Abstract
E2A (TCF3) is a multifunctional basic helix loop helix (bHLH), transcription factor. E2A regulates transcription of target genes by homo- or heterodimerization with cell specific bHLH proteins. In general, E2A promotes cell differentiation, acts as a negative regulator of cell proliferation in normal cells and cancer cell lines and is required for normal B-cell development. Given the diverse biological pathways regulated/ influenced by E2A little is known about its expression in cancer. In this study we investigated the expression of E2A in prostate cancer. Unexpectedly, E2A immuno-histochemistry demonstrated increased E2A expression in prostate cancer as compared to normal prostate. Silencing of E2A in prostate cancer cells DU145 and PC3 led to a significant reduction in proliferation due to G1 arrest that was in part mediated by increased CDKN1A(p21) and decreased Id1, Id3 and c-myc. E2A silencing in prostate cancer cell lines also resulted in increased apoptosis due to increased mitochondrial permeability and caspase 3/7 activation. Moreover, silencing of E2A increased sensitivity to doxorubicin induced apoptosis. Based on our results, we propose that E2A could be an upstream regulator of Id1 and c-Myc which are highly expressed in prostate cancer. These results for the first time demonstrate that E2A could in fact acts as a tumor promoter at least in prostate cancer.
Keywords: Prostate, cancer, bHLH, E2A, TCF3
INTRODUCTION
E2A (TCF3) belongs to the larger basic helix loop helix (bHLH) family of transcription factors. The conserved bHLH domain is involved in homo- or hetero-dimerization to form a functional transcriptional unit that binds to the canonical E- Box response element (CANNTG) found in the promoter of many genes [1].
E2A gene codes for two alternatively spliced transcription factors E12 and E47 [2]. E12 and E47 primarily serve as partners for dimerization with tissue specific bHLH proteins such as MyoD, NeuroD and MASH to promote cell specific differentiation [1]. E2A is regulated primarily at post-transcriptional level through protein-protein interactions: interaction of E2A with dominant negative HLH proteins, the inhibitor of differentiation family (Id) appears to be the primary mechanism that regulates E2A activity [3]. The Id proteins neutralize E2A through HLH mediated dimerization; however the lack of the basic domain in Id proteins renders the dimer transcriptionally inactive. The HLH domain of E2A also interacts with non-bHLH proteins (see supplemental Fig. 1 for known E2A interaction partners) such as ubiquitin-conjugating enzyme UBC9/UbcE2A and histone acetyl transferases: p300, CBP and PCAF. The interaction with UBC9 is required for ubiquitination and subsequent proteosomal degradation of E2A [4]. E2A proteins also extend the half-life of its primary interaction partners, the Id and MyoD proteins [5].
E2A is essential for lymphopoiesis as reveled in E2A null mice [6]. Evidence suggests that E2A proteins act as general negative regulator of cell proliferation in several normal cells and cancer cell lines [7,8,9]. The growth inhibition by E2A occurs at multiple levels involving both bHLH dependent and independent mechanisms. Primary among these are the transcriptional up-regulation of multiple cyclin dependent kinase inhibitors CDKN1A (p21), p15INK4B and p16INK4B [7,8,9,10]. Ectopic expression of E2A also promotes apoptosis in E2A deficient lymphomas, independent of an arrest in cell-cycle progression [7,8].
Contrary to its well established role as an inhibitor of proliferation, E2A expression is also observed in cells undergoing rapid proliferation in the rat embryo [11] and in proliferating periventricular neuroepithelial cells in the developing brain and in centroblasts within germinal centers [12]. E2A also promotes epithelial to mesenchymal transitions due to direct inhibition of E-cadherin expression at the promoter level [13], a mechanism central to cancer progression.
During our investigations to understand the mechanism of action of Id proteins in prostate cancer (PCa), we observed that E2A expression was low to negligible in PCa cell line LNCaP [14] but high in two aggressive PCa cell lines DU145 and PC3. These preliminary observations prompted us to investigate the association of E2A with PCa. Data mining demonstrated that E2A expression directly correlated with PCa. These observations were validated by E2A immuno-histochemical analysis in PCa specimens in this study. At the molecular level, ablation of E2A leads to apoptosis and G1 arrest dependent proliferation block in PCa cell lines. Taken together, our results demonstrate for the first time that E2A acts as a potential tumor promoter which is contrary to its well established role as a tumor suppressor.
MATERIALS AND METHODS
Cell culture and E2A Gene silencing
Human PCa cell lines DU145 and PC3 were obtained from ATCC (Rockville, MD) and were cultured as described previously [15]. The expression of E2A in PCa cell lines was temporarily silenced using E2A siRNA (sc-35245, Santa Cruz Biotechnology). The non-silencing control siRNA (siRNA-A: sc-37007, Santa Cruz Biotechnology) or un-transfected cells were used as controls. Transient transfections (Fugene) with siRNA were performed according to the manufacturers' recommendations on approximately 3 × 105 PCa cells in six-well plates. All the analysis including gene expression studies were performed 48–72 h after transfection.
Proliferation Assay
Cell proliferation was determined using CyQUANT NF Assay (Molecular Probes) according to the manufacturer's protocol. Briefly, PCa cells/well in 96-well plate (in triplicates, 50% confluency) were transiently transfected with E2A or non-silencing (NS) control siRNA. Hundred µl of green-fluorescent CyQUANT NF dye was added to the wells after a media change at 48hrs and incubated for 30 min at 37°C. The fluorescence intensity was then measured using BioTek Synergy HT fluorescence reader with excitation at 485nm and emission at 530nm.
Cell Cycle analysis and Apoptosis Assay
Cell cycle distribution was determined by staining DNA with propidium iodide (PI, Calbiochem). Briefly, transiently E2A silenced PCa cells were fixed in 70% ethanol and cell pellets were suspended in PI at 37° C for 30 min. For apoptosis assay, the cells were washed and then stained with PI and Alexa Fluor 488 conjugated Annexin V (Molecular Probes). The PI or PI+Annexin V cells were analyzed with Accuri C6 flow cytometer.
Measurement of caspase-3/7 activation and mitochondrial membrane potential
Caspase-3/7 activation and changes in mitochondrial membrane potential were detected using the dualsensor MitoCasp (Cell Technology), according to the manufacturers’ protocol. Stained cells were quantitated in Accuri C6 cytometer.
RT-PCR and Real Time RT-PCR
RNA (2 µg) was reverse transcribed in a final volume of 25 µl as per standard protocols [15]. Reverse transcribed RNA from cells was used for real time quantitative PCR based on the TaqMan chemistry using gene specific primers. The validated real time PCR probes for respective genes were obtained from Applied Biosystems. Amplification of target sequences was detected with ABI7900HT sequence detection system (Applied Biosystems, Foster City, CA). The cycle threshold (Ct) was used to calculate relative amounts of target mRNA. Standard reverse transcribed polymerase chain reaction was performed using gene specific primers (not shown).
Prostate tissue Microarray Slides
E2A expression was investigated in two different prostate tissue microarray slides obtained from Cybrdi Inc. (MD): Slide 1 (Cat # CC19-11, Lot # CC19-11-001) – consisted of 30 core biopsies representing 11 individual patient samples (3 Benign Prostatic Hyperplasia (BPH), 3 adenocarcinoma and 5 normal prostate) and Slide 2 (Cat # CC19- 11, Lot# CC19-11-007) consisted of core biopsies in duplicate from 40 individual patients (34 cases of Cancer, 1 case of adjacent normal and PCa and 5 cases of normal/ benign prostate tissue). The histo-pathology for all samples was pathologist certified and PCNA (slide 1) or PSA (slide 2) confirmed for immuno-histochemistry and stage.
Immuno-histochemistry (IHC) of Tissue Microarray slides
Tissue microarray slides were processed through standard protocols. Following antigen retrieval (autoclave in 0.01 M sodium citrate buffer pH 6.0 at 121C/20psi for 30min), the peroxidase activity was blocked in 3% H2O2 and non-specific binding sites blocked in 10% Goat serum. The blocked sections were incubated overnight at 4C with E2A antibody (sc-349, Santa Cruz Biotechnology) followed by incubation with secondary antibody (SA1-9510, HRP- goat anti-rabbit, Thermo Scientific) for 1 hour. The slides were stained with DAB for 2 min, counterstained with hematoxylin and mounted with Immuno-mount (Thermo Scientific), examined and photo-micrographs taken using the Zeiss microscope with an AxoimCam version 4.5 imaging system.
Data Analysis
The flow cytometry data was analyzed with CFlow Plus (Accuri Cytometers, Inc.) and FlowJo (Tree Star Inc.) softwares. Student’s t-test was used to compare gene expression data. The non-parametric Kruskal-Wallis test (distribution free) followed by post-hoc Dunn’s test was used to compare the E2A expression data in tissue microarray studies.
RESULTS
E2A expression in Prostate Cancer Tissue
The E2A antibody used in this study was extensively validated using cell lysates from PCa cell lines [14] (supplemental Fig. 2). These results confirmed that the antibody is mono-specific for E2A.
The age matched normal (65.9± 3.46 years, range: 36–78 years) and cancer samples (65.44±2.05 years. n=38, range: 20–82 years) used for E2A expression analyses were as follows: 11 grade I, 13 grade II, 2 grade III and 6 Grade IV, and 6 samples with Gleason Score (G+S) between 4–9. Samples with G+S=9 were assigned grade IV and those with less than 9 were assigned as grade III for statistical analysis.
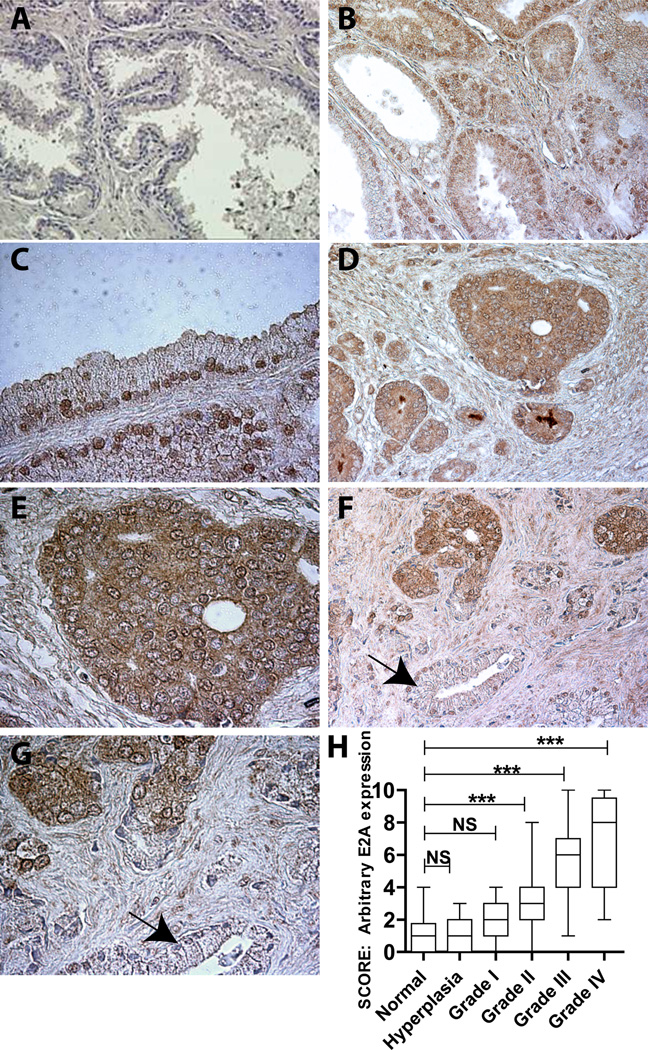
The E2A immuno-reactivity was low to undetectable in the majority of ductal epithelium of the normal/ BPH (11/14) prostate or normal adjacent prostate tissue (Fig. 1A). Prostate lobe specific analysis (2 triplicates from posterior lobe and 3 triplicates middle lobe) also indicated the lack of E2A expression (data not shown). The E2A expression pattern in BPH was similar to that of the normal tissue (data not shown). Unlike normal/BPH samples, high E2A expression was observed in PCa. E2A expression was primarily nuclear in Grade I PCa (Fig. 1B and C) but weak cytoplasmic expression, was also observed (Fig. 1B and C). The cytoplasmic staining became more pronounced with increasing grade (Fig. 1D thru G). Interestingly, E2A staining was absent in adjacent normal tubules (Fig. 1F and H, indicated by arrow). These results further support the observations that increased E2A expression is a specific cancer associated event.
Figure 1.
Prostate cancer tissue microarrays were used to investigate E2A expression by IHC. E2A expression (brown staining) was negligible in normal prostate tissue (A, 200x). E2A expression increased with increasing grade of prostate cancer (B: Grade II, D: grade III). B (200x) and C (400x): Grade I human prostate adenocarcinoma showing prominent nuclear localization of E2A. D (200x) and E (400x): Grade III human prostate adenocarcinoma showing higher E2A expression compared to Grade II with cytoplasmic localization. F (200x) and G (400x): Comparison of E2A expression between adenocarcinoma and adjacent normal prostate tissue (arrow). H: Semi-quantitative analysis of E2A expression in prostate cancer. No significant (NS) difference between E2A expression in normal vs. BPH and normal versus grade I was observed. Significant differences were observed when normal samples where compared with Grade II, Grade III and Grade IV (P<0.001: ***).
The E2A expression intensity in PCa specimens was scored by two independent observers (correlation coefficient r=0.94) and graded from 0-10 on a linear scale (0: Absent, 10: highest) and subjected to non-parametric statistical tests. A significant difference in the median between 6 groups was observed (at P<0.0001, Kruskal-Wallis test statistics H=80.41, Fig. 1H). The post hoc Dunn’s test also indicated a progressive and significant increase in E2A expression in various grades of PCa (Fig. 1H) compared to normal/BPH specimens.
Transcriptome wide analysis based on NCBI GEO datasets also indicated increased E2A expression PCa: GDS1439 (probes 213811_x_at, 210776_x_at, 215260_s_at, 209152_s_at, 209152_s_at and 213730_x_at targeting a unique region of the E2A gene) and GDS2545 (probe 1374_g_at). Collectively, our immuno-histochemical and public databases support increased E2A expression in PCa.
E2A silencing blocks proliferation
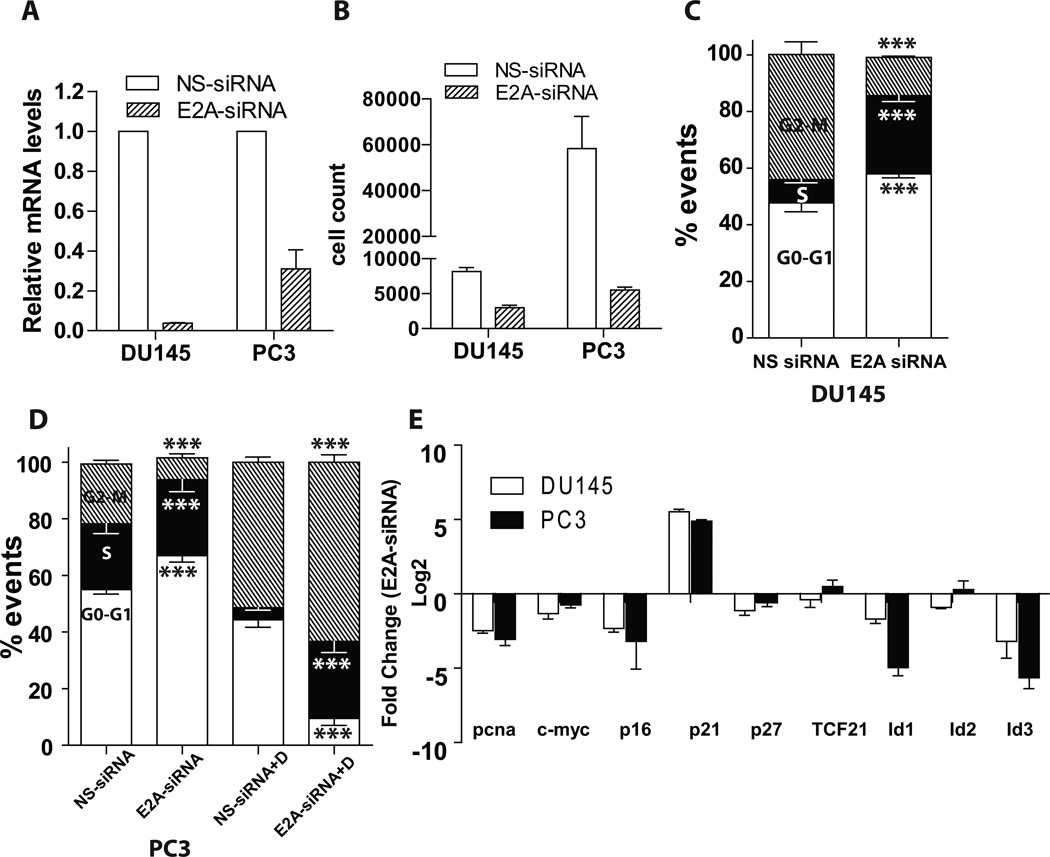
SiRNA based silencing (E2A-siRNA) led to a significant decrease in E2A transcript (Fig. 2A) in DU145 (25 fold) and PC3 (4 fold) cells as compared to non-silencing controls (NS-siRNA) as determined by qtPCR. Both these cell lines constitutively express E2A [14].
Figure 2.
E2A silencing represses prostate cancer cell proliferation. (A): E2A silencing (E2A-siRNA) was determined by real-time quantitative RT-PCR in human prostate cancer cell lines DU145 and PC3 and compared to cells transfected with non-specific siRNA (NS-siRNA). The values are relative to GAPDH. (B): E2A was silenced in DU145 and PC3 and cell proliferation was determined by CyQUANT cell proliferation assay. Each sample was assayed in triplicate in two biologically independent experiments. C and D: DU145 and PC3 cells, respectively, were silenced with E2A siRNA, fixed, and stained with PI and DNA content was determined by flow cytometer. Histogram shows the percentage of cells in G0-G1, S, and G2-M phases. The figure depicts the result of three biologically independent experiments. (E): DU145 and PC3 cells were silenced with E2A siRNA and quantitative real time PCR was performed on putative E2A target genes. The data in log2 scale reflects decrease in expression as negative and increase as positive as compared to non-silencing controls. The data represents mean+SEM of three replicates from three different experiments. Except TCF21 and Id2 (in PC3 cells) all changes were statistically significant at P<0.001.
Silencing of E2A led to significant reduction in the proliferation of DU145 and PC3 cells (Fig. 2B). Upon E2A silencing, the fraction of cells in G1 phase increased significantly as compared to controls (Fig. 2C and D). The increase in fraction of cells in G1 phase was associated with a corresponding decrease in cells in G2/M phase in both cell lines with E2A siRNA. The results suggest a G1 arrest in E2A-siRNA transfected cells that was also accompanied by decrease in the proliferation marker PCNA (Fig. 2E).
To uncover the potential mechanism of G1 arrest in E2A-siRNA cells, we investigated the expression of cyclin dependent kinase inhibitors CDKN2A (p16), CDKN1A (p21) and CDKN1B (p27) by qtPCR. As expected, loss of E2A was associated with an increase in the expression of CDKN1A (Fig. 2E). Surprisingly, CDKN2A and CDKN1B expression decreased in PCa cells following E2A silencing. E2A transcriptionally regulates CDKN2A and CDKN1B by binding to E box sequence in their proximal promoter. Thus, down-regulation of CDKN2A and CDKN1B confirms successful silencing of E2A in PCa cells but is counter-intuitive to its role as a tumor promoter. Increase in CDKN1A in E2A silenced cells though consistent with cell cycle data is unexpected because like CDKN2A and CDKN1B, it is also an E2A transcriptional target [8].
CDKN1A is one of the major targets of c-Myc repression. A significant decrease in c-Myc expression following E2A silencing (Fig. 2E) could lead to increased CDKN1A expression and G1 arrest in DU145 and PC3 cells. The G1 arrest in E2A-siRNA PCa cells was also associated with a decrease in dominant negative HLH and E2A dimerization partners Id1 and Id3. Both Id1 and Id3 regulate and integrate multiple cell cycle regulatory pathways notably involving c-Myc that promote G1/S and G2/M progression.
E2A silencing promotes apoptosis
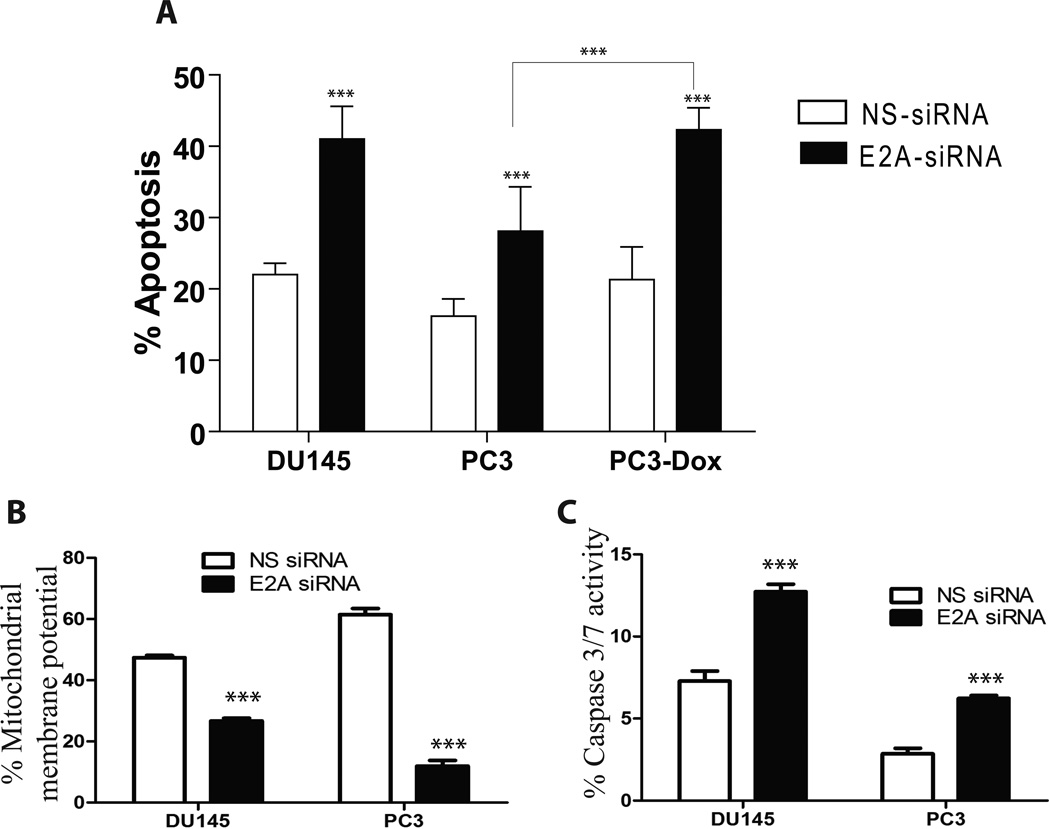
Silencing of E2A in PCa cell lines significantly increased apoptosis (Fig. 3A and supplemental Fig. 3) associated with increased cytochrome c release (Fig. 3B). Consistent with decreased mitochondrial potential (increased cytochrome c release), a significant increase in caspase 3/7 activity was observed in E2A-siRNA PCa cells (Fig. 3C and supplemental figure 3). A clear correlation between apoptosis and its mediators between E2A silenced DU145 and PC3 cells was however not observed, possibly due to number of altered signaling and apoptosis effector molecules between DU145 and PC3 cells (e.g. PC3 are null for p53 and DU145 harbor mutant p53).
Figure 3.
A: Effect of E2A on apoptosis in prostate cancer cell lines DU145 and PC3. The data shows the effect of NS-siRNA and E2A-siRNA on apoptosis in DU145 and PC3 cells after staining the cells with Annexin V–Alexa 488 and PI. The graph represents mean+SEM from three replicates and at least three independent experiments. The figure also demonstrates the effect of 500nm doxorubicin on PC3 cells transfected with either NS-siRNA or E2A-siRNA. Apoptosis in PC3 cells treated with Doxorubicin (500nm) was not statistically different from untreated cells whereas a significant increase was observed in PC3 cells transfected with E2A-siRNA. (***: P<0.001 by student’s t-test). B and C: E2A regulates apoptosis through cytochrome c (B) and caspase 3/7 (C) dependent pathways. The cells were stained with a cationic dye for detection of mitochondrial membrane potential (MMP) and a carboxyfluorescein (FAM) labeled fluoromethyl ketone (FMK) -peptide inhibitors of caspases and quantitated with flow cytometer. Values (mean+SEM) that are statistically significant to p < 0.001 are indicated with asterisk (***, student’s t-test: duplicates from three different experiments)
E2A silencing enhances sensitivity to Doxorubicin
Based on the dose response curve (0.1nm–10um, data not shown), we selected 500nm doxorubicin as the minimum dose that did not significantly increased apoptosis over a 24hr treatment period in PC3 and DU145 cells as compared to untreated cells (PC3 results shown in Fig. 3A, DU145 results not shown but essentially similar to PC3). However at this dose a typical doxorubicin dependent G2/M arrest was observed (Fig. 2D). In the absence of E2A, doxorubicin induced a significant increase in apoptosis as compared to cells silenced only with E2A-siRNA (Fig. 3A). These results clearly demonstrated that E2A prevents cells against doxorubicin induced cell death and that lack of E2A promotes sensitivity to doxorubicin. Collectively, our results provide direct evidence that E2A protects PCa cells against apoptosis and promotes proliferation.
DISCUSSION
E2A is a multi-functional transcription factor that interacts and modulates the activity of a large repertoire of proteins including transcription factors (supplemental Fig. 1). Dimerization with E2A is also essential for bHLH transcription factor activity. In general, E2A acts as a general growth inhibitor, however a positive correlation between E2A expression and PCa as demonstrated in this study, supports its pro-tumor role in PCa.
Low to undetectable E2A in the normal human prostate in our study and the transcriptome wide expression analysis is striking. Restricted E2A expression is also observed in human pituitary, kidney, adrenals and liver [12]. Lack of E2A in some tissues such as prostate suggest that bHLH transcription factors or at least those involving E2A as a hetero/homo-dimer have negligible role in maintaining normal prostate function.
Our immuno-localization studies overwhelmingly demonstrate that E2A expression is associated with increasing grade of PCa. These results are consistent with transcriptome wide profiling studies. The lack of E2A expression within the epithelium of the normal, adjacent normal and BPH further suggests that increased E2A is a cancer specific event and not associated with benign proliferative disease. These observations are noteworthy in context of reports that show E2A expression in populations of cells associated with high proliferative activity such as B cells. The effect of E2A on proliferation is cell-lineage dependent and involve participation of different dimerization partners. For example, whereas heterodimerization between E2A and myogenic bHLH proteins is anti-proliferative in fibroblasts [9], formation of E47 homodimers in B cells promotes proliferation [16] based on increased cyclin D2/ D3 expression [17] and decreased Cdk6 activity [18]. In fact Zhao et al [17] demonstrated that suppression of E47 decreased cell proliferation while its induction promoted cell proliferation of serum deprived B-cells and non B-cells.
Decrease in proliferation and G1 arrest in E2A ablated PCa cells are inconsistent with the majority of observations that support the role of E2A as a negative regulator of cell proliferation in normal, immortalized and cancer cell lines [10]. The tumor-suppressor activity of E2A is mediated at least in part by promoting E-Box dependent expression of CDKN2A and CDKN1B. Thus down-regulation of CDKN2A and CDKN1B in absence of E2A supports this regulatory pathway but not consistent with the G1 arrest. We speculate that low levels of CDKN2A and CDKN1B could promote the assembly of cyclinD-cdk4 complex for efficient cell cycle progression. The down-regulation of Id1 and Id3 in PCa E2A suppression model is also a novel observation. Id1 and to a lesser extent Id3 are known tumor promoters in PCa [15,19] and are required for G1 progression [9]. Mechanistically, the E2A-Id1 cross-talk appears to be transcriptional but lack of E2A could also promote proteasome mediated degradation of Id1/Id3 [5] that could further block G1 progression [9]. We are currently testing the hypothesis that cytoplasmic expression of E2A in higher grade PCa could in-fact protects degradation of Id1/Id3.
The increase in CDKN1A following E2A-siRNA PCa cells provides a possible mechanism for cell cycle arrest. Similar to CDKN2A and CDKN1B, CDKN1A is also an E2A transcriptional target but its regulation appears to be cell specific. Unlike CDKN2A and CDKN1B, CDKN1A does not appear to be a direct E2A transcriptional target at least in PCa cells. Instead, CDKN1A could be regulated by c-Myc: c-Myc is a transcriptional repressor of CDKN1A and promotes proliferation [20] and induce or sensitize cells to apoptosis [21]. Thus a decrease in c-Myc expression in the E2A silenced cells tends to support our observations.
The caspase 3/7 dependent apoptosis due to increased mitochondrial permeability is consistent with reports demonstrating spontaneous apoptosis in CLL following E2A silencing [22] and alternatively, protection of apoptosis in B- and non-B cells through an E2A dependent suppression of caspase-3 activation [23]. Interestingly, meta-analysis suggested E47 as a possible doxorubicin resistance-associated gene [24] which was experimentally validated in this study. These results are clinically significant that could explain resistance to doxorubicin based treatment regimens. Although we did not investigate the potential downstream effectors of E47 dependent mediators of doxorubicin resistance but candidate genes such as c-myc [25] and Id family of transcriptional regulators could be involved.
In conclusion, we provide multiple lines of evidence that support the role of E2A as a tumor promoter. At the molecular level, at least three potential E2A regulated mechanisms in PCa can be predicted: 1) promoting the expression of c-Myc: the oncogene c-Myc is itself highly expressed in PCa and regulates a large repertoire of genes and pathways involved in cancer progression including Id family of proteins[26]; 2) protection of Id1, a well-established marker of PCa progression [19,27], against ubiquitin-proteasome mediated degradation [5] and 3) the likelihood of another interacting bHLH protein, possibly with oncogenic properties such as Tal1. In this last scenario E12/E47 could still act as a tumor suppressor whose activity is inhibited by an oncogene similar to TAL1/SCL [28].
E2A, considered as a tumor suppressor is highly expressed in prostate cancer
Silencing of E2A attenuates cell proliferation and promotes apoptosis
E2A regulates c-myc, Id1, Id3 and CDKN1A expression
Loss of E2A promotes doxorubicin dependent apoptosis in prostate cancer cells.
Results suggest that E2A acts as a tumor promoter at least in prostate cancer
Supplementary Material
Acknowledgements
The work was supported by NIH/NCI CA128914 (JC) and in part by NIH/NCRR/RCMI G12RR03062.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Slattery C, Ryan MP, McMorrow T. E2A proteins: regulators of cell phenotype in normal physiology and disease. Int J Biochem Cell Biol. 2008;40:1431–1436. doi: 10.1016/j.biocel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 3.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huggins GS, Chin MT, Sibinga NE, Lee SL, Haber E, Lee ME. Characterization of the mUBC9- binding sites required for E2A protein degradation. J Biol Chem. 1999;274:28690–28696. doi: 10.1074/jbc.274.40.28690. [DOI] [PubMed] [Google Scholar]
- 5.Lingbeck JM, Trausch-Azar JS, Ciechanover A, Schwartz AL. E12 and E47 modulate cellular localization and proteasome-mediated degradation of MyoD and Id1. Oncogene. 2005;24:6376–6384. doi: 10.1038/sj.onc.1208789. [DOI] [PubMed] [Google Scholar]
- 6.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, van der Valk M, te Riele HPJ, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 7.Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc Natl Acad Sci U S A. 1999;96:996–1001. doi: 10.1073/pnas.96.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peverali FA, Ramqvist T, Saffrich R, Pepperkok R, Barone MV, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. Embo J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagliuca A, Gallo P, De Luca P, Lania L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors' promoter activity and negatively affect cell growth. Cancer Res. 2000;60:1376–1382. [PubMed] [Google Scholar]
- 11.Roberts VJ, Steenbergen R, Murre C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc Natl Acad Sci U S A. 1993;90:7583–7587. doi: 10.1073/pnas.90.16.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutherford MN, LeBrun DP. Restricted expression of E2A protein in primary human tissues correlates with proliferation and differentiation. Am J Pathol. 1998;153:165–173. doi: 10.1016/S0002-9440(10)65557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- 14.Asirvatham AJ, Carey JP, Chaudhary J. ID1-, ID2-, and ID3-regulated gene expression in E2A positive or negative prostate cancer cells. Prostate. 2007;67:1411–1420. doi: 10.1002/pros.20633. [DOI] [PubMed] [Google Scholar]
- 15.Asirvatham AJ, Schmidt MA, Chaudhary J. Non-redundant inhibitor of differentiation (Id) gene expression and function in human prostate epithelial cells. Prostate. 2006;66:921–935. doi: 10.1002/pros.20366. [DOI] [PubMed] [Google Scholar]
- 16.Baker SJ, Reddy EP. B cell differentiation: role of E2A and Pax5/BSAP transcription factors. Oncogene. 1995;11:413–426. [PubMed] [Google Scholar]
- 17.Zhao F, Vilardi A, Neely RJ, Choi JK. Promotion of cell cycle progression by basic helix-loop-helix E2A. Mol Cell Biol. 2001;21:6346–6357. doi: 10.1128/MCB.21.18.6346-6357.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz RI, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY. Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res. 2004;10:2044–2051. doi: 10.1158/1078-0432.ccr-03-0933. [DOI] [PubMed] [Google Scholar]
- 20.Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci U S A. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 22.Kardava L, Yang Q, St Leger A, Foon KA, Lentzsch S, Vallejo AN, Milcarek C, Borghesi L. The B lineage transcription factor E2A regulates apoptosis in chronic lymphocytic leukemia (CLL) cells. Int Immunol. 2011;23:375–384. doi: 10.1093/intimm/dxr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kee BL, Bain G, Murre C. IL-7Ralpha and E47: independent pathways required for development of multipotent lymphoid progenitors. Embo J. 2002;21:103–113. doi: 10.1093/emboj/21.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyorffy A, Vasarhelyi B, Szoke D, Dietel M, Tulassay T, Gyorffy B. Comparative promoter analysis of doxorubicin resistance-associated genes suggests E47 as a key regulatory element. Anticancer Res. 2006;26:2971–2976. [PubMed] [Google Scholar]
- 25.Grassilli E, Ballabeni A, Maellaro E, Del Bello B, Helin K. Loss of MYC confers resistance to doxorubicin-induced apoptosis by preventing the activation of multiple serine protease- and caspase-mediated pathways. J Biol Chem. 2004;279:21318–21326. doi: 10.1074/jbc.M313532200. [DOI] [PubMed] [Google Scholar]
- 26.Swarbrick A, Akerfeldt MC, Lee CS, Sergio CM, Caldon CE, Hunter LJ, Sutherland RL, Musgrove EA. Regulation of cyclin expression and cell cycle progression in breast epithelial cells by the helix-loop-helix protein Id1. Oncogene. 2005;24:381–389. doi: 10.1038/sj.onc.1208188. [DOI] [PubMed] [Google Scholar]
- 27.Forootan SS, Wong YC, Dodson A, Wang X, Lin K, Smith PH, Foster CS, Ke Y. Increased Id-1 expression is significantly associated with poor survival of patients with prostate cancer. Hum Pathol. 2007;38:1321–1329. doi: 10.1016/j.humpath.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Park ST, Sun XH. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J Biol Chem. 1998;273:7030–7037. doi: 10.1074/jbc.273.12.7030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.