Abstract
It seems quite clear that microRNAs play important roles in neuro-oncology, as they do across perhaps all areas in biology. With recent advances in detecting and quantifying microRNAs in tissue and serum, it appears increasingly likely that microRNAs will be clinically useful as biomarkers for brain tumors and other cancers. Applying microRNAs for treatment of brain tumors poses greater hurdles by far, however, and despite promising in vitro results this may never become a reality. This review fits recent advances into a framework for considering the potential of microRNAs for brain tumor therapy, considering the power of individual microRNAs, delivery issues, and indirect microRNA-based therapies.
Keywords: MicroRNA, Glioblastoma, Glioma, Delivery
Introduction
The treatment of malignant brain tumors likely represents one of the greatest challenges confronting modern medicine. Their sensitive and inaccessible location, invasiveness, and relative resistance to standard therapies such as radiation and chemotherapy have forced an urgent search for more sophisticated treatment modalities. This is especially true for glioblastoma (GBM), the most aggressive brain tumor and the most common in adults. As we have come to better understand the genetic lesions and pathways that drive GBMs, it has become possible to consider targeted therapies that block specific genes and pathways. This is frequently attempted through the use of small-molecule drugs, which can have major advantages such as oral bioavailability and ability to penetrate the blood–brain barrier. However, they typically act by blocking enzymatic activity, leaving the target protein intact. An alternative approach under development, RNA interference, can circumvent this limitation by dramatically reducing expression of target proteins. MicroRNA represents an endogenous form of RNA interference that is generating considerable excitement, both for its roles in GBM biology and also for its therapeutic potential. But is that therapeutic potential realistic? In this review we will systematically address this question from different angles.
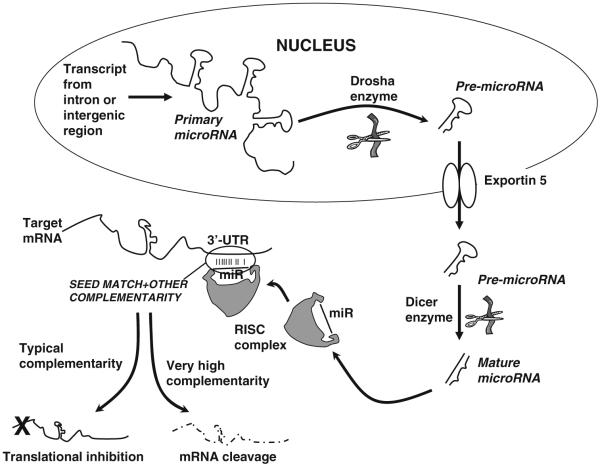
RNA interference was first discovered when C. elegans worms were fed short double-stranded RNAs mimicking regions of host mRNAs, which led to the surprising result that those mRNAs were then down-regulated [1]. This led to subsequent findings in many other organisms, including humans, that delivery into cells of such short double-stranded RNAs, termed siRNAs for small (or short) interfering RNAs, also caused degradation of the target mRNAs containing the siRNA sequences [2-4]. Sometimes in parallel but usually lagging behind, discoveries were made of endogenous analogs of siRNAs in eukaryotic cells from plant to human [5-8]. These “microRNAs” are formed as hairpins in transcribed RNAs that form via the annealing of adjacent partially complementary regions. These harpins are found in loci not coding for proteins, either in introns or intergenic areas, and the human genome is estimated to hold approximately 1,000 [9, 10]. After the transcription of the primary microRNA (pri-miRNA), the hairpin (the pre-miRNA) is spliced out in the nucleus by the Drosha/DGCR8 complex [11]. The pre-miRNA is then exported to the cytoplasm by the exportin-5 protein, where it is processed to the final mature miRNA by the Dicer protein [12, 13]. One strand of this mature miRNA is then utilized by RISC (RNA-induced silencing complex) to target mRNAs whose 3′-untranslated regions (3′-UTRs) have one or more regions of partial complementarity to the miRNA strand [14]. Typically this region of partial complementarity needs to include a “seed match”—an area of perfect complementarity to bases 2–8 or 1–7 of the miRNA [15]. MicroRNA/target interactions typically lead to suppressed translation of target mRNAs [16, 17], though sometimes when there is higher complementarity it leads to degradation of target mRNAs [18-20]. Importantly, each miRNA has many targets, and genes are commonly targeted by multiple miRNAs. The microRNA life cycle is shown in Fig. 1.
Fig. 1.
MicroRNA synthesis and mechanism
MicroRNAs have had a dramatic impact across biology, and major developments continue to come at a rapid pace in reports on both the plant and animal kingdoms. Despite their small size, microRNAs powerfully regulate the human proteome. MiRNAs appear to play especially important roles in the nervous system and in oncology, so it is unsurprising that a growing number of publications have linked miRNAs to brain tumors. Many of these reports demonstrate biological roles for individual miRNAs in neuro-oncology, and some have even suggested the clinical potential of delivering tumor-suppressive miRNAs or inhibitors of oncogenic miRNAs. But is it realistic to think we might be able to treat brain tumors with such miRNA-based therapies? This review will address the key questions subsumed within this larger one: (1) Can individual microRNAs have a powerful enough role in GBM that effective delivery of a single miRNA or an inhibitor could have therapeutic effect? (2) Can adequate delivery of miRNAs or miRNA inhibitors for therapeutic effect be achieved? (3) Regardless of whether direct delivery of miRNAs or miRNA inhibitors is feasible, are there indirect miRNA-based therapies that might have a strong chance of success?
Can individual microRNAs have a major impact on brain tumors?
Several microRNAs, termed oncomiRs, have been shown to act in an oncogenic fashion in diverse cancers, including GBM. miR-21 was the first oncomiR to be demonstrated in GBM [21, 22], and is the most deeply investigated to date. In some settings its overexpression is driven by the transcription factors STAT3 and AP-1 [23, 24], and its relevant targets appear to include tumor-suppressive genes such as PDCD4, PTEN, TPM1, RECK, and TIMP3 [25-28]. Importantly, inhibiting miR-21 with a short complementary oligonucleotide modified for stability decreases glioma cell viability [22, 29]. Delivery of an oncomiR inhibitor such as this is one means to directly incorporate miRs into GBM therapy.
Other microRNAs have also shown oncogenic character in GBM. Like miR-21, the analogous microRNAs miR-221 and miR-222 are over-expressed in numerous cancers, including GBM, and appear to boost cell growth and survival [30-33]. The key downstream targets are unclear, but the cell cycle inhibitors p27 and p57 have both been proposed [34, 35]. miR-10b is another that has been shown to be up-regulated in both GBM and other cancers [30, 36,37]. In breast cancer miR-10b has been found to migration invasion via RhoC, and in GBM its expression has been found to up-regulate both uPAR (urokinase plasminogen activator receptor) and RhoC [36, 38]. Recently an oncogene cluster was discovered in GBM that includes a putative oncomiR, miR-26a, along with the genes CDK4 and CENTG1 [39].
A number of miRNAs with tumor-suppressive properties have also been identified, some of which seem to be shared with other cancers and some which may be unique to glioblastoma. The let-7 miRNAs are potently tumor-suppressive miRNAs and tend to be highly expressed in normal tissues but not most cancers, and act to inhibit key oncogenes such as K-Ras and Myc [40-43]. While not well-explored in GBM, forced expression of let-7 miRs has been shown to reduce proliferation of GBM cells [44]. miR-34a is another well-established tumor suppressor found to be an important downstream mediator of p53 [45-48], and recent reports establish a role for it in GBM [49,50]. A number of important targets have been identified for miR-34a, including Notch family members and the c-met oncogene [50]. An early report on miRs in GBM identified miR-124 and miR-137 as potential tumor suppressor miRs [37]. miR-7 demonstrates characteristics of a tumor suppressor in GBM, and it inhibits expression of the epidermal growth factor receptor (EGFR) and members of the Ras and Akt pathways [51]. miR-128 inhibits the Bmi-1 oncogene and can act as a differentiating tumor suppressor in GBM as well [52]. Recently, miR-326 was found to act in a feedback loop with the Notch pathway and to have tumor-suppressive characteristics in GBM [53].
miRNAs have been linked specifically with a number of key features of GBMs. In the area of metabolism, long known to be aberrant in cancer but increasingly seen as critical in oncology, an exciting report recently showed miR-451 to be regulated in GBM by glucose levels and to affect cell proliferation and migration [54]. Angiogenesis is critical in most solid tumors, notably including GBM, and miR-296 has been demonstrated to be up-regulated in GBM-associated endothelial cells [55]. This miRNA promotes angiogenesis by down-regulating HGS (hepatocyte growth factor-regulated tyrosine kinase substrate), an inhibitor of pro-angiogenic receptors VEGFR2 and PDGFRβ (vascular endothelial growth factor and platelet-derived growth factor receptor beta, respectively). This study raised the prospect of anti-angiogenic therapy of GBM by delivery of a miR-296 inhibitor. Immunosuppression is another trait allowing GBMs to develop, and miRs have been shown to play a role here as well; miR-222 and miR-339 were found to promote GBM immune evasion by down-regulating the immunologic molecule ICAM-1 [56]. Treatment resistance is one of the principal challenges in GBM therapy, and early results hint at miRNA involvement in this phenomenon. A temozolomide-resistant GBM cell line was found to have several up-regulated miRNAs versus the parental line, including miR-195, miR-455-3p, and miR-10a* [57]. miR-181a, down-regulated in GBM, may influence its sensitivity to radiation [58] (Table 1).
Table 1.
MicroRNAs implicated in glioblastoma
| Category | MicroRNAs | Relevant targets | References |
|---|---|---|---|
| Oncogenic miRs | |||
| miR-21 | PDCD4, PTEN, TPM1, RECK, TIMP3 |
[21-28] | |
| miRs-221/222 | p27, p57 | [30-35] | |
| miR-10b | Up-regulates uPAR, RhoC | [30, 36-38] | |
| miR-26a | Unknown | [39] | |
| Tumor-suppressive miRs | let-7 family | K-Ras, Myc | [40-44] |
| miR-34a | c-met, Notch family | [49, 50] | |
| miR-124a | Unknown | [37] | |
| miR-137 | Unknown | [37] | |
| miR-7 | EGFR; Akt pathway | 51] | |
| miR-128 | Bmi-1 | [52] | |
| miR-326 | Notch family | [53] | |
| Metabolism | |||
| miR-451 | CAB39 | [54] | |
| Angiogenesis | |||
| miR-296 | HGS | [55] | |
| Immune escape | |||
| miR-22, miR-339 | ICAM-1 | [56] | |
| Treatment resistance | |||
| miR-195, miR-455-3p, miR-10a* |
Unknown | [57] | |
| miR-181a | Unknown | [58] |
Some previous reports show effective in vitro suppression of GBM cell viability with delivering the above-mentioned tumor-suppressive miRNAs or inhibitors of oncomiRs. Restoring expression in glioma cells of tumor-suppressive microRNAs such as miR-34a or miR-7 can strongly inhibit vital oncogenic pathways such as c-Met, Ras, and Akt. This can selectively slow cell division or even kill glioma cells while sparing normal cells. Similar effects in glioma cells can be achieved by delivering short oligonucleotide inhibitors of oncomiRs such as miR-21 or miR-221. While it is already clear from these studies that individual miRNAs may serve as potential therapeutic targets in GBM, no doubt many more remain to be discovered.
One important question for the development of RNA interference as therapy is whether it would be preferable to manipulate gene expression with delivery of miRNAs/miRNA inhibitors or siRNAs. siRNAs allow specific knockdown of individual gene targets, while microRNAs result in a broad swath of gene expression being affected. However, the ability of individual miRNAs to target multiple genes/pathways could be a major advantage, especially given studies indicating the therapeutic necessity of simultaneously targeting multiple pathways in GBM. siRNA advocates counter that a cocktail of a few siRNAs could be combined, but a similar cocktail of miRNAs would again allow more pathways to be targeted. The off-target effects of the miRNAs would likely be much higher, but on the other hand miRNAs are all expressed endogenously and will likely tend to be safe for normal cells. One issue to be considered with cocktails of either miRNAs or siRNAs is the ability to overload the RNA interference system, a possibility that has already been demonstrated [59].
Delivery—the principal problem
The thorniest hurdle in developing miRNA-based therapies for brain tumors is doubtless that of effective delivery, as is the case for most prospective therapies that embody RNA interference. However, brain tumors such as GBM present special issues relating to delivery. Due to the blood-brain barrier (BBB), it is unlikely that a systemically-administered vector could deliver adequate miRNA or miRNA inhibitor to a brain tumor. The BBB tends to be disrupted at the core of most glioblastomas and this may allow some delivery of a microRNA-based therapy via the circulation, but the BBB is intact in the surrounding brain where GBM cells have infiltrated. In a few strategies this may not be relevant, as in a vasculature-directed therapy such as delivery of miR-296 inhibitor. However, for most miRNA-based brain tumor therapies, it will be necessary to use local delivery techniques. Even with efficient local delivery the task might seem impossible, given that direct miRNA-based therapies would appear to act only on the cells receiving the miRNAs/miRNA inhibitors and to lack bystander effects. A few approaches may not require local delivery to be near-perfect, such as those employing miRNAs or miRNA inhibitors to up-regulate the anti-GBM immune response. But even for straightforward approaches in which miRNA or inhibitor must reach nearly all the cancer cells, certain phenomena may work in favor of such therapies for brain tumors.
Unlike most cancers, GBM rarely metastasizes outside the brain, giving local delivery at least a chance to succeed. Furthermore, in the last decade advances in local delivery to the brain have made exciting progress, particularly via the technique of convection-enhanced delivery (CED) (recently reviewed in Passirani et al. [60]). This entails prolonged low-pressure infusion via catheter, creating a pressure wave that drives the infusate to replace brain interstitial fluid over what can become a large field around the catheter tip. Even relatively large particles such as viruses can be delivered to large brain regions with some success. However, there remain substantial challenges still being addressed with CED, such as poorer delivery of infusate into the high-pressure environment of many brain tumors and backflow of infusate around catheters [61]. Despite its current limitations, CED likely represents the most promising modality for local delivery to the brain for most applications.
If local delivery with methods such as CED does not prove successful, other strategies may increase the chances of successful systemic delivery via the circulation. A number of agents have been proposed or tested that can disrupt the BBB [62, 63], which can be coupled with intraarterial infusion to increase delivery to the brain. Peptides have also been identified that can be conjugated to a payload and allow its transfer across the BBB [64], and such peptides could be used for delivery of modified tumor-suppressive microRNAs or inhibitors of oncomiRs.
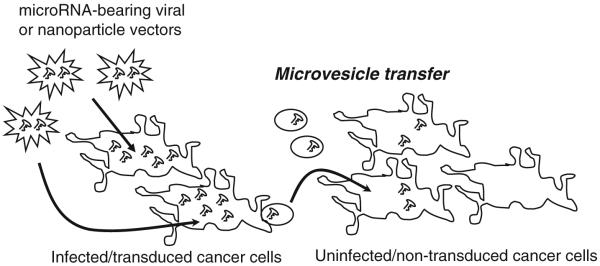
One recent discovery that may have dramatic implications for this field is that GBM cells bud off microvesicles that deliver cytoplasmic contents to nearby cells [65]. Thus mRNAs, microRNAs, proteins, and other cytoplasmic molecules may be “shared” by GBM cells with surrounding GBM and normal cells (Fig. 2), and this should certainly apply to transfected siRNAs/miRNAs/miRNA inhibitors that are present at high concentrations in the affected cells. This phenomenon could markedly reduce the threshold efficiency for effective delivery of a miRNA-based therapy to a fraction of what would otherwise be required. Some recent evidence—mostly unpublished—supports this possibility [66-68]. If this is borne out by further studies, it could radically improve the chances for miRNA therapies to succeed in neuro-oncology.
Fig. 2.
MicroRNA transfer between infected and uninfected cells
The vector used for miRNA/inhibitor delivery will no doubt strongly influence the chances for a viable therapy, and this is a field that is advancing rapidly. At the present time, the most suitable vectors for miRNA delivery are viruses encoding miRNAs/inhibitors or liposomal nanoparticles. Viruses may pose more worrisome safety issues, but offer the potential for prolonged high expression, as opposed to the brief pulse that would occur with nanoparticle delivery. However, new developments in nanoparticle technology are reported frequently, and these modalities are promising as well. One of the most interesting technologies involves nanoparticles with ferromagnetic cores, allowing MRI imaging or magnetic manipulation of the infused particles [60]. Possible viral vectors include adenovirus, lentivirus, and adeno-associated virus (AAV). Adenoviral delivery would yield a relatively short period of high expression, while lentiviral or AAV-based delivery would give incorporation into the genome with long-term lower expression. Thus adenovirus may be better-suited to miRNA delivery for GBM treatment, if the aim is to achieve a brief period with high expression of a lethal miRNA. There may also be particular issues with certain viruses. A recent report indicated that lentiviral delivery of shRNAs resulted in good knockdown of target with minimization of off-target effects [69]. However, siRNA/shRNA off-target effects tend to occur via inadvertent seed matches and microRNA-like activities; therefore the report hints that lentiviral delivery may be well-suited to siRNAs but might somehow interfere with miRNA activity.
Indirect miRNA-based therapies
In addition to direct delivery of miRNAs and miRNA inhibitors to GBMs, there may also be indirect ways to apply miRNAs to the challenge of GBM therapy. One intriguing approach involves using differential miRNA expression in target and non-target tissues to regulate expression of vector payloads. This was first established by the Naldini group in non-cancer settings [70-72], but it is eminently applicable to cancer therapy. At its most basic, the technique entails delivery of a suicide gene with an artificial 3′-UTR containing target sites for one or more miRNAs with high expression in normal cells but minimal expression in cancer cells. In normal cells, the miRNA(s) will suppress expression of the suicide gene, but cancer cells will maintain transgene expression and therefore die. An obvious contender as miRNA regulator is the let-7 family, which is highly expressed in most normal cells but down-regulated in cancer cells. A let-7-regulated cancer-specific virus has already been reported [73]. A similar virus designed to kill GBM cells has also been described, incorporating sites for miRNAs-31, -127, and -143 that were found to be highly expressed in astrocytes but not GBM cells [74]. An analogous strategy could be used to regulate a key viral gene to create a conditionally-replicating cancer-targeted virus, and the feasibility of this has been demonstrated for a hepatocyte-sparing adenovirus [75]. Such miRNA-regulated transgene approaches can be combined with other targeting strategies such as cancer-specific promoters, with the potential for elegantly multi-targeted therapies.
miRNAs may also have therapeutic impact by guiding therapeutic choices. One of the critical tasks now emerging in oncology is to match new targeted therapies to the cancer subsets most sensitive to them. This requires extensive profiling of each cancer to identify these distinct subsets, and evidence suggests that miRNA profiling may have advantages over traditional gene expression micro-arrays [76]. In the future we may be customizing cancer treatments not only by sequencing the genome of each patient’s cancer, but also by determining its microRNA and gene expression patterns.
Conclusion
With the advent of each new approach to cancer treatment, it seems there are bursts of wild optimism and disappointed pessimism before the emergence of a more realistic and cautiously hopeful middle ground. One hopes that with miRNA-based cancer therapies we may go directly to the final stage. While the challenges facing miRNA-based therapies in neuro-oncology are daunting, there are some hopeful new developments. microRNA “sharing” by microvesicles has the potential to reduce the efficiency required for delivery, perhaps the most intimidating hurdle. Numerous technologies are progressing, such as CED and nanoparticles. In the meantime, our knowledge of microRNAs in GBM and other brain tumors is advancing quickly. Combining all these factors, miRNA-based therapy for brain tumors does not appear to be such stuff as dreams are made on; the prognosis is one of guarded optimism.
References
- 1.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Weinstein EG, Rhoades MW, et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner G, Zamore PD. A microRNA in a multipleturnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 13.Lund E, Guttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 14.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 15.Lai EC, Tam B, Rubin GM. Pervasive regulation of Dro-sophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Valencia-Sanchez MA, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 18.Baek B, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:44–45. [Google Scholar]
- 19.Selbach M, Schwanhausser B, Thierfelder N, et al. Wide-spread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 20.Yu Z, Jian Z, Shen SH, et al. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krichevsky AM, King KS, Donahue CP, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 23.Fujita S, Ito T, Mizutani T, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Loffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 25.Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 26.Gabriely G, Wurdinger T, Kesari S, et al. MiR-21 promotes glioma invasion by targeting MMP regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu S, Wu H, Wu F, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 29.Corsten MF, Miranda R, Kasmieh R, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 30.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 31.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 32.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 33.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 35.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 37.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasayama T, Nishihara M, Kondoh T, et al. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Huang W, Jiang X, et al. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glio-blastoma survivorship. Proc Natl Acad Sci USA. 2010;107:2183–2188. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 41.Johnson SM, Grosshans H, Shingara J, et al. RAS is reg-ulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Bur-kitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 43.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 44.Lee ST, Chu K, Oh HJ, et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol. 2010 doi: 10.1007/s11060-010-0286-6. doi:10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 45.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 47.Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 49.Guessous F, Zhang Y, Kofman A, et al. MicroRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kefas B, Godlewski J, Comeau L, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 52.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 53.Kefas B, Comeau L, Floyd DH, et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–15168. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godlewski J, Nowicki MO, Bronisz A, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wurdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ueda R, Kohanbash G, Sasaki K, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci USA. 2009;106:10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ujifuku K, Mitsutake N, Takakura S, et al. miR-195, miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–248. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Slaby O, Lakomy R, Fadrus P, et al. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–269. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 59.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 60.Allard E, Passirani C, Benoit JP. Convection-enhanced delivery of nanocarriers for the treatment of brain tumors. Biomaterials. 2009;30:2302–2318. doi: 10.1016/j.biomaterials.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Bidros DS, Liu JK, Vogelbaum MA. Future of convection-enhanced delivery in the treatment of brain tumors. Future Oncol. 2010;6:117–125. doi: 10.2217/fon.09.135. [DOI] [PubMed] [Google Scholar]
- 62.Liu HL, Hua MY, Chen PY, et al. Blood-brain barrier disruption with focused ultrasound enhances delivery of chemotherapeutic drugs for glioblastoma treatment. Radiology. 2010;255:415–425. doi: 10.1148/radiol.10090699. [DOI] [PubMed] [Google Scholar]
- 63.Black KL, Ningaraj NS. Modulation of brain tumor capillaries for enhanced drug delivery selectively to brain tumor. Cancer Control. 2004;11:165–173. doi: 10.1177/107327480401100304. [DOI] [PubMed] [Google Scholar]
- 64.Thomas FC, Taskar K, Rudraraju V, et al. Uptake of ANG1005, a novel paclitaxel derivative, through the blood-brain barrier into brain and experimental brain metastases of breast cancer. Pharm Res. 2009;26:2486–2494. doi: 10.1007/s11095-009-9964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq601. doi:10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 68.Yuan A, Farber EL, Rapoport AL, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klinghoffer RA, Magnus J, Schelter J, et al. Reduced seed region-based off-target activity with lentivirus-mediated RNAi. RNA. 2010;16:879–884. doi: 10.1261/rna.1977810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown BD, Cantore A, Annoni A, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- 71.Brown BD, Venneri MA, Zingale A, et al. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 72.Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 73.Edge RE, Falls TJ, Brown CW, et al. A let-7 microRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- 74.Wu C, Lin J, Hong M, et al. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol Ther. 2009;17:2058–2066. doi: 10.1038/mt.2009.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ylosmaki E, Hakkarainen T, Hemminki A, et al. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific microRNA. J Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]