing the Iμ promoter (primer pair 2 in Figure 5). Immediately downstream of the SμTR the levels of Pser5 RNAPII in both resting and stimulated B cells are similar. In stimulated SμTR −/− splenic B cells, the patterns of Pser5 RNAPII appear to be similar to those in WT B cells, although the 3 kb deletion does result in higher RNAPII density levels in the Cμ region (Fig. 5B).
Figure 5. Enrichment of RNAPII at the Immunoglobulin heavy chain μ locus in splenic B cells.
(A) Diagram of the Ig heavy chain μ locus as in Fig. 2. The solid box represents the Iμ exon. The hatched box represents the region containing the Iμ splice site as well as other promoters and splice sites that are sufficient to provide significant CSR in mutant mice that lack either the Iμ promoter (Bottaro et al., 1998) or the Iμ splice site (Kuzin et al., 2000). (B) Association of RNAPII Phos serine 5 (transcriptional initiation) at the IgH μ locus in WT resting splenic B cells (n=3), WT splenic B cells stimulated with LPS and IL4 for 48 hours (n=2), and SμTR−/− splenic B cells stimulated in with LPS and IL4 for 48 hours, (n=2). (C) Association of RNAPII Phos serine 2 (transcriptional elongation) at the IgH μ locus in WT resting splenic B cells (n=2), WT splenic B cells stimulated in culture with LPS and IL4 for 48 hours (n=3), and SμTR−/− splenic B cells stimulated in culture with LPS and IL4 for 48 hours, (n=3). Chromatin Immunoprecipitation was performed on freshly isolated B cells or after LPS/IL4 stimulation for 48 hours with antibody specific for RNAPII phosphorylated on serine 5 or serine 3. Analyses and data presentation are similar to Fig. 2. * Indicates p-value < 0.05, ** indicates p-value < 0.01. Asterisks (*) indicate significant differences between WT B cells LPS/IL4 and WT resting B cells within the same DNA segment.
The Pser2 RNAPII enrichment levels in stimulated WT B cells appear to decrease in the region upstream of SμTR compared to unstimulated controls (Figure 5C). Pser2 RNAPII enrichment levels are not significantly affected by the removal of the SμTR. In total, our data are consistent with an increase in transcriptional initiation in the Iμ promoter region upon the induction of class switching and confirm some previous reports.
Pser5 RNAPII increases due to B cell stimulation are also observed in sequences upstream and downstream of the Iμ promoter (Fig. 5B). We do not know the origin of these increases. Because primer pairs 1 and 2 are close, some of the primer pair 1 increases might result from CHIP DNA fragments that span the Iμ and primer pair 1 sequences. However, the distance between primer pairs 2 and 3 is relatively large, suggesting that B-cell stimulation is increasing Pser5 RNAPII loading in these sequences downstream of the Iμ promoter. Other reports have suggested that RNAPII is pausing in sequences downstream of the Iμ promoter in B cells that are induced to undergo CSR and that this pausing is important for interactions with AID that target AID-activity for S-region CSR. Our results are consistent with these suggestions. However, the elevated levels of Pser5 RNAPII that we observe in the primer pair 3 region could also reflect transcriptional initiation at promoters that are located in this region and that appear to be sufficient to allow significant CSR in mice that lack the Iμ promoter (Bottaro et al., 1998).
4. Discussion
We have mapped chromatin histone modifications within the μ and γ1 IgH switch region sequences in resting and activated B cells from WT and SμTR−/− mutant mice. We find that AcH3 and tri-MeH3K4 histone modifications are most highly enriched in WT Sμ and Sγ1 DNA region sequences that are targets for CSR, suggesting that these might regulate the accessibility of S-regions to CSR. Our results are consistent with studies showing that impairing tri-MeH3K4 modification reduces CSR (Daniel et al., 2010; Schwab et al., 2011; Stanlie et al., 2010). In Sγ1, the pattern of di-MeH3K4 follows the AcH3 and tri-MeH3K4 patterns but, in Sμ, the peak of Sμ di-MeH3K4 enrichment is found at the 3′ boundary of the CSR region. This location could suggest a role for di-MeH3K4 in establishing the Sμ 3′ CSR boundary. SμTR−/− mice show patterns of histone modifications in the JH-Cμ intron that are remarkably similar to the patterns in WT mice, even though the underlying sequences have shifted relative to the histone modification pattern. Although our studies indicate no changes in Sμ histone modification patterns when resting WT B cells are stimulated to undergo CSR, we do find that RNAPII loading in the Eμ/Iμ region is increased upon B-cell activation. This result is consistent with the possibility that RNAPII may promote entry of AID into accessible Sμ DNA target regions and lead to CSR.
Comparing our results from SμTR−/− mutant mice with recent studies of a related Sμ mouse mutant designated as SμΔ/Δ (Wang et al., 2009) suggests some new insights into CSR targeting. SμΔ/Δ mice have a larger Sμ deletion than SμTR−/−, lacking additional sequences upstream and downstream of the SμTR region. Analyses of CSR recombination sites in SμΔ/Δ indicate that they cluster near the Iμ promoter (Khamlichi et al., 2004) and do not appear to occur within the Cμ region where 20–30% of SμTR−/− CSR sites are found (Min et al., 2005). On the other hand, histone trimethylation is elevated at a single Cμ site that has recently been analyzed in SμΔ/Δ B cells (Wang et al., 2009), suggesting that the accessible Sμ chromatin domain in SμΔ/Δ B cells might extend into the Cμ region, similar to our finding for SμTR−/−. This indicates that the correlations between histone modifications and CSR sites observed in SμTR−/− mice do not occur in SμΔ/Δ, indicating that the additional sequences that are absent in the SμΔ/Δ deletion might play a role in regulating the region within the accessible domain that is available for CSR. Perhaps this regulation reflects the differences in Sμ RNA:DNA complex formation found when comparing SμTR−/− and SμΔ/Δ B cells (Huang et al., 2007). The elements responsible for this regulation could be located in the sequences between the Iμ and SμTR regions, although we cannot rule out possible elements between SμTR and Cμ1.
Other features of the Sμ region that are induced by B-cell stimulation might also contribute to recruiting the CSR machinery. Some studies have indicated that AID might complex with the RNA transcription machinery to provide access to single-stranded DNA during passage of RNAPII (Chaudhuri et al., 2004; Nambu et al., 2003). However, our results and other studies have indicated that the levels of Iμ transcripts remain constant before and after B-cell stimulation (Rajagopal et al., 2009). Nevertheless, comparing resting and stimulated B cells, we find 2–4 fold increases of polymerase loading in the DNA regions just downstream of the Eμ/Iμ region, but no significant increases just downstream of the SμTR. Our results confirm reports of increased RNAPII loading upstream of the SμTR in WT B cells (Rajagopal et al., 2009; Wang et al., 2009). We also find increased RNAPII in this region in SμTR−/− B cells, suggesting that the increased RNAPII loading is independent of the Sμ tandem repeats.
Polymerase density increases near Iμ could indicate an induction of transcriptional initiation from this promoter when B cells are stimulated for CSR. It is also possible that increased Eμ/Iμ polymerase loading may reflect pausing of RNAPII after initiation (Wang et al., 2009). A recent report shows that a factor associated with paused RNAPII can also recruit AID to DNA, suggesting that pausing could play an important role in CSR targeting (Pavri et al., 2010). The similarity of RNAPII patterns that we find in WT and SμTR−/− mice suggests that the SμTR is not needed for pausing. On the other hand, an analysis of RNAPII loading in SμΔ/Δ mice (Rajagopal et al., 2009; Wang et al., 2009) indicates only lower densities throughout the Iμ, Sμ, and Cμ regions, suggesting that elements just upstream or downstream of SμTR might regulate the RNAPII patterns that are seen in WT and SμTR−/− mice. Upstream elements seem most likely because the DNA sequences between Iμ and SμTR that exhibit increased RNAPII loading in activated B cells are partly missing in SμΔ/Δ mice. Perhaps this regulation is also related to differences between Sμ RNA:DNA complex formation found in stimulated SμTR−/− and SμΔ/Δ B cells; the sequences thought important for initiating RNA:DNA complex formation are also located within the Iμ/SμTR region that is deleted in SμΔ/Δ mice (Huang et al., 2007).
The sequences accessible to CSR in S regions associated with different isotypes exhibit substantial variations in length. This variation could reflect the different repeat region lengths in different S regions. However, we find SμTR sequences are not needed for establishing the length of DNA exhibiting enriched histone modifications, and the length of the CSR region is also independent of the SμTR (Luby et al., 2001; Min et al., 2005). Perhaps CSR target length regulation differs between S regions; this could be related to the different di-MeH3K4 patterns in Sμ and Sγ1 that we observe. Analyses of length mutants for other S-region loci might shed light on this issue.
Highlights.
Histone H3 acetylation and trimethylation mark CSR accessible regions in B cells.
Sμ histone H3 marks depend on flanking but not underlying DNA sequences.
Histone H3 dimethylation marks match with downstream boundaries of Sμ CSR.
Increases in RNAPII loading in the Eμ/Iμ region are linked to initiation of CSR.
Sequences just upstream of the Sμ tandem repeats control Sμ RNAPII loading.
Acknowledgments
This research was supported by National Institutes of Health (ES and MAO) and Eshe Foundation (ES) grants.
Abbreviations used in this paper
- AcH3
histone H3 acetylation
- AID
activation-induced cytidine deaminase
- CH region
antibody heavy (H) – chain constant region
- ChIP
Chromatin Immunoprecipitation
- CSR
class switch recombination
- di-MeH3K4
histone H3 dimethylation
- GLT
germline transcript
- PST
post switched transcript
- RNAPII
RNA polymerase II
- S
switch
- S region
switch region
- SμTR
Sμ tandem repeat
- tri-MeH3K4
histone H3 trimethylation
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakawa H, Iwasato T, Hayashida H, Shimizu A, Honjo T, Yamagishi H. The complete murine immunoglobulin class switch region of the alpha heavy chain gene-hierarchic repetitive structure and recombination breakpoints. J Biol Chem. 1993;268:4651–5. [PubMed] [Google Scholar]
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–96. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt FW. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. Embo J. 1994;13:665–74. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro A, Young F, Chen J, Serwe M, Sablitzky F, Alt FW. Deletion of the IgH intronic enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int Immunol. 1998;10:799–806. doi: 10.1093/intimm/10.6.799. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–8. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 2003;18:229–41. doi: 10.1016/s1074-7613(03)00030-x. [DOI] [PubMed] [Google Scholar]
- Chowdhury M, Forouhi O, Dayal S, McCloskey N, Gould HJ, Felsenfeld G, Fear DJ. Analysis of intergenic transcription and histone modification across the human immunoglobulin heavy-chain locus. Proc Natl Acad Sci U S A. 2008;105:15872–7. doi: 10.1073/pnas.0808462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–23. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–8. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25:1511–25. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–32. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–7. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Miyata T, Honjo T. Repetitive sequences in class-switch recombination regions of immunoglobulin heavy chain genes. Cell. 1981;23:357–68. doi: 10.1016/0092-8674(81)90131-8. [DOI] [PubMed] [Google Scholar]
- Khamlichi AA, Glaudet F, Oruc Z, Denis V, Le Bert M, Cogne M. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood. 2004;103:3828–36. doi: 10.1182/blood-2003-10-3470. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kuang FL, Luo Z, Scharff MD. H3 trimethyl K9 and H3 acetyl K9 chromatin modifications are associated with class switch recombination. Proc Natl Acad Sci U S A. 2009;106:5288–93. doi: 10.1073/pnas.0901368106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin, Ugine GD, Wu D, Young F, Chen J, Bottaro A. Normal isotype switching in B cells lacking the I mu exon splice donor site: evidence for multiple I mu-like germline transcripts. J Immunol. 2000;164:1451–7. doi: 10.4049/jimmunol.164.3.1451. [DOI] [PubMed] [Google Scholar]
- Li Z, Luo Z, Scharff MD. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc Natl Acad Sci U S A. 2004;101:15428–33. doi: 10.1073/pnas.0406827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby TM, Schrader CE, Stavnezer J, Selsing E. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J Exp Med. 2001;193:159–68. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Rothlein LR, Schrader CE, Stavnezer J, Selsing E. Shifts in targeting of class switch recombination sites in mice that lack mu switch region tandem repeats or Msh2. J Exp Med. 2005;201:1885–90. doi: 10.1084/jem.20042491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–82. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowatt MR, Dunnick WA. DNA sequence of the murine gamma 1 switch segment reveals novel structural elements. J Immunol. 1986;136:2674–83. [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–40. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Nakai S, Honjo T. Switch region of immunoglobulin Cmu gene is composed of simple tandem repetitive sequences. Nature. 1981;292:845–8. doi: 10.1038/292845a0. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Yamawaki-Kataoka Y, Honjo T. Nucleotide sequences of switch regions of immunoglobulin C epsilon and C gamma genes and their comparison. J Biol Chem. 1982;257:7322–9. [PubMed] [Google Scholar]
- Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, San-Martin BR, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci U S A. 2002;99:7003–8. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–44. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–8. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Mol Cell Biol. 2011;31:1503–11. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–12. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–68. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc Natl Acad Sci U S A. 2010;107:22190–5. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Radcliffe G, Lin YC, Nietupski J, Berggren L, Sitia R, Severinson E. Immunoglobulin heavy-chain switching may be directed by prior induction of transcripts from constant-region genes. Proc Natl Acad Sci U S A. 1988;85:7704–8. doi: 10.1073/pnas.85.20.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. Embo J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurek P, Petrini J, Dunnick W. Complete nucleotide sequence of the murine gamma 3 switch region and analysis of switch recombination sites in two gamma 3-expressing hybridomas. J Immunol. 1985;135:620–6. [PubMed] [Google Scholar]
- Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–72. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J Exp Med. 2006;203:215–26. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–30. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–89. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–22. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, Alt FW. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. Embo J. 1986;5:3259–66. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–51. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–81. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]