SUMMARY
The natural product CJ-15,801 is an inhibitor of Staphylococcus aureus, but not other bacteria. Its close structural resemblance to pantothenic acid, the vitamin precursor of coenzyme A (CoA), and its Michael acceptor moiety suggest that it irreversibly inhibits an enzyme involved in CoA biosynthesis or utilization. However, its mode of action and the basis for its specificity have not been elucidated to date. We demonstrate that CJ-15,801 is transformed by the uniquely selective S. aureus pantothenate kinase, the first CoA biosynthetic enzyme, into a substrate for the next enzyme, phosphopantothenoylcysteine synthetase, which is inhibited through formation of a tight-binding structural mimic of its native reaction intermediate. These findings reveal CJ-15,801 as a vitamin biosynthetic pathway antimetabolite with a mechanism similar to that of the sulfonamide antibiotics, and highlight CoA biosynthesis as a viable antimicrobial drug target.
INTRODUCTION
The antibiotic CJ-15,801 (1; Figure 1A) was discovered in 2001 by a Pfizer research team when it was isolated from fermenting cultures of a Seimatosporium sp. fungus (Sugie, et al., 2001). Structural analysis showed the compound to resemble pantothenic acid (vitamin B5, 2; Figure 1B), the natural precursor of the essential metabolic cofactor coenzyme A (CoA, 3) (Strauss, 2010), with the notable exception of a trans-substituted double bond in the β-alanine moiety. This feature imparts an N-acyl vinylogous carbamic acid functionality to 1, an uncommon motif that is also present in the anti-cancer agent palytoxin and lipopeptide antibiotics such as enamidonin (Han, et al., 2004; Nicolaou and Mathison, 2005). Subsequently, CJ-15,801 was shown to inhibit the growth of drug-resistant strains of Staphylococcus aureus (MRSA) with minimum inhibitory concentration (MIC) values ranging between 6.25–50 μg/mL (30–230 μM). Interestingly, other bacteria, including Escherichia coli, Haemophilus influenza and several Streptococcus species, were not inhibited. In a later study, Saliba and Kirk determined that CJ-15,801 also inhibits the intraerythrocytic growth stage of the malaria parasite Plasmodium falciparum with an IC50 value of 39 μM, while leaving rat hepatoma HTC cells unaffected (Saliba and Kirk, 2005). Moreover, the malarial inhibition was reversed when the concentration of pantothenic acid in the medium was increased. Taken together, these findings all pointed to CJ-15,801 targeting CoA biosynthesis, or an enzyme or process dependent on this cofactor (Spry, et al., 2008; Spry, et al., 2010). However, such a proposal has not been confirmed experimentally in any of the organisms sensitive to CJ-15,801, nor does it provide a satisfactory explanation for this antibiotic’s peculiar selectivity.
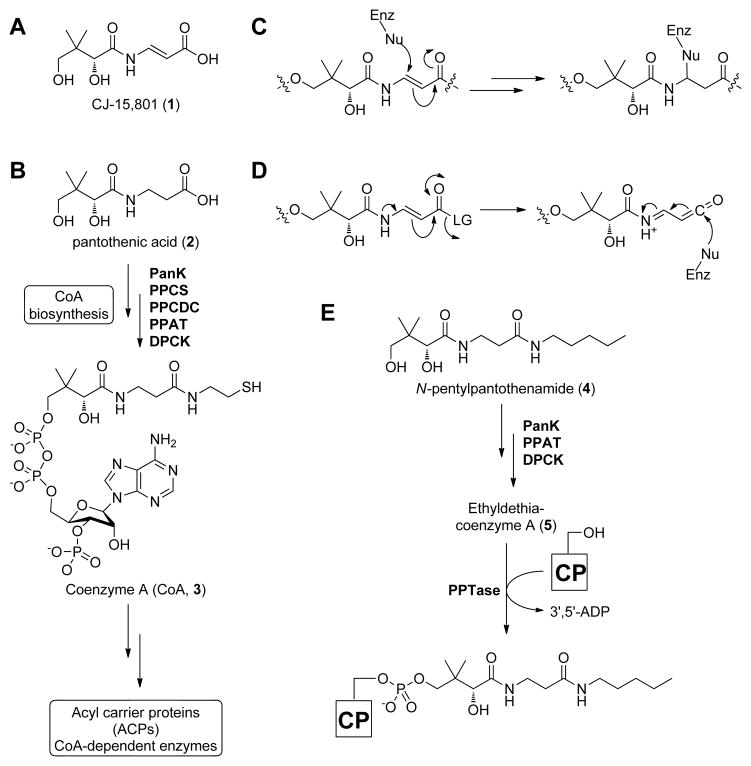
Figure 1. Structures of the antibiotic CJ-15,801 (1), selected coenzyme A biosynthetic intermediates, antimetabolites, and intermediate mimics.
(A) Structure of the CJ-15,801 (1).
(B) Biosynthesis of CoA (3) from pantothenic acid (2). PanK, pantothenate kinase; PPCS, phosphopantothenoylcysteine synthetase; PPCDC, phosphopantothenoylcysteine decarboxylase; PPAT, phosphopantetheine adenylyltransferase; DCPK, dephospho-coenzyme A kinase.
(C) Proposed mechanism for irreversible inhibition by CJ-15,801 acting as an electrophilic trap.
(D) Proposed mechanism for irreversible inhibition by CJ-15,801 after its transformation into a ketene intermediate.
(E) Biotransformation of the pantothenic acid analogue N-pentylpantothenamide (4) to the CoA antimetabolite ethyldethia-CoA (5), which has the catalytically essential thiol of the cofactor replaced by a propyl group. The antimetabolite subsequently serves as donor in the phosphopantetheinyl transferase (PPTase)-catalyzed post-translational modification of acyl and peptidyl carrier proteins (CPs), which results in similarly inactive crypto-CPs.
The presence of the N-acyl vinylogous carbamic acid functionality in CJ-15,801, combined with a knowledge of the chemical and biological reactivity of other inhibitors with similar reactive moieties, suggests that 1 most probably acts as an irreversible inhibitor of its biological target. Two mechanisms for such inhibition can be proposed: first, the α,β-unsaturated carbonyl moiety can act as a Michael-acceptor that would trap an active site nucleophile (Figure 1C); such a mechanism of action is seen in the structurally-related dapdiamide family of antibiotics (Hollenhorst, et al., 2011; Kucharczyk, et al., 1990), and has also been exploited in the development of irreversible inhibitors of CoA disulfide reductase (van der Westhuyzen and Strauss, 2010). Second, if the carboxylate of CJ-15,801 were functionalized in an enzymatic transformation with an appropriate leaving group, this may provide sufficient activation for the formation of a reactive ketene intermediate that could similarly act as an electrophilic trap (Figure 1D). Importantly, both of these mechanisms would remain equally viable if the 4′-OH group of CJ-15,801 is phosphorylated or otherwise functionalized, suggesting it could be transformed by the CoA biosynthetic enzymes into an inhibitory CoA analogue with a target downstream of the pathway. Such a mechanism of action has been demonstrated for the pantothenamides, a class of pantothenic acid analogues in which its carboxylate has been functionalized by amidation (Clifton, et al., 1970; van Wyk and Strauss, 2008). For example, N-pentylpantothenamide (N5-Pan, 4), which acts as a bacteriostatic agent in both E. coli and S. aureus (Choudhry, et al., 2003), is transformed by three of the five CoA biosynthetic enzymes into ethyldethia-CoA (5), a CoA antimetabolite that lacks the thiol required for the cofactor’s acyl carrier functions (Figure 1E) (Strauss and Begley, 2002). This analogue subsequently serves as donor in the post-translational modification of the apo-acyl carrier protein (ACP), leading to the formation of catalytically inactive crypto-ACP (Leonardi, et al., 2005; Mercer and Burkart, 2007; Zhang, et al., 2004). The resulting loss of function and subsequent impact on fatty acid metabolism is believed to be the major cause for bacteriostasis, although a recent study pointed to an impact on CoA biosynthesis as well (Thomas and Cronan, 2010).
Interestingly, like CJ-15,801, the pantothenamides also exhibit organism-based specificity. While the basis for this specificity may partially reflect differences in the targeted organism’s cell permeability and/or the nature of potential drug efflux pumps, the most important factor relates to the nature of the pantothenate kinase (PanK) enzyme present in the cell. PanK catalyzes the first step of CoA biosynthesis, namely the ATP-dependent phosphorylation of pantothenic acid (2) to form 4′-phosphopantothenic acid (P-Pan, 6), (Figure 3A). This reaction is unique because it is catalyzed by three distinct types of PanK, referred to as type I, type II and type III respectively. These different PanK types can be distinguished based on their sequence, structure, the extent to which they experience feedback inhibition by CoA, and importantly, their ability to act on pantothenamides as alternative substrates (Brand and Strauss, 2005; Hong, et al., 2006; Strauss, 2010; Strauss, et al., 2010; Yang, et al., 2008). Only type I and II PanKs (PanKI and PanKII) phosphorylate pantothenamides, thereby allowing the formation of CoA antimetabolites. Organisms like Pseudomonas aeruginosa that have type III PanK (PanKIII) enzymes that exclude these compounds from their active sites are therefore refractory to the effects of N-pentylpantothenamide and similar analogues (Balibar, et al., 2011). This suggests that the basis for CJ-15,801’s unique specificity for S. aureus and P. falciparum may also be based on the type-specificity of their PanKs, both of which have been characterized as atypical type II enzymes (Hong, et al., 2006; Leonardi, et al., 2005; Spry, et al., 2010).
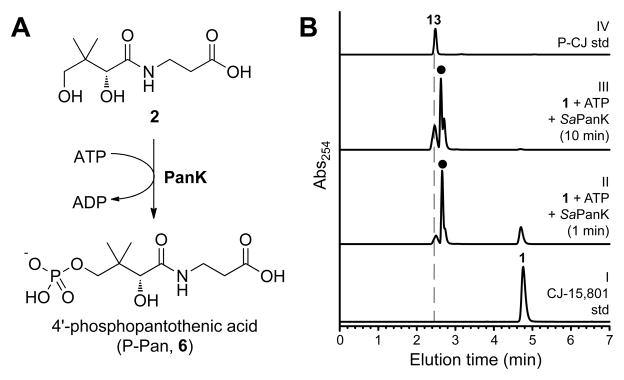
Figure 3. Evaluation of PanK as a potential gatekeeper to CJ-15,801’s inhibitory action.
(A) The ATP-dependent phosphorylation of pantothenic acid (2)catalyzed by PanK enzymes.
(B) HPLC analysis of the putative ATP-dependent phosphorylation of CJ-15,801 by SaPanK. The traces represent: I, a synthetically prepared standard of CJ-15,801 (1); II, a reaction mixture containing CJ-15,801, ATP and SaPanK that had been incubated for 1 minute; III, the same after 10 minutes of incubation; and IV, a synthetic P-CJ (13) standard. The peak labeled with (●) represents ATP and ADP. See also Table S2.
In this study we set out to identify the target of CJ-15,801’s antibiotic action in bacteria, to elucidate its mechanism of action, and to establish the basis for its specificity. Our results reveal that CJ-15,801 acts as an antimetabolite by using the first enzyme of the CoA biosynthesis as gateway to the pathway, after which it inhibits the second CoA biosynthetic enzyme, phosphopantothenoylcysteine synthetase (PPCS). This mode of action is reminiscent of the sulfonamide antibiotics, which block folic acid biosynthesis using a similar strategy. However, contrary to expectations, detailed analysis of the inhibition mechanism failed to provide any evidence of irreversible inhibition. Instead, our results show that CJ-15,801 is transformed into a tight-binding mimic of the PPCS enzyme’s natural reaction intermediate, a mechanism also utilized by other known natural product inhibitors of adenylating enzymes, such as ascamycin and mupirocin (May, et al., 2005; Pope, et al., 1998). Taken together, these findings not only provide insight into the basis of CJ-15,801’s antibiotic action, but also suggest a new and potentially general strategy for the development of synthetase inhibitors with in vivo efficacy.
RESULTS
Synthesis of CJ-15,801 and its esters
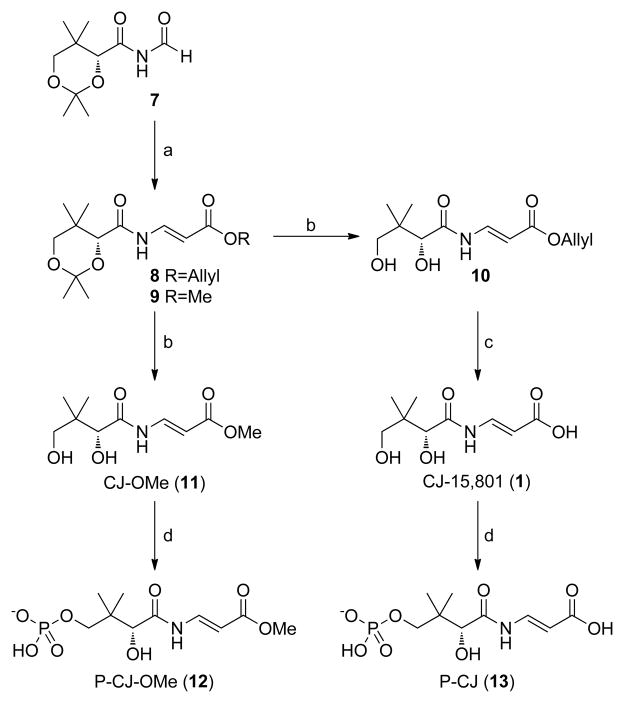
The synthesis of CJ-15,801 and its related analogs presents several challenges, the first of which relates to the preparation of the N-acyl vinylogous carbamic acid moiety. In fact, several groups have used the synthesis of CJ-15,801 to showcase new methodologies developed specifically for the preparation of this functionality (Han, et al., 2004; Lee, et al., 2006; Nicolaou and Mathison, 2005). However, we found these methods to be unsuitable for this study as they are either optimized for small scale synthetic preparations, or led to the preferential formation of the unwanted (Z)-double bond configuration. We therefore utilized the more recent method of Villa et al., which installs the required N-acyl vinylogous carbamic acid moiety mainly with an (E)-configuration via Wittig-type chemistry, for preparation of the protected esters 8 and 9 (Figure 2) (Villa, et al., 2007). A second challenge derives from the sensitivity of the N-vinyl amide moiety to both acid and base, which severely restricts the options for the protection and deprotection of the synthetic intermediates. In this case, established protocols that make use of neutral conditions were used to remove the acetal protecting groups from 8 and 9 to respectively obtain CJ-15,801’s allyl (10) and methyl (11) esters, and to remove the allyl ester from 10 to provide CJ-15,801 itself. The (Z)-configured analogue of CJ-15,801 (Z-1) was obtained by similar deprotection of the minor constituent of the Wittig coupling reaction product mixture.
Figure 2. Synthesis of CJ-15,801, its methyl ester and their respective phosphorylated versions.
The respective reaction conditions are: (a) Allyl- or methylacetyltriphenylphosphonium bromide, Et3N, toluene, 90°C, 75%.(Villa, et al., 2007) (b) BiCl3, H2O, CH3CN, rt, 5h, 75%. (c) Pd(PPh3)4, pyrollidine, THF, 81%. (d) SaPanK, ATP, MgCl2, Tris-HCl, pH 7.5, 98% (for 12); 79% (for 13). See also Figure S3.
Recently, an updated report of the Wittig-based synthesis of CJ-15,801 appeared in which several other options for deprotection of its carboxyl group are also provided (Sewell, et al., 2011).
A bacterium’s susceptibility to CJ-15,801 correlates with its PanK type
The original Pfizer discovery group found that in standard susceptibility tests conducted using cation adjusted Mueller-Hinton broth as growth medium CJ-15,801 is uniquely and peculiarly selective for S. aureus, with none of the other organisms tested showing inhibition at 100 μg/mL (460 μM) (Sugie, et al., 2001). These resistant organisms included both Gram-positive (Staphylococcus, Enterococcus and Streptococcus spp. ) and Gram-negative ( H. influenzae, Moraxella catarrhalis and E. coli) bacteria, indicating that cell envelope type alone does not determine selectivity. However, the panel of test organisms also comprised bacteria expressing all three known pantothenate kinase (PanK; Figure 3A) types: the enterococci, streptococci, H. influenzae and E. coli are known or predicted to have PanKI enzymes, while M. catarrhalis is predicted to have a PanKIII (Yang, et al., 2006). Importantly, the staphylococci are the only known bacteria to have an active PanKII, suggesting that the presence of this PanK type is a prerequisite for inhibition by CJ-15,801.
To test this hypothesis, we repeated the CJ-15,801 susceptibility tests against a panel of organisms representing all combinations of cell envelope and PanK type. Since the original tests were conducted in growth medium rich in pantothenic acid, these experiments were conducted in 1% tryptone (that contains essentially no pantothenic acid) instead to exclude any potential antagonistic effects that the presence of the vitamin could have on inhibition. The results show that the PanKII-containing S. aureus (MRSA strain TCH1516) was the most sensitive to the inhibitory effects of CJ-15,801, showing an MIC of 15 μM (Table S1). This value is two-fold lower than the lowest MIC reported in the previous susceptibility tests, suggesting that pantothenate does affect inhibition similar to what was seen in the experiments with malaria parasites (Saliba and Kirk, 2005). Moreover, under these test conditions some of the other organisms (most notably P. aeruginosa) also showed low levels of inhibition (MIC ~60 μM).
To determine which aspects of CJ-15,801’s structure are required for inhibition, we evaluated the inhibitory activity of the (Z)-configured analogue (Z-1) and CJ-15,801’s methyl ester (CJ-OMe, 11) against S. aureus. Neither compound showed any inhibition, indicating that the configuration of the double bond and the availability of the free carboxylate of CJ-15,801 are both important determinants for its mechanism of inhibition.
Only PanKII enzymes accept CJ-15,801 as substrate
To confirm that the observed inhibition results can indeed be correlated with PanK type and activity, we performed in vitro activity analyses using purified PanK enzymes. Based on the inhibition results we first used the most likely candidate, the PanKII enzyme from S. aureus (SaPanKII), to determine if it converts CJ-15,801 to phospho-CJ,15,801 (P-CJ, 13) when incubated with ATP. HPLC analysis of the reaction mixture confirmed this to be the case, based on the time-dependent formation of P-CJ (Figure 3B). Next, the activity of the PanKI from E. coli (EcPanKI), SaPanKII and the PanKIII from P. aeruginosa (PaPanKIII) toward CJ-15,801 were fully characterized kinetically. The resulting data indicate that EcPanKI and PaPanKIII only show activity towards pantothenic acid and not CJ-15,801; moreover, neither enzyme was inhibited by 100 μM CJ-15,801 (Table S2). However, SaPanKII shows little distinction between CJ-15,801 and pantothenic acid, exhibiting specificity constants (kcat/Km) of 13.8±4.2 mM−1.s−1 and 30.2±8.7 mM−1.s−1 for the two compounds respectively, with the difference being mainly due to an elevated Km value. The enzyme also acts on CJ-OMe, indicating that the lack of inhibition seen for this analogue is due to effects downstream of PanK.
Taken together, these results indicate that PanK acts as a gatekeeper to the inhibitory effects of CJ-15,801 in S. aureus.
CJ-15,801’s antistaphylococcal activity is affected by pantothenic acid and pantetheine
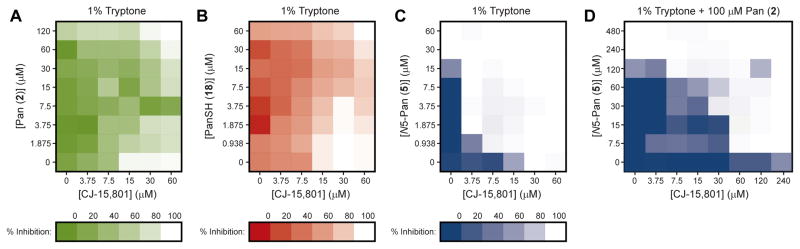
The observation that S. aureus’s susceptibility to inhibition by CJ-15,801 is apparently affected by pantothenic acid (2), as well as the finding that SaPanKII accepts both the inhibitor and the vitamin as substrates, led us to perform checkerboard assays to quantify these effects (Figure 4A). The same assay was also conducted with pantetheine (17), the precursor to the CoA salvage pathway that bypasses the PPCS and PPCDC enzymes (Strauss, 2010; Strauss, et al., 2010) (Figure 4B). The results show that the presence of either compound reduces the potency of CJ-15,801, most likely through competition with SaPanKII. However, their interaction with the natural product is complex, since increasing the concentration of either compound above a certain level (~7.5 μM for 2, and ~15 μM for 17) leads to inhibition being restablished. While the basis for this observation is currently unknown, the results confirm that the point of CJ-15,801’s inhibitory action is a process dependent on pantothenic acid (or pantetheine).
Figure 4. Synergism in the antibacterial action of CJ-15,801 and N5-Pan.
(A) Checkerboard assay of the growth inhibition of S. aureus by CJ-15,801 (1) when grown in the presence of pantothenic acid (2) in 1% tryptone shows that increasing the amount of vitamin (up to a certain concentration) reverses the inhibition. MIC value for CJ-15,801 is 15 μM.
(B) As for (A), but with pantetheine (17) added instead of pantothenic acid.
(C) Checkerboard assay of the growth inhibition of S. aureus grown in 1% tryptone by CJ-15,801 (1) and N5-Pan (4) demonstrates the synergism between these compounds (FICI<0.5). MIC value for N5-Pan is 30 μM.
(D) As for (C), but determined in the presence of 100 μM pantothenic acid (2), demonstrating that the combination of CJ-15,801 and N5-Pan can overcome the antagonism caused by an excess of the natural substrate of the CoA biosynthetic pathway, but at the expense of converting the synergism to an additive effect (0.5<FICI<1.0). See also Tables S1 and S3.
Pantothenamides increase the potency of CJ-15,801’s antistaphylococcal activity
Previous studies have shown that treatment of S. aureus with N5-Pan (and its analogue, N-heptylpantothenamide, N7-Pan) results in growth inhibition by formation of inactive carrier proteins (Figure 1E) (Leonardi, et al., 2005; Zhang, et al., 2004). To determine if the points of action of the pantothenamides and CJ-15,801 overlap, a checkerboard assay with N5-Pan (4) and CJ-15,801 was performed in 1% tryptone media (Figure 4C). The results show that when combined, CJ-15,801 and N5-Pan exhibited significantly reduced MICs of 3.75 μM and ~1.0 μM respectively, with a fractional inhibitory concentration index (FICI) of 0.25, indicating synergism (Table S3). Importantly, when the same assay was performed in the presence of 100 μM pantothenic acid, the inhibition was not as adversely affected as in the case of the individual compounds (Figure 4D). Instead, the MIC values were slightly elevated and the previously observed synergism was converted to an additive inhibitory effect (FICI ~0.5–1.0) (Table S3). This finding suggests that CJ-15,801 and N5-Pan does not have the same mode of action, and indicates that in combination these compounds can counter the antagonism caused by pantothenic acid.
Synthesis of the 4′-phosphates of CJ-15,801 and its methyl ester
The discovery that SaPanK phosphorylates both CJ-15,801 and CJ-OMe (11) provided us with a convenient biocatalytic method by which their 4′-phosphorylated versions could be prepared. P-CJ (13) and its methyl ester, 12 (P-CJ-OMe), were therefore obtained by milligram-scale biotransformations using recombinant SaPanK and ATP, followed by purification by either preparative HPLC (for P-CJ) or solid-phase extraction (SPE) (for P-CJ-OMe) (Figure 2).
PPCS accepts P-CJ as substrate – and is then inhibited by it
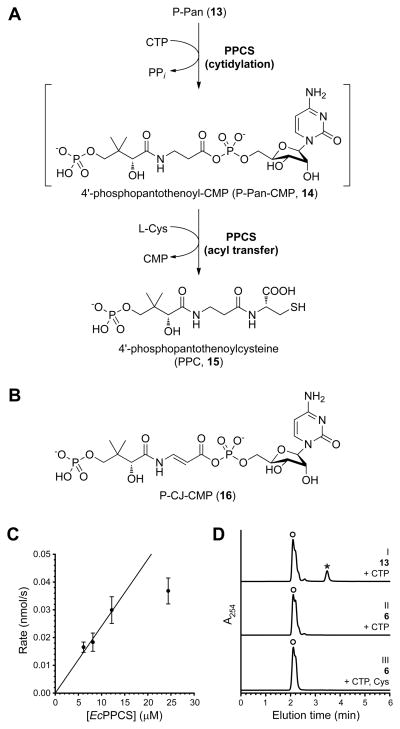
With the knowledge that CJ-15,801 can be converted to P-CJ within S. aureus, we set out to determine if PPCS, the next enzyme in the pathway, is the target for the antibiotic’s inhibitory action. PPCS catalyzes the condensation of P-Pan (6) with L-cysteine to form 4′-phosphopantothenoylcysteine (PPC, 15) using a two-step acyl transfer mechanism common to most synthetase (C–N ligase) enzymes, including all the amino acid tRNA synthetases and the adenylation domains of the modular non-ribosomal peptide synthetases (NRPSs) (Figure 5A) (Schimmel, et al., 1998; Sieber and Marahiel, 2005). While all known PPCSs follow this general scheme, bacterial enzymes are unique in that they use CTP instead of ATP as nucleotide source for the activation reaction, and therefore form 4′-phosphopantothenoyl-CMP (P-Pan-CMP, 14) as intermediate (Kupke, 2004; Kupke, 2002; Manoj, et al., 2003; Stanitzek, et al., 2004; Strauss, 2010; Strauss, et al., 2001).
Figure 5. Evaluation of PPCS activity with P-CJ (13) as substrate.
(A) The reaction catalyzed by PPCS, showing the two steps of its mechanism and the P-Pan-CMP (14) reaction intermediate.
(B) The structure of P-CJ-CMP (16) resulting from the putative cytidylation of P-CJ (13) by PPCS.
(C) The rate of pyrophosphate release from a reaction mixture containing P-CJ (13), CTP and EcPPCS correlates with enzyme concentration. Experiments were run in duplicate; the shown data points are the average of duplicate assays (error bars indicate the standard deviation, SD).
(D) HPLC analysis of a reaction mixture containing P-CJ (13), CTP and EcPPCS (24 μM) (trace I) show the formation of a new peak (indicated with an asterisk, *) that represents P-CJ-CMP (16) based on LCMS analysis. This peak is not present when P-CJ is replaced with P-Pan (6) (trace II) or in native reaction mixtures containing P-Pan, CTP and L-cysteine. Note that the retention time of pure P-CJ is ~2.4 min (see Figure 2B). The broad peak labeled with (○) represents CTP and CMP. See also Figure S1.
In most bacteria PPCS activity is located on one domain of the bifunctional CoaBC protein (which also carries PPCDC activity), although monofunctional PPCS enzymes do occur in certain enterococci (such as E. faecalis (Yao, et al., 2009)) and streptococci. Among all bacterial PPCS enzymes the PPCS activity of E. coli’s bifunctional CoaBC protein is by far the best studied: its mechanism has been established through trapping and isolation of P-Pan-CMP (14), and the structure of the N210D mutant with the trapped intermediate 14 bound in the active site has been determined (Kupke, 2004; Kupke, 2002; Stanitzek, et al., 2004). Moreover, many of these studies were performed using the separate PPCS domain (EcPPCS) expressed and purified on its own, showing that the two activities of the bifunctional CoaBC protein are independent. This conclusion is also supported by kinetic isotope studies of the PPCDC activity of E. coli’s CoaBC (Strauss and Begley, 2001). We therefore decided to use the EcPPCS domain to perform the initial tests on the interaction of PPCS and P-CJ.
To determine if P-CJ is accepted as an alternate substrate by PPCS and forms the corresponding P-CJ-CMP intermediate 16 (Figure 5B), mixtures of P-CJ and CTP were incubated with increasing concentrations of EcPPCS. A clear correlation between the rate of pyrophosphate release and enzyme concentration was evident in such reactions (Figure 5C). Moreover, HPLC analysis of the reaction mixture containing the highest EcPPCS concentration (24 μM) showed the formation of a peak absent in native reaction mixtures, or in mixtures containing P-Pan (6) instead of P-CJ, suggesting the formation of P-CJ-CMP (Figure 5D). Note that P-Pan-CMP is usually not observed under these conditions, as was previously reported (Kupke, 2004; Kupke, 2002). LCMS analysis was subsequently conducted on the same mixture. While the P-CJ-CMP-derived molecular ion was not observed, the corresponding mass spectrum did show mass peaks that are in agreement with the fragmentation of P-CJ-CMP (Figure S1). The same experiment was subsequently repeated with the bifunctional CoaBC protein from S. aureus (SaCoaBC). A similar result was obtained, although the apparent rate of conversion of P-CJ into P-CJ-CMP was significantly slower than for EcPPCS (Figure S1).
In combination, these results indicate that P-CJ is accepted as an alternate substrate by PPCS, which cytidylylates its carboxylate to form P-CJ-CMP.
PPCS transform P-CJ into P-CJ-CMP, which inhibits its activity
Subsequent activity analysis of PPCS in the presence of P-CJ, CTP and L-cysteine failed to show catalytic turnover. Moreover, when P-CJ was added to PPCS reaction mixtures containing the enzyme’s native P-Pan substrate, the rate of the reaction was reduced compared to mixtures without it, suggesting inhibition by P-CJ. To confirm that such inhibition is dependent on the reaction catalyzed by PPCS, an inhibition screen was performed in which either CJ-15,801, CJ-OMe, P-CJ or P-CJ-OMe was added to native PPCS reaction mixtures at a concentration of 100 μM. The results showed that among these compounds, only P-CJ inhibited EcPPCS (data not shown). The lack of inhibition by P-CJ-OMe indicated that the free carboxylate group is required for inhibition, while the inability of CJ-15,801 to affect enzyme activity highlights the necessity of the phosphate group for binding. Taken together with the results described above, these findings reveal PPCS as CJ-15,801’s target of inhibitory action, which is achieved through formation of P-CJ-CMP 16 as an apparent dead-end inhibitor.
Kinetic characterization reveals P-CJ-CMP as a tight-binding inhibitor of PPCS
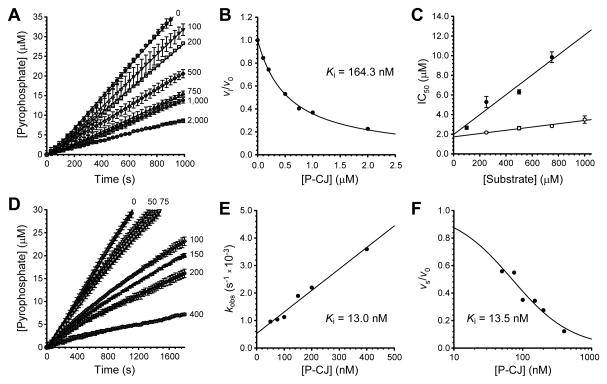
Inhibition by P-CJ-CMP could potentially occur by either of the proposed irreversible inhibition mechanisms (Figures 1C and 1D), or by a completely unrelated mechanism. To elucidate the actual mechanism of inhibition, detailed kinetic analyses were performed. First, dose-response curves were used to evaluate the inhibition of the cytidyl-transfer reaction catalyzed by EcPPCS. The resulting IC50 value of 2.66±0.16 μM, determined using saturating concentrations of CTP and 100 μM P-Pan (note that EcPPCS experiences substrate inhibition, see Figure S2), was less than 10-fold the concentration of enzyme used in these assays (240 nM). This indicated that P-CJ-CMP acts as either a tight-binding or irreversible inhibitor of EcPPCS (Copeland, 2000). The inhibition of EcPPCS was next studied by the progress curve method. The resulting curves increased in a linear fashion, showing a decreased rate at increased concentrations of P-CJ (Figure 6A). Surprisingly, this indicated that the inhibition was tight-binding and not irreversible (which would have led to the steady state rates approaching zero) as predicted (Copeland, 2005). This finding was confirmed by showing that removal of the inhibitor by gel-filtration was sufficient to restore the activity of a sample of EcPPCS that had been pre-treated with P-CJ and CTP.
Figure 6. Kinetic characterization of the CJ-15,801-based inhibition of PPCS.
(A) Progress curves analysis for the inhibition of EcPPCS in the presence of increasing concentrations of P-CJ (13), indicated in nM next to each curve.
(B) Plot of the fractional activity (initial velocity vi relative to the inhibited velocity v0), as determined from the progress curves in (a), against inhibitor concentration. The indicated Ki value is determined by fitting the data to the Morrison and Cheng-Prusoff equation.
(C) Determination of the inhibition modality of EcPPCS by P-CJ (13) by calculation of the IC50 values in the presence of increasing concentrations of P-Pan (6) (●), and CTP (○).
(D) Progress curves analysis for the inhibition of SaCoaBC in the presence of increasing concentrations of P-CJ (13), indicated in nM next to each curve.
(E) Plot of the first order rate of inactivation constants (kobs), as determined from the data presented in (D), against inhibitor concentration. The indicated Ki value is determined by fitting the data to the equation for a straight line.
(F) Plot of the fractional activity (steady state velocity vs relative to the inhibited velocity v0), as determined from the progress curves in (D), against inhibitor concentration. The indicated Ki value is determined by fitting the data to a Langmuir isotherm.
All progress curve analyses were performed in triplicate (error bars represent the SD). See Supplemental Experimental Procedures for details on calculations, as well as Figure S2 for the determination of EcPPCS and SaCoaBC kinetic parameters.
The Kiapp (apparent inhibition constant) was subsequently calculated by plotting the fractional activity vs. inhibitor concentration, fitting the data to the Morrison equation (Figure 6B), and converting the obtained value to the true Ki by applying the appropriate Cheng-Prusoff equation (Copeland, 2005; Copeland, 2000). The mode of inhibition was established using a method specifically developed for tight-binding inhibitors in which IC50 values are determined at a fixed enzyme concentration, but in the presence of increasing concentrations of either P-Pan (6) or CTP (Figure 6C). A linear increase in IC50 values with increasing concentration of P-Pan but not CTP, indicates that inhibition is competitive towards P-Pan but non-competitive towards CTP (Copeland, 2005; Copeland, 2000). This observation agrees with previous studies of the E. faecalis and human PPCS enzymes that determined an ordered Bi Uni Uni Bi Ping-Pong kinetic mechanism with CTP binding first, followed by P-Pan (6) (Yao and Dotson, 2009; Yao, et al., 2009). In this manner a Ki of 164.3 nM was determined for P-CJ’s inhibition of EcPPCS.
While EcPPCS is a convenient and well-studied model to use for characterization of CJ-based inhibition, E. coli is not affected by CJ-15,801 due to the gatekeeping activity of its PanK (Tables S1 and S2). We therefore performed the same kinetic analysis on SaCoaBC, which is expected to be the natural target for CJ-15,801’s observed anti-staphylococcal action. A progress curve analysis similar to that conducted on EcPPCS showed that SaCoaBC experiences time-dependent inhibition, since the curves showed a fast initial velocity (vi) phase and a slower steady state velocity (vs) phase (Figure 6D). The first order rate of inactivation constants (kobs) were subsequently determined and plotted against the inhibitor concentration. The resulting plot (Figure 6E) showed a linear relationship between kobs and [I], which points to a one-step inhibition mechanism (Copeland, 2005). However, it is more likely that the true mechanism consists of two steps, with P-CJ 13 binding first in a reversible manner, followed by the slow formation of P-CJ-CMP. Such a mechanism would be kinetically indistinguishable from a one-step mechanism.
The Ki value for the inhibition of SaCoaBC by P-CJ-CMP can be determined from the slope and y-intercept of the linear plot shown in Figure 6E. The Ki value can also be calculated by using the progress curves to determine the steady state velocity (vs) at each inhibitor concentration; a plot of the fractional activity against inhibitor concentration yields an isotherm curve with the Ki at its midpoint (Figure 6F) (Copeland, 2005). In this manner, Ki values of 13.0 and 13.5 nM were determined respectively. These values is nearly an order of magnitude lower than that determined for EcPPCS, clearly showing that the nature of the actual target protein also plays an important role in the extent of inhibition. Taken together, these data show that P-CJ-CMP acts as a tight-binding inhibitor of PPCS enzymes.
Analysis of the mechanistic basis for the tight-binding inhibition of P-CJ-CMP
The close structural homology between P-Pan-CMP (14) and P-CJ-CMP (16), and the fact that both compounds contain reactive acyl phosphate moieties, raises a question regarding the mechanistic basis for the inhibition by P-CJ-CMP. We considered three possible explanations: first, inhibition is based on differences in the electrophilicity of the carbonyl carbon of the acyl phosphate moieties of P-Pan-CMP and P-CJ-CMP respectively; second, inhibition is caused by the two compounds having different binding modes in the active site, due to the structural nature of the planar N-acyl vinylogous carbamate functionality of P-CJ-CMP; and third, inhibition is caused by P-Pan-CMP and P-CJ-CMP having different effects on any conformational changes that occurs during catalysis through stabilization of the protein’s quaternary structure. While the significant synthetic effort that is required to prepare P-Pan-CMP and P-CJ-CMP precluded a direct experimental comparison of their respective reactivities, we were able to perform tests to assess whether the latter two proposed mechanisms contribute to inhibition.
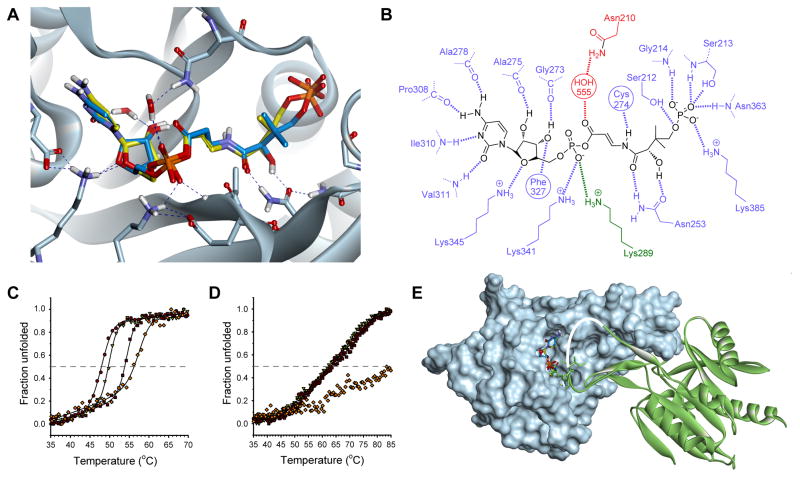
To establish if P-Pan-CMP (14) and P-CJ-CMP (16) have different binding modes, we used the only available structure of a PPCS enzyme with its intermediate bound, that of the EcPPCS Asn210Asp mutant with P-Pan-CMP bound in the active site (PDB ID: 1U7Z). This mutant, which accumulates sufficient amounts of the intermediate to have allowed direct confirmation of its identity and structure (Kupke, 2004; Stanitzek, et al., 2004), is apparently unable to catalyze the second step of the PPCS reaction. However, the molecular basis for this catalytic disability remains speculative since no PPCS structure with cysteine bound has been determined thus far. Nonetheless, the mutation clearly results in tightly-bound P-Pan-CMP in the enzyme’s active site.
We constructed a model of the native enzyme by reversing the Asn210Asp mutation in the crystal structure, and used it to dock P-CJ-CMP (16) in the active site using 150 starting conformations to ensure sufficient conformational space was sampled considering the complexity of the molecule. The highest scoring docked pose of P-CJ-CMP very closely mimics the pose of the co-crystalized P-Pan-CMP in the crystal structure (Figures 7A), and facilitates all the same hydrogen bonding interactions while maintaining a near planar geometry across the N-acyl vinylogous carbamate moiety (Figure 7B). This suggests that the basis for inhibition by P-CJ-CMP is not due it having a different binding mode, or taking on a potentially unreactive conformation.
Figure 7. Structural basis for the tight-binding inhibition of the CJ-15,801-derived inhibitor.
(A) Structure of the P-CJ-CMP inhibitor 16 (stick structure with carbon atoms in yellow) modeled and overlaid on that of the P-Pan-CMP intermediate 14 (stick structure with carbon atoms in blue) bound in the active site of native EcPPCS. The model was created from the crystal structure of EcPPCS Asn210Asp with co-crystalized P-Pan-CMP 14 bound (pdb: 1U7Z) by reversing the mutation, and docking P-CJ-PMP. The highest scoring pose of P-CJ-CMP closely resembles that of P-Pan-CMP and similarly facilitates all hydrogen bonding interactions observed for P-Pan-CMP. Only selected residues are shown for clarity. See also Figure S4.
(B) Schematic view of polar interactions between P-CJ-CMP and EcPPCS. Residues from monomer A are in blue and those from monomer B in green. The key interaction between Asn210 and the acyl phosphate carbonyl, mediated via a bridging water molecule, is shown in red.
(C) Heat-induced protein melting curves for EcPPCS. The four curves represent from left to right: the free protein (●); protein with MgCTP (▽); protein with MgCTP and P-Pan (2) (■); and protein with MgCTP and P-CJ (13) (◇). The curves were determined by following changes in the protein’s secondary structure by circular dichroism.
(D) Heat-induced protein melting curves for SaCoaBC determined as for EcPPCS in (B).
(E) Structure of the EcPPCS dimer (pdb: 1U7Z) with one monomer shown as a solvent-accessible surface (in cyan) and the other as ribbon structure (in green). The overlayed P-Pan-CMP and P-CJ-CMP stick structures are shown docked in the active site. Note that residues 291 to 298, which form part of a loop (shown in white) that cover the active site, is normally disordered and is absent in all crystal structures of PPCS enzymes (Manoj, et al., 2003; Stanitzek, et al., 2004).
Members of the ANL superfamily of adenylating enzymes, such as the adenylation domains of the NRPSs, use a large domain rotation event to facilitate the catalysis of their two-step reactions (Gulick, 2009). While it is currently unknown whether PPCS enzymes, which belong to the ribokinase family, undergo similar large conformational changes during catalysis, the determined crystal structures of the E. coli and human proteins show that they occur as dimers, with the dimer interface partially occluding the active site (Manoj, et al., 2003; Stanitzek, et al., 2004). To evaluate the effect of different ligands on the enzyme’s overall stability, the melting temperatures (Tm) of the EcPPCS protein in the presence of its native substrates or the inhibitor were determined by following the heat-induced denaturation of these mixtures by circular dichroism (CD). The results show that the Tm increases from 47.7 °C (for the free enzyme) to 49.7 °C (for the enzyme bound to CTP) to 54.3 °C (for the enzyme bound to P-Pan-CMP, formed in situ from P-Pan and CTP), highlighting the many stabilizing interactions the protein has with its native ligands (Figure 7C). Importantly, the Tm of EcPPCS bound to P-CJ-CMP (formed in situ from P-CJ and CTP) shows an even larger increase to 57.3 °C. When the same experiment was conducted with the bifunctional SaCoaBC, the protein showed increased stability compared to EcPPCS (CoaBC proteins exists as higher order oligomers, usually dodecamers (Manoj, et al., 2003)), but only small differences in the Tm values for the protein in the absence and presence of its native ligands. However, the Tm showed an impressive improvement from ~65 °C to 85 °C for the proteins bound to P-Pan-CMP and P-CJ-CMP respectively (Figure 7D). Taken together, these findings indicate that the binding of the conformationally more rigid ligand 16 results in an increased stabilization of the PPCS oligomers and the trapping of the ligand in the active site. This stabilization, which may be due to interactions between the ligand and residues of the adjacent monomer (such as Lys289, see Figure 7B) contained in a disordered loop that partially covers the entrance to the active site (Figure 7E), is therefore most likely an important contributor to the mechanism of inhibition by P-CJ-CMP.
DISCUSSION
One of the biggest obstacles to elucidation of the CJ-15,801 mode of action is scarcity of the natural product. The original discoverers shared the bulk of the isolated material with the authors of a malaria parasite inhibition study (Spry, et al., 2008), but new stocks are unlikely to be obtained in this manner since the CJ-producing Seimatosporium strain has since been lost (Y. Sugie to E. Strauss, personal communication). New developments in the synthesis of CJ-15,801 have certainly improved the situation, but the unique sensitivity of the N-acyl vinylogous carbamic acid functionality still presents a significant challenge to the synthesis of this antibiotic.
The original finding that CJ-15,801 is highly selective in its antibiotic action, affecting only S. aureus strains among a range of other Gram-positive bacteria including Streptococcus and Enterococcus spp. (Sugie, et al., 2001), provoked curiosity in regards to its mechanism of action. Here we show that the primary reason for this selectivity is the substrate specificity of the host organism’s PanK enzyme. Only the PanKII of S. aureus accepts CJ-15,801 and phosphorylates it, thereby allowing it to enter the CoA biosynthetic pathway. Interestingly, SaPanKII is the only known active bacterial representative of a type II PanK; all other examples occur in eukaryotes. However, an earlier study showed that CJ-15,801 does not inhibit rat hepatoma cells, suggesting that typical eukaryotic PanKs do not act as gateways to its antibiotic action (Saliba and Kirk, 2005). Such a conclusion is supported by the results of a structure-activity relationship study of the inhibition of EcPanKI and the PanKIIs from Aspergillus nidulans, Mus musculus (mouse) and S. aureus by the pantothenamides, which showed a clear differentiation between eukaryotic PanKIIs and SaPanKII, and even between the eukaryotic PanKIIs themselves (Virga, et al., 2006). The varied selectivity of PanKIIs have clearly not been fully explored, since P. falciparum (which is also sensitive to inhibition by CJ-15,801) is also predicted to have a PanKII (Spry, et al., 2010).
The evolution of CJ-15,801 as a natural product that specifically targets the S. aureus CoA biosynthetic pathway may reflect this organism’s unique redox physiology. Like some other Gram-positive bacteria, including Bacillus anthracis (Nicely, et al., 2007), S. aureus does not contain glutathione but instead relies on the sacrificial oxidation of CoA and a specific NADPH- and flavin-dependent CoA disulfide reductase (CoADR) enzyme to maintain its intracellular redox balance (delCardayre and Davies, 1998; Mallett, et al., 2006). This causes S. aureus to maintain high intracellular levels of reduced CoA (Newton, et al., 1996), which imparts an additional unique characteristic to SaPanKII: unlike other eukaryotic PanKIIs, it is refractory to feedback inhibition by CoA and its thioesters (Choudhry, et al., 2003; Hong, et al., 2006; Leonardi, et al., 2005). The combination of S. aureus’s distinctive reliance on CoA biosynthesis and the unique characteristics of its SaPanKII enzyme have seemingly created an ideal target for antibacterial action that was eventually exploited through the production of CJ-15,801.
The interaction between CJ-15,801 and pantothenic acid and its analogues is complex, and warrants further study (Figure 4A & B). The observation that inhibition by CJ-15,801 is alleviated in the presence of either pantothenic acid or pantetheine is expected in light of its determined mode of action; however, it is unclear why the inhibition returns when the concentrations of the vitamin is increased beyond a certain level. It is possible that the susceptibility to CJ-15,801 under these circumstances is due to the high concentrations of pantothenic acid stimulating CoA biosynthesis, as this would result in increased amounts of P-CJ-CMP being formed. The low levels of inhibition seen in the absence of pantothenic acid in organisms such as P. aeruginosa that cannot form P-CJ (and which therefore should not experience any inhibition by the mechanism proposed here) is probably due to similar effects on the regulation of pantothenic acid biosynthesis and/or utilization. However, such effects are clearly weak, as they are completely abolished in the presence of increased concentrations of the vitamin.
The finding that CJ-15,801 and members of the pantothenamide class of antibiotics work in a synergistic fashion supports most studies that have indicated that the pantothenamides have a target downstream of CoA biosynthesis (Figure 4C) (Leonardi, et al., 2005; Mercer, et al., 2009; Thomas and Cronan, 2010; Zhang, et al., 2004). Moreover, such a combination can apparently maintain its inhibitory effects even in the presence of pantothenic acid (Figure 4D). This indicates that it is possible to increase the modest antistaphylococcal activity of CJ-15,801 and even counteract the antagonism caused by pantothenic acid by combining it with other appropriate inhibitors, especially those that also affect CoA-based metabolism.
The finding that P-CJ-CMP acts as a tight-binding inhibitor of the PPCS enzyme can be rationalized in light of the findings of previous studies that have shown that non-hydrolyzable mimics of acyl nucleotidylate intermediates are often potent (tight-binding) inhibitors of enzyme activity (Cisar and Tan, 2008; Kim, et al., 2003; Schimmel, et al., 1998). In fact, this strategy was also successfully applied to PPCS in a study in which the phosphodiester analogue of P-Pan-CMP was shown to be a potent inhibitor of the PPCS of E. faecalis (Patrone, et al., 2009). However, the finding that P-CJ-CMP does not undergo attack by cysteine in spite of having an activated acyl phosphate moiety remains surprising. Of the three possible mechanisms that were considered for the inhibition by P-CJ-CMP, the most likely is that the introduction of the conjugated system in the β-alanine moiety reduces the reactivity of the acyl phosphate carbonyl towards nucleophilic attack. Such a reduced activity could be based on electronic effects, or be due to the potential tautomerization of the carbonyl enamine function of the inhibitor to an enol imine. Such a modification in reactivity has been noted in similar systems, as in the case of the vinylogous carbamate ester that forms as a result of the mechanism of action of the β-lactamase inhibitor clavulanate (and penam sulfones), and which has been reported to be stable towards hydrolysis (Imtiaz, et al., 1993). However, structural studies have indicated that this stability is partly due to interactions with active site residues, and that it can be enhanced by manipulation of these interactions (Padayatti, et al., 2005; Padayatti, et al., 2006). Our model of P-CJ-CMP docked in the active site of EcPPCS does not show the formation of any new active site-ligand interactions that are not present with P-Pan-CMP, suggesting that such factors are not at play in the inhibition of PPCS by P-CJ-CMP.
Our experiences with the synthetic preparation of CJ-15,801 (Figure 2) indicated that the conjugation provided by the unsaturated β-alanine moiety does deactivate this center. Specifically, all chemical attempts to hydrolyze the methyl esters 9 and 11 en route to 1 proved difficult and inefficient, which necessitated the use of an allyl ester protection strategy that deprotects via non-hydrolytic mechanisms. However, direct evidence for the difference in reactivities between P-CJ-CMP and P-Pan-CMP will have to be obtained through experimentation with model systems; these studies are currently underway.
Nonetheless, the results of the temperature melting curve analyses clearly show that P-CJ-CMP also significantly improves the overall stabilization of PPCS oligomers compared to the natural intermediate P-Pan-CMP, indicating that the stabilization of certain conformations of the enzyme could also be an important factor causing inhibition. The finding that the extent of apparent stabilization is much larger for SaCoaBC than EcPPCS (which is in agreement with the significantly lower Ki observed in the case of the former protein) could be due to the increase in stability being compounded in the bifunctional CoaBC proteins that usually form dodecamers instead of the dimers formed by most PPCS enzymes. We conclude that this stabilization prevents further reaction by cysteine by protecting the potentially reactive P-CJ-CMP intermediate, resulting in its trapping in the active site and the observed inhibition. This analysis is supported by the available EcPPCS structures, which show significant differences in the active site architectures of the CTP (PDB: 1U7W), P-Pan-CMP (PDB: 1U7Z) and CMP-bound (PDB: 1U70) forms of the enzyme, indicating that the enzyme undergoes significant movement during catalysis.
In conclusion, we have demonstrated that the natural product CJ-15,801 hijacks the CoA biosynthetic pathway, and inhibits its second enzyme by forming a tight-binding inhibitor in situ within the active site. This mechanism is highly reminiscent of that of the sulfonamide antibiotics, which inhibit the biosynthesis of another vitamin—folic acid—in a similar manner. The mechanism is also complementary to that employed by a adenosine sulfamate inhibitor of ubiquitin-like protein conjugation pathways, which also forms a non-hydrolyzable mimic of the its target’s reaction intermediate in situ (Brownell, et al., 2010; Chen, et al., 2011). Taken together, these findings provide important new insights into inhibitors that target CoA biosynthesis and CoA-utilizing enzymes. They also highlight an alternative strategy for inhibitor development—one based on the stabilization of certain protein conformations—that may also hold significant potential for drug design efforts targeting other medicinally-relevant synthetase enzymes.
SIGNIFICANCE
The antibiotic CJ-15,801 was found to be a selective inhibitor of CoA biosynthesis in S. aureus due to the unique selectivity of SaPanK, the first enzyme of the CoA pathway, which currently is the only known example of an active bacterial PanKII enzyme. This demonstrates that the diversity of PanK enzymes can be exploited for selective inhibition or, as in this case, specific gatekeeping functions. The phospho-CJ-15,801 product that forms as a result of the phosphorylation by PanK subsequently acts as a substrate for PPCS, which transfers a cytidyl group to its carboxylate to form P-CJ-CMP. Surprisingly, this reaction does not result in the formation of an electrophilically-reactive intermediate; instead P-CJ-CMP acts as a potent tight-binding inhibitor of the enzyme. The results of temperature-dependent stability studies indicate that this inhibition is at least partly due to stabilization of the interactions between the adjacent monomers in the PPCS dimer, which seemingly prevents the reaction of cysteine—the second substrate in the PPCS reaction—with the activated acyl group of P-CJ-CMP. These findings indicate that the inhibition of synthetase enzymes by non-reactive analogues of their intermediates may have an alternative mechanistic basis in some cases, which is the stabilization of certain structural conformations, and that this should be considered as a potential new strategy for the inhibition of these enzymes. The findings also reveal CJ-15,801 as an antimetabolite of a vitamin biosynthetic pathway, similar to the sulfonamide antibiotics. Finally, we show that the potency of CJ-15,801 is significantly improved in combination with a pantothenamide, a known CoA antimetabolite precursor, confirming that these antimicrobials have different points of action and establishing the potential of drug development strategies that are focused on enzymes involved in CoA biosynthesis and utilization.
EXPERIMENTAL PROCEDURES
See Supplemental Experimental Procedures for full details of all synthetic procedures (including characterization data for all compounds), antibacterial activity assays, procedures for the preparation of proteins, enzyme assays and accompanying data analyses, and protein melting temperature determinations and analysis.
Synthesis of CJ-15,801 and its analogues
The inhibitor and its analogues were prepared by modification of published procedures (Figure S3) (Nicolaou and Mathison, 2005; Sewell, et al., 2011; Villa, et al., 2007).
Inhibition assays and analysis
The activity of CJ-15,801 against test strains of S. aureus, S. agalactiae, E. coli, P. aeruginosa and B. subtilis was assessed by MIC assays performed by microbroth dilution in 96-well microtiter plates and turbidometric analysis at OD600. The effect of pantothenate and its analogues on the antibiotic activity of CJ-15,801 against S. aureus was assessed in a checkerboard assay to calculate fractional inhibitory concentration index (FICI) indicative of synergy, additivity, indifference, or antagonism(Orhan, et al., 2005).
PanK assay and data analysis
Pantothenate kinase activity was determined using a continuous spectrophotometric assay that coupled the production of ADP to the consumption of NADH as described previously (Brand and Strauss, 2005; Strauss and Begley, 2002). All enzyme assays were based on decrease of NADH concentration, as monitored by change in absorbance at 340 nm. An extinction coefficient of 6220 M−1.cm−1 was used for NADH. Assays were performed in 96-well UV transparent plates (Greiner Bio) and monitored for between 10 minutes and one hour at 25 °C using a Perkin-Elmer HTS 7000 Bio-Assay Reader or a Thermo Scientific Varioskan Multimode Reader. For EcPanKI and SaPanKII each 320 μL reaction contained ATP (1.5 mM), MgCl2 (10 mM), KCl (20 mM), NADH (0.3 mM), phosphoenolpyruvate (0.5 mM), pyruvate kinase (5 units), and lactate dehydrogenase (5 units) in 50 mM HEPES buffer (pH 7.5). For PaPanKIII each 300 μl contained ATP (5.0 mM), MgCl2 (1 mM), NH4Cl (60 mM), NADH (0.5 mM), phosphoenolpyruvate (2.0 mM), pyruvate kinase (2 units), and lactate dehydrogenase (2.75 units) in 100 mM HEPES buffer (pH 7.5). The concentration of pantothenate, CJ-15,801, and other substrate analogues was varied between 6.25 and 200 μM. Reactions were initiated by addition of SaPanKII (3 μg), EcPanKI (3.5 μg), or PaPanKIII (5 μg). Initial velocities were calculated for each substrate concentration and fitted to the Michaelis-Menten equation using Prism software (Graphpad) or SigmaPlot 11.0 (Systat software). All measurements were obtained in triplicate.
PPCS assay and data analysis
PPCS assays were performed according to the published procedure (Patrone, et al., 2009; Yao, et al., 2009). The PPCS reaction was observed in the forward reaction via an enzyme-linked assay in which the pyrophosphate which is released during transfer of CMP from CTP to the substrate is continuously detected; this is achieved using the commercially available pyrophosphate reagent from Sigma-Aldrich (Cat. # P7275). Each vial of the pyrophosphate reagent was resuspended in 4.5 mL of dH2O. The assays were performed on a Biotek PowerWave microplate spectrophotometer at 37 °C. All curve fitting analyses were performed using SigmaPlot 11.0 (Systat software).
Molecular modeling
Modeling studies were performed using Accelrys Discovery Studio 3.1 (DS 3.1). The co-ordinates for EcPPCS Asn210Asp containing P-Pan-CMP as a co-crystalized ligand were obtained from the protein data bank (PDB code: 1U7Z). The structure was optimized prior to any receptor-ligand calculations (full details of the receptor preparation are given in the Supplemental Experimental Procedures). Docking of P-CJ-CMP was accomplished using CDocker (Wu, et al., 2003), launched from within DS 3.1. Deviations from the default settings included generating 150 starting conformations of the ligands to adequately sample conformational space and 50 poses were selected for simulated annealing. The top scoring pose (CDOCKER_Energy) was visually inspected and found to mimic the binding of the co-crystalized P-Pan-CMP. To ensure that the optimized water bridging the connection between the Asn210 amide side chain and the phosphate of P-Pan-CMP was not biasing the pose found in the docking, the exercise was repeated without this water molecule, which resulted in a nearly identical pose being found. Similarly, docking of P-CJ-CMP in the original structure of the EcPPCS Asn210Asp mutant (1U7Z) again showed that P-CJ-CMP very closely mimics the binding pose of the P-Pan-CMP molecule co-crystallized in the active site (Figure S4).
Protein melting temperature determinations and analysis
Protein melting curves were determined by measuring the heat-induced unfolding of the PPCS proteins by circular dichroism (CD) at 220 nm in a 0.5 mm cuvette using an Applied Photophysics Chirascan-Plus CD spectrometer. Samples contained the PPCS protein (10 μM) and MgCl2 (1 mM) in 50 mM Tris-HCl buffer (pH 7.6) and either CTP (150 μM), CTP and P-Pan (150 μM each) or CTP and P-CJ (150 μM each).
Supplementary Material
HIGHLIGHTS.
CJ-15,801 is a natural product inhibitor of coenzyme A (CoA) biosynthesis
Its unique selectivity is due to the gatekeeping action of pantothenate kinase
The first two CoA biosynthetic enzymes convert it into a tight-binding inhibitor
Pantothenamides (precursors of known CoA antimetabolites) increase its potency
Acknowledgments
We thank the following persons for assistance with various experiments: Leisl Brand for the preparation of the SaCoaBC-expression vector, Andrew Mercer for expression of the PanK biosynthetic enzymes, Leanne Barnard for PanK assays and Andrew Smith for synthesis of CJ-15,801. This project was funded by grants from the National Research Foundation (NRF; FA2007041600013) and Medical Research council (MRC) of South Africa to E.S., and from National Institutes of Health to V.N. (GM084350), J.C.H. (T32CA009523) and M.D.B (R01GM086225). R.v.d.W. was supported by a scarce skills scholarship from the NRF.
Footnotes
Author contributions: R.v.d.W. prepared the proteins, performed all the PPCS enzyme activity assays and inhibitor characterization studies, and prepared P-CJ-OMe; J.C.H. and J.L.M. prepared all other compounds; J.L.M performed PanK kinetic assays; S.H and V.N. performed the inhibition studies and analyzed the resulting data; W.J.M. performed the melting temperature determinations; S.C.P. was responsible for the modeling studies; M.D.B. and E.S. designed and directed the research, and E.S. wrote the paper with input from all the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balibar CJ, Hollis-Symynkywicz MF, Tao J. Pantethine Rescues Phosphopantothenoylcysteine Synthetase and Phosphopantothenoylcysteine Decarboxylase Deficiency in Escherichia coli but Not in Pseudomonas aeruginosa. J Bacteriol. 2011;193:3304–3312. doi: 10.1128/JB.00334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LA, Strauss E. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J Biol Chem. 2005;280:20185–20188. doi: 10.1074/jbc.C500044200. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Molecular Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Tsu CA, Gavin JM, Milhollen MA, Bruzzese FJ, Mallender WD, Sintchak MD, Bump NJ, Yang X, Ma J, et al. Mechanistic Studies of Substrate-assisted Inhibition of Ubiquitin-activating Enzyme by Adenosine Sulfamate Analogues. J Biol Chem. 2011;286:40867–40877. doi: 10.1074/jbc.M111.279984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry AE, Mandichak TL, Broskey JP, Egolf RW, Kinsland C, Begley TP, Seefeld MA, Ku TW, Brown JR, Zalacain M, et al. Inhibitors of pantothenate kinase: Novel antibiotics for staphylococcal infections. Antimicrob Agents Chemother. 2003;47:2051–2055. doi: 10.1128/AAC.47.6.2051-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar JS, Tan DS. Small molecule inhibition of microbial natural product biosynthesis-an emerging antibiotic strategy. Chem Soc Rev. 2008;37:1320–1329. doi: 10.1039/b702780j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton G, Bryant SR, Skinner CG. N1-(substituted) pantothenamides, antimetabolites of pantothenic acid. Arch Biochem Biophys. 1970;137:523–528. doi: 10.1016/0003-9861(70)90470-4. [DOI] [PubMed] [Google Scholar]
- Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. Hoboken, NJ: John Wiley & Sons, Inc; 2005. Tight Binding Inhibitors; pp. 179–213. [Google Scholar]
- Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. New York: Wiley-VCH; 2000. Tight Binding Inhibitors; pp. 305–317. [Google Scholar]
- delCardayre SB, Davies JE. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J Biol Chem. 1998;273:5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- Gulick AM. Conformational Dynamics in the Acyl-CoA Synthetases, Adenylation Domains of Non-ribosomal Peptide Synthetases, and Firefly Luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Shen R, Su S, Porco JA., Jr Copper-Mediated Synthesis of N-Acyl Vinylogous Carbamic Acids and Derivatives: Synthesis of the Antibiotic CJ-15,801. Org Lett. 2004;6:27–30. doi: 10.1021/ol0360041. [DOI] [PubMed] [Google Scholar]
- Hollenhorst MA, Ntai I, Badet B, Kelleher NL, Walsh CT. A Head-to-Head Comparison of Eneamide and Epoxyamide Inhibitors of Glucosamine-6-Phosphate Synthase from the Dapdiamide Biosynthetic Pathway. Biochemistry. 2011;50:3859–3861. doi: 10.1021/bi2004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong BS, Yun MK, Zhang YM, Chohnan S, Rock CO, White SW, Jackowski S, Park HW, Leonardi R. Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties. Structure. 2006;14:1251–1261. doi: 10.1016/j.str.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Imtiaz U, Billings E, Knox JR, Manavathu EK, Lerner SA, Mobashery S. Inactivation of class A. beta.-lactamases by clavulanic acid: the role of arginine-244 in a proposed nonconcerted sequence of events. J Am Chem Soc. 1993;115:4435–4442. [Google Scholar]
- Kim S, Lee SW, Choi EC, Choi SY. Aminoacyl-tRNA synthetases and their inhibitors as a novel family of antibiotics. Appl Microbiol Biotechnol. 2003;61:278–288. doi: 10.1007/s00253-003-1243-5. [DOI] [PubMed] [Google Scholar]
- Kucharczyk N, Denisot MA, Le Goffic F, Badet B. Glucosamine-6-phosphate synthase from Escherichia coli: determination of the mechanism of inactivation by N3-fumaroyl-L-2,3-diaminopropionic derivatives. Biochemistry. 1990;29:3668–3676. doi: 10.1021/bi00467a012. [DOI] [PubMed] [Google Scholar]
- Kupke T. Active-site residues and amino acid specificity of the bacterial 4′-phosphopantothenoylcysteine synthetase CoaB. Eur J Biochem. 2004;271:163–172. doi: 10.1046/j.1432-1033.2003.03916.x. [DOI] [PubMed] [Google Scholar]
- Kupke T. Molecular Characterization of the 4′-Phosphopantothenoylcysteine Synthetase Domain of Bacterial Dfp Flavoproteins. J Biol Chem. 2002;277:36137–36145. doi: 10.1074/jbc.M206188200. [DOI] [PubMed] [Google Scholar]
- Lee JM, Ahn DS, Jung DY, Lee J, Do Y, Kim SK, Chang S. Hydrogen-Bond-Directed Highly Stereoselective Synthesis of Z-Enamides via Pd-Catalyzed Oxidative Amidation of Conjugated Olefins. J Am Chem Soc. 2006;128:12954–12962. doi: 10.1021/ja0639315. [DOI] [PubMed] [Google Scholar]
- Leonardi R, Chohnan S, Zhang YM, Virga KG, Lee RE, Rock CO, Jackowski S. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J. Biol. Chem. 2005;280:3314–3322. doi: 10.1074/jbc.M411608200. [DOI] [PubMed] [Google Scholar]
- Mallett TC, Wallen JR, Karplus PA, Sakai H, Tsukihara T, Claiborne A. Structure of Coenzyme A-Disulfide Reductase from Staphylococcus aureus at 1.54 Å Resolution. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoj N, Strauss E, Begley TP, Ealick SE. Structure of Human Phosphopantothenoylcysteine Synthetase at 2.3 Å Resolution. Structure. 2003;11:927–936. doi: 10.1016/s0969-2126(03)00146-1. [DOI] [PubMed] [Google Scholar]
- May JJ, Finking R, Wiegeshoff F, Weber TT, Bandur N, Koert U, Marahiel MA. Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium’s susceptibility to antibiotics that target the cell wall. FEBS J. 2005;272:2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- Mercer AC, Burkart MD. The ubiquitous carrier protein-a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- Mercer AC, Meier JL, Torpey JW, Burkart MD. In Vivo Modification of Native Carrier Protein Domains. ChemBioChem. 2009;10:1091–1100. doi: 10.1002/cbic.200800838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton G, Arnold K, Price M, Sherrill C, Delcardayre S, Aharonowitz Y, Cohen G, Davies J, Fahey R, Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, Claiborne A. Structure of the Type III Pantothenate Kinase from Bacillus anthracis at 2.0 Å Resolution: Implications for Coenzyme A-Dependent Redox Biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou KC, Mathison CJN. Synthesis of imides, N-acyl vinylogous carbamates and ureas, and nitriles by oxidation of amides and amines with Dess-Martin periodinane. Angew Chem, Int Ed. 2005;44:5992–5997. doi: 10.1002/anie.200501853. [DOI] [PubMed] [Google Scholar]
- Orhan G, Bayram A, Zer Y, Balci I. Synergy Tests by E Test and Checkerboard Methods of Antimicrobial Combinations against Brucella melitensis. J Clin Microbiol. 2005;43:140–143. doi: 10.1128/JCM.43.1.140-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padayatti PS, Helfand MS, Totir MA, Carey MP, Carey PR, Bonomo RA, van den Akker F. High Resolution Crystal Structures of the trans-Enamine Intermediates Formed by Sulbactam and Clavulanic Acid and E166A SHV-1 β-Lactamase. J Biol Chem. 2005;280:34900–34907. doi: 10.1074/jbc.M505333200. [DOI] [PubMed] [Google Scholar]
- Padayatti PS, Sheri A, Totir MA, Helfand MS, Carey MP, Anderson VE, Carey PR, Bethel CR, Bonomo RA, Buynak JD, et al. Rational Design of a β-Lactamase Inhibitor Achieved via Stabilization of the trans-Enamine Intermediate: 1.28 Å Crystal Structure of wt SHV-1 Complex with a Penam Sulfone. J Am Chem Soc. 2006;128:13235–13242. doi: 10.1021/ja063715w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrone JD, Yao J, Scott NE, Dotson GD. Selective Inhibitors of Bacterial Phosphopantothenoylcysteine Synthetase. J Am Chem Soc. 2009;131:16340–16341. doi: 10.1021/ja906537f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope AJ, Moore KJ, McVey M, Mensah L, Benson N, Osbourne N, Broom N, Brown MJB, O’Hanlon P. Characterization of Isoleucyl-tRNA Synthetase from Staphylococcus aureus. J Biol Chem. 1998;273:31691–31701. doi: 10.1074/jbc.273.48.31691. [DOI] [PubMed] [Google Scholar]
- Saliba KJ, Kirk K. CJ-15,801, a fungal natural product, inhibits the intraerythrocytic stage of Plasmodium falciparum in vitro via an effect on pantothenic acid utilization. Mol Biochem Parasitol. 2005;141:129–131. doi: 10.1016/j.molbiopara.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Schimmel P, Tao J, Hill J. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J. 1998;12:1599–1609. [PubMed] [Google Scholar]
- Sewell AL, Villa MVJ, Matheson M, Whittingham WG, Marquez R. Fast and Flexible Synthesis of Pantothenic Acid and CJ-15,801. Org Lett. 2011;13:800–803. doi: 10.1021/ol103114w. [DOI] [PubMed] [Google Scholar]
- Sieber SA, Marahiel MA. Molecular Mechanisms Underlying Nonribosomal Peptide Synthesis: Approaches to New Antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- Spry C, Kirk K, Saliba KJ. Coenzyme A biosynthesis: an antimicrobial drug target. FEMS Microbiol Rev. 2008;32:56–106. doi: 10.1111/j.1574-6976.2007.00093.x. [DOI] [PubMed] [Google Scholar]
- Spry C, van Schalkwyk DA, Strauss E, Saliba KJ. Pantothenate utilization by plasmodium as a target for antimalarial chemotherapy. Infect Disord Drug Targets. 2010;10:200–216. doi: 10.2174/187152610791163390. [DOI] [PubMed] [Google Scholar]
- Stanitzek S, Augustin MA, Huber R, Kupke T, Steinbacher S. Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase. Structure. 2004;12:1977–1988. doi: 10.1016/j.str.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Strauss E. Coenzyme A Biosynthesis and Enzymology. In: Mander L, Liu H-W, editors. Comprehensive Natural Products II Chemistry and Biology. Oxford: Elsevier; 2010. pp. 351–410. [Google Scholar]
- Strauss E, Begley TP. The antibiotic activity of N-pentylpantothenamide results from its conversion to ethyldethia-coenzyme A, a coenzyme A antimetabolite. J Biol Chem. 2002;277:48205–48209. doi: 10.1074/jbc.M204560200. [DOI] [PubMed] [Google Scholar]
- Strauss E, Begley TP. Mechanistic studies on phosphopantothenoylcysteine decarboxylase. J Am Chem Soc. 2001;123:6449–6450. doi: 10.1021/ja016020y. [DOI] [PubMed] [Google Scholar]
- Strauss E, de Villiers M, Rootman I. Biocatalytic Production of Coenzyme A Analogues. ChemCatChem. 2010;2:929–937. [Google Scholar]
- Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP. Phosphopantothenoylcysteine synthetase from Escherichia coli Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J Biol Chem. 2001;276:13513–13516. doi: 10.1074/jbc.C100033200. [DOI] [PubMed] [Google Scholar]
- Sugie Y, Dekker KA, Hirai H, Ichiba T, Ishiguro M, Shiomi Y, Sugiura A, Brennan L, Duignan J, Huang LH, et al. CJ-15,801, a novel antibiotic from a fungus, Seimatosporium sp. J Antibiot. 2001;54:1060–1065. doi: 10.7164/antibiotics.54.1060. [DOI] [PubMed] [Google Scholar]
- Thomas J, Cronan JE. Antibacterial activity of N-pentylpantothenamide is due to inhibition of coenzyme A synthesis. Antimicrob Agents Chemother. 2010;54:1374–1377. doi: 10.1128/AAC.01473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Westhuyzen R, Strauss E. Michael acceptor-containing coenzyme A analogues as inhibitors of the atypical coenzyme A disulfide reductase from Staphylococcus aureus. J Am Chem Soc. 2010;132:12853–12855. doi: 10.1021/ja106204m. [DOI] [PubMed] [Google Scholar]
- van Wyk M, Strauss E. Development of a method for the parallel synthesis and purification of N-substituted pantothenamides, known inhibitors of coenzyme A biosynthesis and utilization. Org Biomol Chem. 2008;6:4348–4355. doi: 10.1039/b811086g. [DOI] [PubMed] [Google Scholar]
- Villa MVJ, Targett SM, Barnes JC, Whittingham WG, Marquez R. An Efficient Approach to the Stereocontrolled Synthesis of Enamides. Org Lett. 2007;9:1631–1633. doi: 10.1021/ol070336e. [DOI] [PubMed] [Google Scholar]
- Virga KG, Zhang YM, Leonardi R, Ivey RA, Hevener K, Park HW, Jackowski S, Rock CO, Lee RE. Structure-activity relationships and enzyme inhibition of pantothenamide-type pantothenate kinase inhibitors. Bioorg Med Chem. 2006;14:1007–1020. doi: 10.1016/j.bmc.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Wu G, Robertson DH, Brooks CL, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER—A CHARMm-based MD docking algorithm. J Comp Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- Yang K, Eyobo Y, Brand LA, Martynowski D, Tomchick D, Strauss E, Zhang H. Crystal structure of a type III pantothenate kinase: insight into the mechanism of an essential coenzyme A biosynthetic enzyme universally distributed in bacteria. J Bacteriol. 2006;188:5532–5540. doi: 10.1128/JB.00469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Strauss E, Huerta C, Zhang H. Structural Basis for Substrate Binding and the Catalytic Mechanism of Type III Pantothenate Kinase. Biochemistry. 2008;47:1369–1380. doi: 10.1021/bi7018578. [DOI] [PubMed] [Google Scholar]
- Yao J, Dotson GD. Kinetic characterization of human phosphopantothenoylcysteine synthetase. Biochim Biophys Acta, Proteins Proteomics. 2009;1794:1743–1750. doi: 10.1016/j.bbapap.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Yao J, Patrone JD, Dotson GD. Characterization and Kinetics of Phosphopantothenoylcysteine Synthetase from Enterococcus faecalis. Biochemistry. 2009;48:2799–2806. doi: 10.1021/bi802240w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Frank MW, Virga KG, Lee RE, Rock CO, Jackowski S. Acyl carrier protein is a cellular target for the antibacterial action of the pantothenamide class of pantothenate antimetabolites. J Biol Chem. 2004;279:50969–50975. doi: 10.1074/jbc.M409607200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.