Abstract
Odorant receptors (Ors) are a unique family of ligand-gated ion channels and the primary mechanism by which insects detect volatile chemicals. Here, we describe 57 putative Ors sequenced from an antennal transcriptome of the cerambycid beetle Megacyllene caryae (Gahan). The male beetles produce a pheromone blend of nine components, and we functionally characterized Ors tuned to three of these chemicals: receptor McOr3 is sensitive to (S)-2-methyl-1-butanol; McOr20 is sensitive to (2S,3R)-2,3-hexanediol; and McOr5 is sensitive to 2-phenylethanol. McOr3 and McOr20 are also sensitive to structurally-related chemicals that are pheromones of other cerambycid beetles, suggesting that orthologous receptors may be present across many cerambycid species. These Ors are the first to be functionally characterized from any species of beetle and lay the groundwork for understanding the evolution of pheromones within the Cerambycidae.
Keywords: Cerambycidae, pheromone, chemoreceptor, transcriptome
1. Introduction
Chemoreception by insects is mediated by a diverse array of receptor proteins that includes the gustatory, odorant, and ionotropic receptors (Benton et al., 2009; Hallem et al., 2006; Rützler and Zwiebel, 2005). Among these groups, the family of odorant receptors (Ors) appears to be the primary mechanism by which insects detect volatile chemicals (Hallem and Carlson, 2006; Wang et al., 2010). This family is an expansion of seven-transmembrane domain proteins, unique to insects, which act as ligand-gated ion channels when bound to the conserved co-receptor Orco (Sato et al., 2008; Vosshall and Hansson, 2011; Wicher et al., 2008).
Ors have been described from several insect species to date, but because of their rapid evolution, annotation has remained almost exclusive to genome projects (e.g., Clyne et al., 1999; Engsontia et al., 2008; Robertson et al., 2010; Vosshall et al., 1999). As such, our understanding of Ors has been limited to the orders Diptera (Drosophila spp., species in the Culicidae), Lepidoptera (Bombyx mori [L.], Helicoverpa zea [Boddie]), and Hymenoptera (Apis mellifera L., Nasonia spp.; see Robertson et al., 2010 for a brief review). Moreover, the function of Ors remains poorly known for all but the model species Drosophila melanogaster Meigen and Anopheles gambiae Giles (Carey et al., 2010; Hallem and Carlson, 2006; Wang et al., 2010). Beetles (Coleoptera), comprise a quarter of known insect species, and hundreds of important pest species, but are represented by only a single set of olfactory receptors described from the genome of Tribolium castaneum (Herbst) (Engsontia et al., 2008). Tribolium castaneum presented a surprisingly diverse array of 341 Ors, of which 111 were detected as transcripts in adult head tissues (Engsontia et al., 2008). However, only the conserved Orco receptor (TcOr1) has been functionally characterized, and no ligands are yet known for any of the TcOrs.
The research summarized in this article advances our understanding of insect olfactory receptors by describing Ors of a second beetle species, and more importantly, by initiating the first functional analysis of coleopteran chemoreceptors. Recent advances in sequencing have resulted in high-throughput techniques that permit extensive genetic surveys of non-model organisms (Toth et al., 2007; Vera et al., 2008), and transcripts of highly expressed receptors may be captured by sequencing RNA from antennal tissues (Ramsdell et al., 2010; Grosse-Wilde et al., 2011). This process is well suited to the extraction of receptors that detect chemicals critical to the life history of the insect, such as host odors or pheromones. Ors for pheromones should be highly sensitive to a small set of chemicals that already have been identified, and thus may offer convenient targets for sequencing and characterization.
Our study species was the cerambycid beetle Megacyllene caryae (Gahan). The Cerambycidae constitute one of the largest insect families, with more than 35,000 described species (Grimaldi and Engel, 2005), and include some of the most destructive pests of forests worldwide (Haack et al., 2010). Larvae of most species feed in the woody tissues of plants, and adults of many species depend on sex pheromones to locate and recognize mates (e.g., Millar et al., 2009; Ray et al., 2011; Rodstein et al., 2009; Silk et al., 2007). Pheromone structures frequently are shared among congeners, and even among more distantly related species (i.e., tribes, or even subfamilies). Known pheromone structures include 2,3-alkanediols and the related 3-hydroxy-2-alkanones (Hanks et al., 2007; Lacey et al., 2009), (E)-6,10-dimethyl-5,9-undecadien-2-ol and the corresponding acetate (Mitchell et al., 2011; Silk et al., 2007), and 3,5-dimethyldodecanoic acid (Barbour et al., 2011). Male M. caryae produce an unusually complex blend for a species in its subfamily (Cerambycinae; see Millar et al., 2009), comprising several compounds that are better known as floral and wood volatiles (see Section 2.1), but also two compounds that are common pheromone components of c erambycids: 2,3-hexanediol and 2-methyl-1-butanol (Lacey et al., 2008; Mitchell, 2012). This diversity of components improved our chances of identifying pheromone receptors, and the commonplace nature of the ligands meant that receptor sequences might yield functionally similar orthologs in other cerambycid species. Here, we describe 57 likely odorant receptors identified from antennal tissue of M. caryae, and the functional characterization of three of those odorant receptors by demonstrating their highly selective responses to several components of the insect’s pheromone blend.
2. Materials and Methods
2.1. Sources of chemical standards and insects
The pheromone of male M. caryae consists of (2S,3R)- and (2R,3S)-2,3-hexanediol, (S)-(−) - limonene, 2-phenylethanol, (−)-α-terpineol, nerol, neral, geranial, and (S)-2-methylbutan-1-ol (Lacey et al., 2008; Mitchell, 2012). Authentic standards of (S)-(−) and (R)-(+)-limonene, 2-phenylethanol, (−)-α-terpineol, and nerol were obtained from Sigma-Aldrich (St. Louis, MO), and (S)-2-methylbutan-1-ol from TCI America (Portland, OR). Because neral and geranial are not available as pure isomers, and readily isomerize, we instead used citral (a ~1:1 mixture of neral and geranial; Sigma-Aldrich). (S)- and (R)-3-hydroxyhexan-2-one were synthesized according to Lacey et al. (2007), in 93.4 and 97.8% ee respectively. Authentic standards of (2R,3R)-, (2S,3S)-, (2S,3R)- and (2R,3S)-2,3-hexanediol and (R)-2-methylbutan-1-ol were not commercially available and so were synthesized as described in the Supplementary Materials.
Adult M. caryae were captured alive with black flight-intercept traps (Panel Trap model, AlphaScents, Portland, OR) during May 2009 at Forest Glen Preserve, Vermilion Co., IL (40°0'51.97"N, 87°34'0.74"W). Traps were baited with plastic polyethylene sachets (Bagettes™ model 14770, 5.1 × 7.6 cm, Cousin Corp., Largo, FL; see Graham et al., 2010) loaded with 50 mg of citral in 1 ml of 95% ethanol. Citral is a blend of the primary components of the pheromone (Lacey et al., 2008) and attracts males and females in similar numbers (Mitchell, 2012). Beetles were maintained on a diet of 10% sugar water for no more than 2 d before antennae were processed. We produced antennal transcriptomes from composite samples of 24 male and 21 female adult M. caryae, respectively. Subsequent assays used RNA extracted from antennae of an additional 10 males and 10 females captured during May 2009, and again in May of 2010 and 2011.
2.2. Isolation of total RNA and construction, sequencing, and assembly of library
Antennae of live adults were cut through the scape and immediately placed on dry ice, then homogenized in 1 ml TRIzol reagent with RNA extracted according to the manufacturer protocol (Invitrogen, Carlsbad, CA). Coupled gas chromatography-electroantennography revealed that antennae of both sexes respond in similar fashion to all components of the pheromone blend (Mitchell, 2012). We therefore combined RNA from males and females and the library was constructed from the pooled sample. Subsequent samples used for cloning were maintained as separate male and female stocks.
We provided 80 µg total antennal RNA to the Roy J. Carver Biotechnology Center High-Throughput Sequencing and Genotyping Unit of the W. M. Keck Center for Comparative and Functional Genomics (University of Illinois, Urbana, IL). Construction of a normalized cDNA library and 454 pyrosequencing were carried out at the Keck Center as described by Lambert et al. (2010). Assembly also was performed at the Keck Center using SeqMan Pro sequence assembly software (DNASTAR Inc., Madison, WI, USA).
2.3. Annotation of odorant receptor genes
We converted the assembled contigs into a searchable database using the formatdb tool of the BLAST+ toolkit (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/). Peptide sequences of Ors from T. castaneum (Engsontia et al., 2008) were used as BLAST queries (tBLASTn, National Center for Biotechnology Information; Altschul et al., 1997) to identify related genes in M. caryae. The encoded proteins of putative Ors were aligned with analogs from T. castaneum and raw reads from the library using CLUSTALX (Thompson et al., 1997) and, when necessary, manually extended or rebuilt from raw reads to create complete ORFs. Some ORFs could not be completed and were designated as “partial Ors”. We did not attempt to extend these sequences through other methods because receptors with low copy numbers were unlikely to be involved in pheromone detection.
Each complete cerambycid Or was used as an additional BLAST query to identify other receptor genes. We used the complete set of Ors described from D. melanogaster (Clyne et al., 1999, Vosshall et al., 1999) and Apis mellifera L. (Robertson et al., 2006) as final queries to search for any subfamilies of Ors not present in T. castaneum. The phylogenetic relationships among the Ors of M. caryae (including partial Ors of at least 200 amino acids) and Ors of T. castaneum were assessed through a distance matrix corrected using the BLOSUM62 exchange matrix in TREEPUZZLE v5.0 (Schmidt et al., 2002). A tree based on these corrected distances was obtained using heuristic search and tree-bisection-and-reconstruction branch swapping in PAUP* (v4.0b10; Swofford, 2001).
2.4. Identification and cloning of candidate pheromone receptors
We chose the initial set of candidate pheromone receptors as Ors that were represented by a high number of reads in the transcriptome, allowing us to build models with high confidence, and because genes with poor coverage were likely uncommon in the initial library and peripheral to our goal. Later candidates were chosen from clades that contained receptors that we had successfully characterized from the initial set.
We designed two pairs of primers to amplify each candidate pheromone receptor. The first pair amplified the exact ORF, whereas a second pair added overhangs with unique endonuclease sites (HindIII, EcoRI, BamH1, or XbaI) for unidirectional cloning. The PCR protocol for both stages was 2 min at 94 °C, 25 cycles of 94 °C for 1 min, 55–62 °C for 1 min (depending on primer TM), and 72 °C for 1 min, and a final incubation for 5 min at 72 °C. Product was digested for 1 h with two enzymes in the appropriate buffer (New England Biolabs, Inc., Beverly, MA) and gel purified (Gel Extraction Kit; Qiagen Corp., Chatsworth, CA). We cloned sequences into the vector pGEMHE using the NEB Quick Ligation Kit (New England Biolabs), and confirmed all clones by sequencing. We also identified and cloned the cerambycid ortholog of the Orco receptor, because odorant receptors of insects will not function in the absence of this protein (Sato et al., 2008; Vosshall and Hansson, 2011).
2.5. Expression of pheromone receptors in Xenopus oocytes
We characterized receptor function as described in earlier publications (Hughes et al., 2010; Wanner et al., 2007). Briefly, we synthesized capped cRNA in vitro from pGEMHE vectors encoding Ors with mMessage mMachine kits (Ambion, Austin, TX). The pGEMHE vector includes 3’ and 5’ UTR of a β-globin gene that facilitates high expression of receptors in oocytes of the frog Xenopus laevis (Daudin) (Liman et al., 1992). Oocytes were surgically removed from anesthetized frogs and follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem) for 2 h at room temperature. Oocytes were co-injected with cRNA of a candidate pheromone receptor and the Orco (McOr1), and incubated for 3–7 d before electrophysiological recording.
2.6. Initial screens for, and functional assay of pheromone receptors
Each receptor was tested against all components of the pheromone blend of M. caryae, as well as enantiomers and the following isomers of pheromone components: (2S,3S)- and (2R,3R)-2,3-hexanediol, (S)- and (R)-3-hydroxyhexan-2-one, (R)-(+)-limonene, and (R)-2-methylbutan-1-ol. We measured odorant-induced currents by two-electrode voltage clamp in an automated parallel electrophysiology system (OpusExpress 6000A; Molecular Devices, Union City, CA). Oocytes were bathed continuously with a buffered saline solution, ND96 (Hughes et al., 2010), and odorants were diluted in ND96 and applied to the oocyte for 20 s at 1.6 ml/min. Micropipettes were filled with 3 M KCl and had resistances of 0.2–2.0 MΩ. The holding potential was −70 mV. Current responses, filtered (4-pole, Bessel, low pass) at 20 Hz (−3 db) and sampled at 100 Hz, were captured and stored with OpusXpress 1.1 software (Molecular Devices, Sunnyvale, CA). We screened a range of concentrations (generally 1–3000 µM, half-log scale) for each odorant that activated a receptor. Data were fit to the following equation that models concentration-response (Prism 5, Graphpad Software, S an Diego, CA): I = Imax/(1+(EC50/X)n), where I = current response at a given concentration of odorant (X), Imax = maximal response; EC50 = concentration of odorant yielding a half-maximal response, and n is the apparent Hill coefficient.
3. Results
3.1. Identification of odorant receptors
The sequencing yielded 583 megabases in 1.56 million reads, with a mean fragment length of 404 bp. These data were assembled into 75,603 contigs with a mean length of 842 bp. We identified the Orco, 30 complete Ors, and 26 partial Ors by querying the database with Ors of T. castaneum and through iterative searches with newly identified cerambycid Ors. No further Ors were identified by queries with receptors of D. melanogaster or A. mellifera. Contigs containing Ors were relatively rare in the database and built from an average of 22 reads and a median of 11 reads (exclusive of the Orco, which was extensively covered by 627 reads). Nucleotide and protein sequences of the receptors are included in the Supplementary Materials.
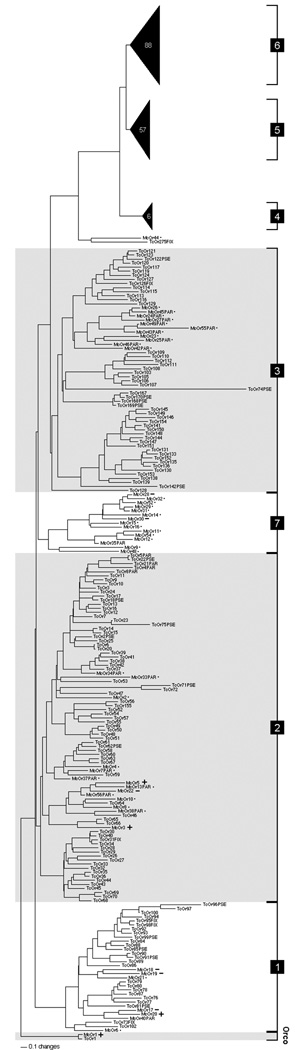
The majority of the expressed Ors of M. caryae were evenly placed among groups 1–3 of the six subfamilies described from T. castaneum (Engsontia et al., 2008; Figure 1). A single cerambycid receptor, McOr44, was placed alongside TcOr275 as an outgroup to groups 4–6. Additionally, a novel expansion of receptors (noted as group 7 on the tree) was recovered from M. caryae that showed no close relationship to any TcOrs. This clade contained almost half (14) of the complete Or transcripts in the library, including the two most common Ors in the library (McOr30 and McOr28). The tree was rooted with Orco proteins from M. caryae and T. castaneum (McOr1, TcOr1), which retained the characteristically high degree of conservation found in the Orco lineage.
Fig. 1.
Phylogram showing peptide sequences of odorant receptors from Tribolium castaneum (TcOrs) and Megacyllene caryae (McOrs). Partial sequences of at least 200 amino acids are included in the tree and denoted by the suffix “PAR”. Receptors are organized into groups 1–6 defined by Engsontia et al. (2008), but including a group described only from M. caryae (7). Receptors from Groups 4–6, not present in the antennal transcriptome of M. caryae, are represented as black triangles to indicate the number of omitted receptors. Receptors of M. caryae that are sensitive to pheromone components (and the Orco) are indicated by “+”, those insensitive to components are indicated by “−”, and those that were not tested (ligands unknown) are indicated by “•”. The tree is rooted with Orco proteins from both species (McOr1, TcOr1).
3.2. Characterization of pheromone receptors
We cloned nine putative odorant receptor genes of M. caryae into pGEMHE, as well as the Orco ortholog, and tested all clones for function in the Xenopus oocyte expression system. We first tested the five most common receptors in the library (McOr30, McOr28, McOr20, McOr3, and McOr19) and then included four additional candidates that were common in the library but also related to confirmed pheromone receptors from the initial group (McOr17, McOr18, McOr22, McOr5; Fig. 1). Unexpectedly, some receptors proved difficult to clone, and were recovered with numerous deletions relative to the gene model. Complete ORFs were eventually cloned from all candidate pheromone receptors.
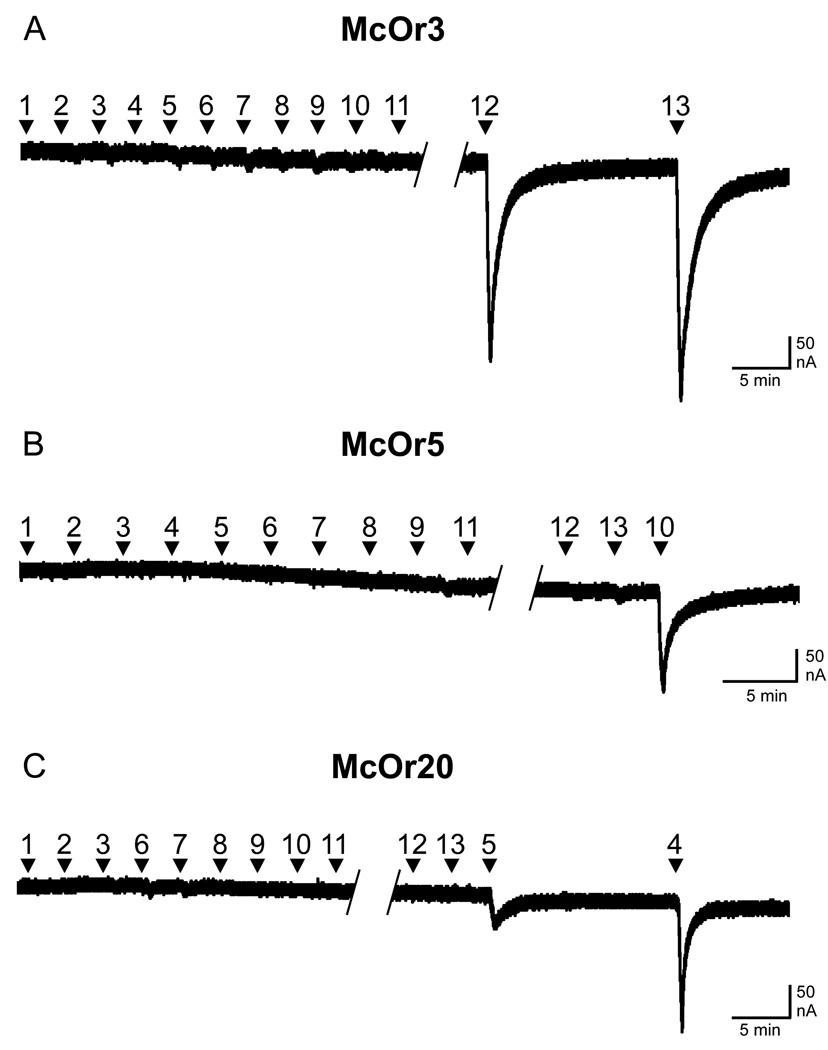
Two receptors from the initial group and one receptor from the subsequent group responded to components of the pheromone blend produced by M. caryae at a concentration of 30 µM (Figures 2–3). In the initial group, McOr3 responded with high sensitivity to both enantiomers of 2-methyl-1-butanol (numbers 12 and 13; Fig. 2A). McOr5, selected in the second group of receptors, was sensitive to 2-phenylethanol (10; Fig. 2B). McOr20 was sensitive to (R)-3-hydroxyhexan-2-one (5) and (2S,3R)-2,3-hexanediol (4; Fig. 2C). In a separate screen performed at 100 µM, this receptor also responded to (2R,3S)- and (2S,3S)-hexanediol (data not shown).
Fig. 2.
Change in electric current induced by candidate odorant receptors, expressed in Xenopus oocytes, in response to pheromone components of Megacyllene caryae. Frog oocytes that expressed receptors A) McOr3, B) McOr5, and C) McOr20 were sequentially challenged with 30 µM of each of the following pheromone components (applied for 20 s): 1. (2R,3R)-2,3-hexanediol, 2. (2S,3S)-2,3-hexanediol, 3. (2R,3S)-2,3-hexanediol, 4. (2S,3R)-2,3-hexanediol, 5. (R)-3-hydroxyhexan-2-one, 6. (S)-3-hydroxyhexan-2-one, 7. (S)-(−)-limonene, 8. (R)-(+)-limonene, 9. (−)-α-terpineol, 10. 2-phenylethanol, 11. citral, 12. (R)-2-methylbutan-1-ol, and 13. (S)-2-methylbutan-1-ol. Diagonal hatching indicates short breaks in recording.
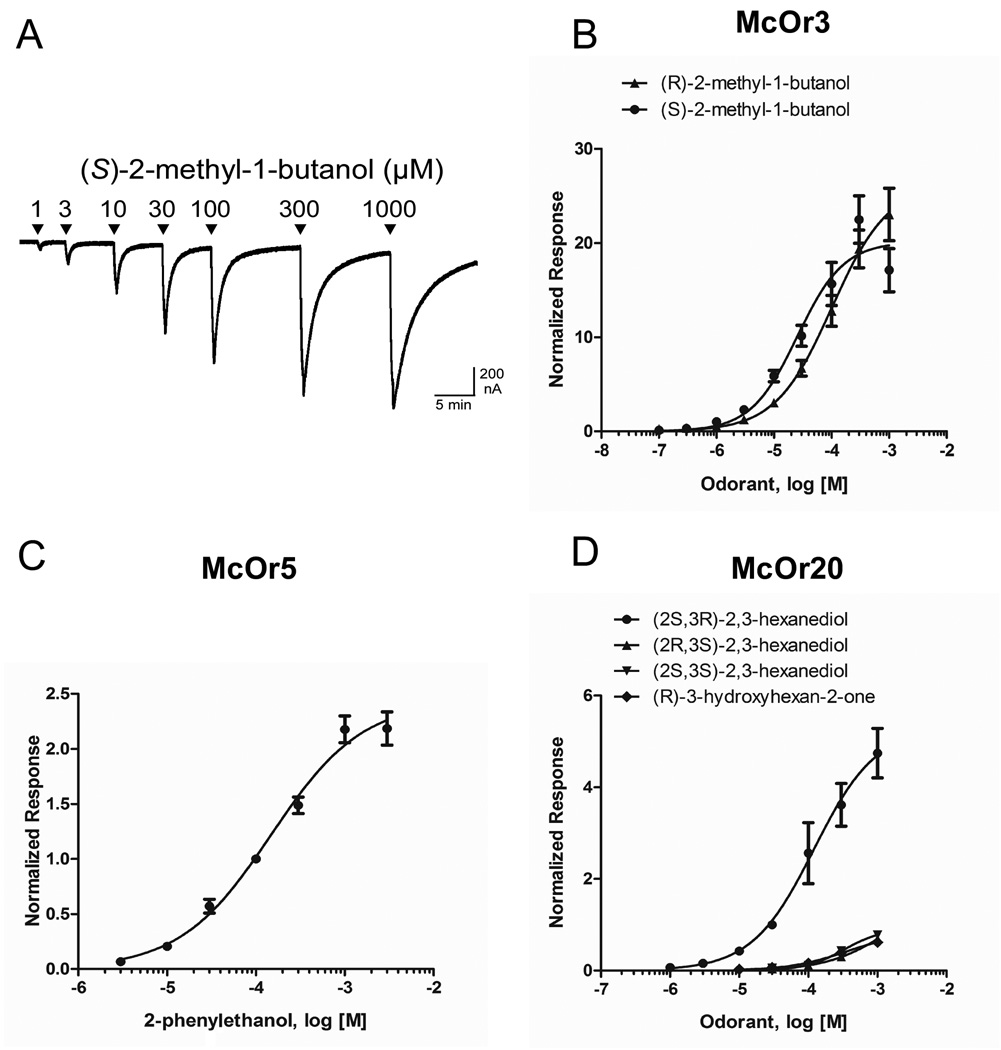
Fig. 3.
Relationships between concentrations of pheromone components and the responses of Xenopus oocytes that are expressing pheromone receptors of Megacyllene caryae. A) An example of a trace used to generate concentration-response curves (described in section 2.6): an oocyte expressing McOr3 is challenged with 20-s applications of (S)-2-methylbutan-1-ol at a range of doses. Concentration-response curves of B) McOr3 challenged with (R)- and (S)-2-methyl-1-butanol (normalized to the response of each ooc yte to 1 µM [S]-2-methylbutan-1-ol; n = 5–15); C) McOr5 challenged with 2-phenylethanol (normalized to 100 µM 2-phenylethanol; n = 6); D) McOr20 challenged with three isomers of 2,3-hexanediol and (R)-hydroxyhexan-2-one (normalized to 30 µM [2S,3R]-2,3-hexanediol; n = 5–7).
McOr3 was sensitive to (S)-2-methyl-1-butanol at a broad range of concentrations (Fig. 3A) with an EC50 of 25 µM (Fig. 3B, Table 1), and less sensitive to (R)-2-methyl-1-butanol at an EC50 of 100 µM (Fig. 3B, Table 1). McOr5 responded to 2-phenylethanol at an EC50 of 150 µM (Fig 3C, Table 1). Finally, McOr20 was sensitive to (2S,3R)-2,3-hexanediol at an EC50 of 120 µM, (R)-3-hydroxyhexan-2-one at an EC50 of 350 µM, and very slightly responsive to (2R,3S)- and (2S,3S)-hexanediol at EC50s 1430 µM and 330 µM, respectively (Fig. 3D, Table 1). None of the remaining six receptors responded to the chemicals over the concentration range tested in our assay.
Table 1.
Cerambycid pheromone components that were detected by odorant receptors of the cerambycid Megacyllene caryae. Receptors were expressed in oocytes of Xenopus laevus and tested against a panel of chemicals in a voltage clamp. Sensitivity is expressed as the concentration of odorant yielding a half-maximal response (EC50 ± SEM).
| Odorant Receptor | Ligand | EC50 (µM) |
|---|---|---|
| McOr3 | (S)-methylbutan-1-ol | 25±7.36 |
| (R)-methylbutan-1-ol | 100±36.3 | |
| McOr5 | 2-phenylethanol | 150±32.5 |
| McOr20 | (2S,3R)-2,3-hexanediol | 120±57.4 |
| (2R,3S)-2,3-hexanediol | 1430±1331 | |
| (2S,3S)-2,3-hexanediol | 330±77.2 | |
| (R)-3-hydroxyhexan-2-one | 350±154.6 |
4. Discussion
The odorant receptors we describe here are only the second published set of receptors from the Coleoptera and include the first Ors functionally characterized from any species of beetle. It is highly unlikely that we have described the full complement of active receptors from adult M. caryae, but the number of described receptors nevertheless compares favorably to those described from genomes of other insect species (Hill et al., 2002; Robertson et al., 2003) and permits a preliminary characterization of receptor evolution in beetles.
The minimal sequence similarity between receptors of T. castaneum and M. caryae is consistent with the overall family of olfactory receptors, which are remarkably divergent among insect groups (Robertson et al., 2010). In fact, the lineages of T. castaneum and M. caryae (Tenebrionoidea and Chrysomeloidea, respectively) are believed to have diverged between 220–236 Mya (Hunt et al., 2007), comparable to the timing of divergence between Drosophila and mosquitoes (i.e, the radiation of Nematocera; Wiegmann et al., 2011). It is thus unsurprising that the relationship between cerambycid and tenebrionid receptors follows a similar pattern of substantial gene loss and radiation (Hill et al., 2002). We did identify a small number of potentially orthologous pairs within the coleopteran receptors (McOr2/TcOr47, McOr8/TcOr64, McOr44/TcOr275) that may bind odorants important to both species. In fact, McOr3 is placed alongside the receptor pair TcOr65/66, suggesting these Tribolium receptors may bind 2-methyl-1-butanol or related chemicals. Such speculation is admittedly risky, given the rapid evolution of chemoreceptors, and receptors must be tested experimentally to verify ligands.
The apparent lack of cerambycid receptors among T. castaneum groups 4–6 supports the hypothesis that these large expansions are recent and specific to Tribolium, or at least to a tenebrionoid lineage (Engsontia et al., 2008). Conversely, the group 7 receptors are not present in Tribolium, and have not been identified from any other insect genome to date. Receptors tested from group 7 did not respond to any components of the pheromone blend, but the large number of Ors in this group suggests an important function in M. caryae, especially because most are strongly represented in the library. This family could instead be sensitive to host plant odors rather than pheromone components (Hanks, 1999; Ginzel and Hanks, 2005).
We were surprised by the high frequency of flawed transcripts characterized by apparent deletions, which were present in the library and recovered in cloned receptors from all pools of RNA used in our experiments. Deletions ranged from a dozen to hundreds of base pairs, and probably stemmed from changed or incorrect splicing of exons, but this mechanism can only be confirmed with a genome sequence. Some genes such as McOr11 were entirely crippled, with complete ORFs unrecoverable, whereas other McOrs were present at considerably reduced frequencies. It is possible that cerambycid Ors are regulated in some manner by alternative splicing, either changing or eliminating their functionality.
We characterized receptors for three pheromone components of M. caryae that were spread across two lineages of Ors (groups 1 and 2). This suggests that pheromone receptors of the Cerambycidae do not share a recent evolutionary origin, in contrast with the pheromone receptors of moths that apparently arose from a single lineage (e.g., Wanner et al., 2010). The most sensitive of the cerambycid receptors, McOr3, detected ligands at an EC50 of 25 µM, a high value relative to other reports (Wanner et al., 2007; Nakagawa et al., 2005), but not unusual for Ors tested in the Xenopus assay. For example, the odorant receptor AmOr11 of A. mellifera was sensitive to the queen substance 9-oxo-2-decenoic acid at an EC50 of 280 nM (Wanner et al., 2007), BmOr 1 of Bombyx mori L. was sensitive to the female-produced pheromone bombykol at 1.5 µM, and BmOr3 was sensitive to bombykal at 260 nM (Nakagawa et al., 2005). Receptors described from the moths Ostrinia nubilalis (Hübner) and Heliothis virescens (F.) responded to pheromone components at EC50 values ranging from 260 nM – 24 µM (Wanner et al., 2010; Wang et al., 2011). However, many of the recently characterized odorant receptors of A. gambiae responded to ligands at EC50 values in the tens or hundreds of µM (Wang et al., 2010). Regardless, the sensitivity of odorant receptors may be highly modulated by odorant binding proteins (Große-Wilde et al., 2007), which were not tested in the present study.
Two of the cerambycid pheromone receptors were sensitive to components that are produced by numerous cerambycid species (2-methyl-1-butanol and 2,3-hexanediol; Hanks et al., 2007; Lacey et al., 2009; RFM, unpub. data). Male M. caryae produce specific stereoisomers of these chemicals, which could impart species specificity to the signal, but the chiral specificity of the receptors was not as exact: McOr3 was responsive to (S)-2-methyl-1-butanol, but also to the unnatural (R)-enantiomer. Moreover, McOr20 was more narrowly tuned to a specific isomer, (2S,3R)-2,3-hexanediol, but also responded to two of the three other stereoisomers, as well as the structurally related (R)-3-hydroxyhexan-2-one. Synthesized standards of (2R,3S)- and (2S,3S)-2,3-hexanediol contained trace amounts of (2S,3R)-2,3-hexanediol, but standards of 3-hydroxyhexan-2-one were free of 2,3-hexanediol (see Supplementary Materials), and (R)-2-methyl-1-butanol was chirally pure, suggesting that the apparent flexibility of these receptors is not due to contamination.
Electroantennographic assays support the dual sensitivity of antennae to the enantiomers of 2-methyl-1-butanol, but antennae of males and females appear highly sensitive to all four isomers of 2,3-hexanediol (RFM, unpub. data). This finding suggests that McOr20 is not the primary receptor for other isomers of hexanediol, but rather that the low affinity for these isomers could be a vestige of an ancestral receptor for the hydroxyalkanones and diols that are common pheromone components in the family. In fact, McOr20 pairs with a similar receptor, McOr17 (45.3% identical peptide sequence; Fig. 1), although McOr17 appeared unresponsive to any isomer of 2,3-hexanediol or 3-hydroxy-2-hexanone.
Functional flexibility of receptors that detect pheromones is consistent with previous findings for the moth Ostrinia nubilalis (Hübner) (Wanner et al., 2010), and suggests that other cerambycids that produce isomers of 2-methyl-1-butanol and 2,3-hexanediol will detect these chemicals with receptors closely related, or orthologous to McOr3 and McOr20. If multiple species rely on a single lineage of similar pheromone receptors, it may be possible to create generic primers that can serve as markers for pheromone sensitivity, and these genes also may inform phylogenetic study of the family. Additionally, many insects produce or respond to 2-methyl-1-butanol (e.g., Matsuura et al., 2010; Pontes et al., 2008) and the sequence of McOr3 may inform study of these species as well. We are currently exploring these potential applications by searching for orthologs of McOr3 and McOr20 in other species of Megacyllene and closely related genera.
In summary, our research has proven that olfactory receptors can be identified through high-throughput sequencing of an antennal transcriptome, and verifies this method as a powerful and efficient means of describing Ors in lieu of a genome sequence. Key receptors for important odorants, such as pheromones, should be represented strongly in the resulting transcriptome and their function can be quickly characterized. It is our hope that this study will encourage research on other non-model species that will improve our understanding of the evolution of chemoreception within the Insecta.
Highlights.
57 odorant receptors sequenced from a longhorned beetle.
Highly expressed receptors tested for sensitivity to the beetle’s pheromone.
Three receptors responded to 2-methyl-1-butanol, 2,3-hexanediol, 2-phenylethanol.
First odorant receptors functionally characterized from a species of beetle.
Supplementary Material
Acknowledgments
We thank Kim Walden and Kevin Wanner for technical advice and assistance. This work was supported by USDA-NIFA grant 2008-35302-18815 (to HMR), USDA-NRI grant 2009-35302-05047 and USDA-APHIS # 10-8100-1422-CA (to LMH, JGM), NIH T32 NS007044 (supporting DTH), NIH R01 DC011091 (to CWL), The Alphawood Foundation (to LMH), and a Francis M. and Harlie M. Clark Research Support Grant (UIUC, to RFM). This research was in partial fulfillment of a doctoral degree for RFM from the University of Illinois at Urbana-Champaign.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour JD, Millar JG, Rodstein J, Ray AM, Alston DG, Rejzek M, Dutcher JD, Hanks LM. Synthetic 3,5-dimethyldodecanoic acid serves as a general attractant for multiple species of Prionus (Coleoptera: Cerambycidae) Ann. Entomol. Soc. Am. 2011;104:588–593. [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–72. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Engsontia P, Sanderson AP, Cobb M, Walden KKO, Robertson HM, Brown S. The red flour beetle's large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Bio. 2008;38:387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Ginzel MD, Hanks LM. Role of host plant volatiles in mate location for three species of longhorned beetles. J. Chem. Ecol. 2005;31:213–217. doi: 10.1007/s10886-005-6735-6. [DOI] [PubMed] [Google Scholar]
- Graham EE, Mitchell RF, Reagel PF, Barbour JD, Millar JG, Hanks LM. Treating panel traps with a fluoropolymer enhances their efficiency in capturing cerambycid beetles. J. Econ. Entomol. 2010;103:641–647. doi: 10.1603/ec10013. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. USA. 2011;108:7449–7454. doi: 10.1073/pnas.1017963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Große-Wilde E, Gohl T, Bouché E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- Haack RA, Herard F, Sun J, Turgeon JJ. Managing invasive populations of Asian longhorned beetle and citrus longhorned beetle: A worldwide perspective. Annu. Rev. Entomol. 2010;55:521–546. doi: 10.1146/annurev-ento-112408-085427. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu. Rev. Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hanks LM. Influence of the larval host plant on reproductive strategies of cerambycid beetles. Annu. Rev. Entomol. 1999;44:483–505. doi: 10.1146/annurev.ento.44.1.483. [DOI] [PubMed] [Google Scholar]
- Hanks LM, Millar JG, Moreira JA, Barbour JD, Lacey ES, McElfresh JS, Reuter FR, Ray AM. Using generic pheromone lures to expedite identification of aggregation pheromones for the cerambycid beetles Xylotrechus nauticus, Phymatodes lecontei and Neoclytus modestus modestus. J. Chem. Ecol. 2007;33:889–907. doi: 10.1007/s10886-007-9275-4. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;289:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J. Chem. Ecol. 2010;36:797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, St. John O, Wild R, Hammond P, Ahrens D, Balke M, Caterino MS, Gómez-Zurita J, Ribera I, Barraclough TG, Bocakova M, Bocak L, Vogler A. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- Lacey ES, Moreira JA, Millar JG, Ray AM, Hanks LM. Male-produced aggregation pheromone of the cerambycid beetle Neoclytus mucronatus mucronatus. Entomol. Exp. App. 2007;122:171–179. [Google Scholar]
- Lacey ES, Moreira JA, Millar JG, Hanks LM. A male-produced aggregation pheromone blend consisting of alkanediols, terpenoids, and an aromatic alcohol from the cerambycid beetle Megacyllene caryae. J. Chem. Ecol. 2008;34:408–417. doi: 10.1007/s10886-008-9425-3. [DOI] [PubMed] [Google Scholar]
- Lacey ES, Millar JG, Moreira JA, Hanks LM. Male-produced aggregation pheromones of the cerambycid beetles Xylotrechus colonus and Sarosesthes fulminans. J. Chem. Ecol. 2009;35:733–740. doi: 10.1007/s10886-009-9633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JD, Chan XY, Spiecker B, Sweet HC. Characterizing the embryonic transcriptome of the snail Ilyanassa. Integr. Comp. Biol. 2010;50:768–777. doi: 10.1093/icb/icq121. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Himuro C, Yokoi T, Yamamoto Y, Vargo EL, Keller L. Identification of a pheromone regulating caste differentiation in termites. Proc. Natl. Acad. Sci. USA. 2010;107:12963–12968. doi: 10.1073/pnas.1004675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JG, Hanks LM, Moreira JA, Barbour JD, Lacey ES. Pheromone chemistry of cerambycid beetles. In: Nakamuta K, Millar JG, editors. Chemical Ecology of Wood-Boring Insects. Japan: Forestry and Forest Products Research Institute, Ibaraki; 2009. pp. 52–79. [Google Scholar]
- Mitchell RF, Graham EE, Wong JCH, Reagel PF, Striman BL, Hughes GP, Paschen MA, Ginzel MD, Millar JG, Hanks LM. Fuscumol and fuscumol acetate are general attractants for many species of cerambycid beetles in the subfamily Lamiinae. Entomol. Exp. Appl. 2011;141:71–77. [Google Scholar]
- Mitchell RF. Ph.D. Dissertation. Urbana, IL: University of Illinois at Urbana-Champaign; 2012. Chemical communication in cerambycid beetles and the molecular basis of olfaction. [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- Pontes G, Bohman B, Unelius R, Lorenzo MG. Metasternal gland volatiles and sexual communication in the triatomine bug, Rhodnius prolixus. J. Chem. Ecol. 2008;34:450–457. doi: 10.1007/s10886-008-9431-5. [DOI] [PubMed] [Google Scholar]
- Ramsdell KM, Lyons-Sobaski SA, Robertson HM, Walden KKO, Feder JL, Wanner K, Berlocher SH. Expressed sequence tags from cephalic chemosensory organs of the northern walnut husk fly, Rhagoletis suavis, including a putative canonical odorant receptor. J. Insect Sci. 2010:10.51. doi: 10.1673/031.010.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray AM, Žunič A, Alten RL, McElfresh JS, Hanks LM, Millar JG. cis-Vaccenyl acetate, a sex attractant pheromone of Ortholeptura valida, a longhorned beetle in the subfamily Lepturinae. J. Chem. Ecol. 2011;37:173–178. doi: 10.1007/s10886-011-9908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Gadau J, Wanner KW. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol. Biol. 2010;19:121–136. doi: 10.1111/j.1365-2583.2009.00979.x. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodstein J, Millar JG, Barbour JD, McElfresh JS, Wright IM, Barbour KS, Ray AM, Hanks LM. Determination of the relative and absolute configurations of the female-produced sex pheromone of the cerambycid beetle Prionus californicus. J. Chem. Ecol. 2011;37:114–124. doi: 10.1007/s10886-010-9890-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rützler M, Zwiebel LJ. Molecular biology of insect olfaction: recent progress and conceptual models. J. Comp. Physiol. A. 2005;191:777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1007. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Silk PJ, Sweeney JD, Wu J, Price J, Gutowski JM, Kettela EG. Evidence for a male-produced pheromone in Tetropium fuscum (F.) and Tetropium cinnamopterum (Kirby) (Coleoptera: Cerambycidae) Naturwissenschaften. 2007;94:697–701. doi: 10.1007/s00114-007-0244-0. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis using Parsimony and other Methods, Version 4. Cambridge: Sinauer Press; 2001. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL−X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AL, Carala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME, Robinson GE. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007;318:441–333. doi: 10.1126/science.1146647. [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Hansson BS. A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses. 2011;36:497–498. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Vásquez GM, Schal C, Zwiebel LJ, Gould F. Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 2011;20:125–133. doi: 10.1111/j.1365-2583.2010.01045.x. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Nichols AS, Walden KKO, Brockmann A, Luetje CW, Robertson HM. A honey bee odorant receptor for the queen substance 9-oxo-2-decanoic acid. Proc. Natl. Acad. Sci. USA. 2007;104:14383–14388. doi: 10.1073/pnas.0705459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner KW, Nichols AS, Allen JE, Bunger PL, Garcynski SF, Linn CE, Robertson HM, Luetje CW. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE. 2010;5:e8685. doi: 10.1371/journal.pone.0008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1012. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, Wheeler BM, Peterson KJ, Pape T, Sinclair BJ, Skevington JH, Blagoderov V, Caravas J, Kutty SN, Schmidt-Ott U, Kampmeier GE, Thompson FC, Grimaldi DA, Beckenbach AT, Courtney GW, Friedrich M, Meier R, Yeates DK. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.