Abstract
Obese breast cancer patients exhibit a higher risk for larger tumor burden and increased metastasis. Molecular effects of obesity on carcinogenesis are mediated by autocrine and paracrine effects of adipocytokine leptin. Leptin participates in tumor progression and metastasis of human breast. We show that leptin induces clonogenicity and migration potential of breast cancer cells. We found that survivin expression is induced in response to leptin. In this study, we examine the role and leptin-mediated regulation of survivin. Leptin treatment leads to survivin upregulation, due in part to the activation of Notch1 and release of transcriptionally active Notch1-intracellular-domain (NICD). ChIP analysis show that NICD gets recruited to survivin promoter at CSL-binding-site in response to leptin treatment. Inhibition of Notch1 activity inhibits leptin-induced survivin upregulation. Leptin-induced transactivation of EGFR is involved in leptin-mediated Notch1 and survivin upregulation showing a novel upstream role of leptin-EGFR-Notch1 axis. We further show that leptin-induced migration of breast cancer cells requires survivin, as overexpression of survivin further increases, whereas silencing survivin abrogates leptin-induced migration. Using a pharmacological approach to inhibit survivin, we show that 3-hydroxy-3-methylglutaryl-coenzyme-A-reductase inhibitors (HRIs), lovastatin, can effectively inhibit leptin-induced survivin expression and migration. Importantly, leptin increased breast tumor growth in nude mice. These data show a novel role for survivin in leptin-induced migration and put forth pharmacological survivin inhibition as a potential novel therapeutic target. This conclusion is supported by in vivo data showing overexpression of leptin and survivin in epithelial cells of high grade ductal carcinoma in situ and high grade invasive carcinoma.
Keywords: Breast Cancer cell migration, Leptin, Notch, EGFR, Survivin
Introduction
Leptin is a multifunctional adipocytokine with a broad range of biological activities ranging from regulating appetite and energy expenditure to modulation of various processes such as reproduction, lactation, hematopoiesis, cell differentiation and importantly carcinogenesis (Huang and Li 2000). Leptin exerts its actions through its specific transmembrane receptors (OB-R) present in a variety of tissues (Ahima and Osei 2004) by activation of various downstream effector molecules. Several epidemiological studies have linked high levels of plasma leptin with increased risk for breast carcinogenesis, but a direct link between leptin signaling and breast cancer was first established in a clinical study showing that leptin receptors were not detectable in normal mammary epithelial cells by immunohistochemistry, whereas in 83% of cases, carcinoma cells showed positive staining for the leptin receptor (Ishikawa, et al. 2004). Importantly, overexpression of leptin was observed in 92% of breast tumors examined but in none of the cases of normal breast epithelium (Ishikawa, et al. 2004). Another noteworthy study showed a positive relationship between blood leptin levels and breast cancer risk. In addition, the degree of leptin mRNA expression in the peritumoral adipose tissue was significantly higher in the breast cancer patients than the control women (Tessitore et al., 2000). A recent study also showed that leptin and leptin receptor are overexpressed in primary and metastatic invasive ductal breast carcinoma compared with non-cancer mammary tissue (Garofalo et al., 2006). These studies suggest the importance of paracrine and autocrine effects of leptin and the importance of leptin receptor in breast carcinogenesis. In recent years many laboratories including ours have shown that leptin increases proliferation of breast, endometrial, hepatocellular and many other cancer cells via multiple signaling pathways including Stat3/ERK/Akt signaling (Housa, et al. 2006; Saxena, et al. 2007a; Saxena, et al. 2008; Saxena, et al. 2007b; Sharma, et al. 2006b). Leptin signaling can induce various molecules implicated in cell growth, adhesion, inflammation and angiogenesis, such as β3 integrin (Gonzalez and Leavis 2001), interleukin-1 (IL-1), IL-1 receptor (Pinteaux, et al. 2007), vascular endothelial growth factor and its receptor type 2 (Gonzalez, et al. 2006). Leptin regulates the cell cycle by inducing cyclin D1 expression modulating its local chromatin structure (Saxena et al. 2007b). Leptin increases E-cadherin expression via enhancing CREB-DNA and Sp1-DNA binding activity to E-cadherin promoter and modulates homotypic tumor cell adhesion and proliferation (Mauro, et al. 2007). Thus, uncontrolled leptin-induced signaling could contribute to various aspects of tumorigenesis and metastasis by modulating key involved genes.
Survivin, a member of inhibitor-of-apoptosis proteins (IAP) family, mainly functions to suppress apoptosis and regulate cell division (Altieri 2004). Although not expressed in normal adult differentiated tissue, it is present in large amounts in fetal tissue and most cancers, regardless of lineage, differentiation and histological type (Ambrosini, et al. 1997). Survivin expression correlates with increased aggressiveness, higher recurrence rate, increased metastasis, unfavorable disease outcome and abbreviated overall survival in colorectal, gastric, breast, ovarian, hepatocellular carcinoma and in neuroblastoma (Adida, et al. 2000; Altieri 2004; Kawasaki, et al. 1998; Satoh, et al. 2001). Elucidation of cancer specific transcription of survivin promoter showed convergence of several oncogenic pathways to up-regulate survivin gene expression in transformed cells. Growth factor receptor signaling, Stat activation, PI3K/Akt signaling, oncogene (Ras) expression, and loss of tumor suppressor molecules, p53, APC and PML (Aoki, et al. 2003; Dan, et al. 2004; Hoffman, et al. 2002; Kim, et al. 2003; Mirza, et al. 2002; O'Connor, et al. 2000; Sommer, et al. 2003; Xu, et al. 2004; Zhang, et al. 2001) are a few oncogenic pathways implicated in survivin regulation in cancer cells. Some downstream effector molecules of leptin signaling (such as Stat3 and Akt) participate in regulation of survivin expression. Increased survivin expression has been associated with more aggressive tumor behavior and parameters of poor prognosis in breast cancer. Survivin is an important IAP owing to its cancer-specific overexpression and its importance in inhibiting cell death and regulating cell division. Overexpression of leptin was observed in 92% of breast tumors examined (Ishikawa, et al. 2004). We hypothesized that elevated expression of leptin in breast tumors may upregulate survivin expression. We found that leptin increases the expression of survivin mRNA and protein.
In the present study, we specifically investigated the effect of leptin-induced survivin on migration of breast cancer cells, and examined the underlying molecular mechanisms by which leptin upregulates survivin expression. Intriguingly, we discovered the involvement of a novel upstream leptin-EGFR-Notch1 axis in survivin regulation in breast cancer cells treated with leptin. We also found that survivin is required for leptin-mediated migration of breast cancer cells, and that pharmacological inhibition of survivin can inhibit this early event of malignant progression.
Materials and Methods
Antibodies
Antibodies for Survivin and Leptin (Ob) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for XIAP, EGFR, pTyr, Notch1 and NICD were purchased from Cell Signaling Technology (Danvers, MA). Antibodies for β-actin were purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture, reagents and treatments
The human breast cancer cell lines, MCF7 and MDA-MB-231 were maintained in DMEM supplemented with 10% fetal bovine serum (Gemini Bioproducts, Woodland, CA) and 2μM L-glutamine (Invitrogen, Carlsbad, CA) (Saxena et al. 2008). For treatment, cells were seeded at a density of 1 × 106 /100-mm tissue culture dish. For leptin treatments, cells were incubated in serum-free media for 24 hours followed by treatment with human recombinant leptin (Sigma-Aldrich, St. Louis, MO) at 100ng/ml (Saxena et al. 2008) for indicated durations. In other sets of experiments, cells were treated with EGFR inhibitor erlotinib at 2.5 μM alone and in combination with leptin. In some experiments, cells were treated with 20-40μM lovastatin (Sigma-Aldrich). g-Secretase inhibitor LY411,575 was kindly provided by Dr. Clodia Osipo (Loyola University). For electric cell-substrate impedance sensing (ECIS) migration assay, ECIS cell culture ware was purchased from Applied Biophysics (Troy, NY).
Clonogenicity assay
Colony formation assay was performed following our previously published protocol (Taliaferro-Smith, et al. 2009). MCF7 and MDA-MB-231 breast cancer cells (single-cell suspension) were plated in 12-well plates at a density of 250 cells per well overnight. The following day, cells were treated with 100ng/ml human recombinant full-length leptin and the medium was replaced with fresh medium containing leptin every 3 days. After a 10 day treatment period, the medium was removed and cell colonies were stained with crystal violet (0.1% in 20% methanol). Colony numbers were assessed visually and colonies containing >50 normal-appearing cells were counted. Pictures were taken using a digital camera. All experiments were performed at least three times in triplicates.
Anchorage-independent growth assay
Anchorage-independent growth of MCF7 and MDA-MB-231 cells was assayed by colony formation on soft agar. Briefly, equal volumes of agar (1.2%) and complete medium were mixed to make 0.6% agar growth medium solution in 6-well tissue culture plates. Cells (2×103 cells/well) were suspended in media with or without treatment followed by mixing with equal volume of agar (0.6%). Cell suspension-agar mix (2 mL) was then added to each well. Plates were incubated at 37°C with 5% CO2 in humidified incubator for 3 weeks, and media with or without treatment were added every 3 days. Colonies were stained with 0.005% crystal violet in PBS for 1 hour at room temperature and observed using Olympus IX50 inverted microscope. Colonies were counted in six randomly selected fields at 10x magnification. Results are expressed as number of colonies counted. All experiments were performed three times in triplicate.
Migration Assay
Migration assays were performed following previously published protocol (Saxena et al. 2007a). Cells were plated into 24-well cell culture plate, pre-coated with human fibronectin (5μg/cm2, Sigma, St. Louis, MO). Cells were allowed to grow in 10% FBS containing DMEM medium to confluence, and then were washed with serum-free medium and serum starved for 16h. A 1-mm wide scratch was made across the cell layer using a sterile pipette tip. After washing with serum-free medium twice, DMEM medium containing 10μg/ml human fibronectin was added to replace matrix depleted with the cells. Plates were photographed immediately after scratching. Cells were treated with human recombinant leptin at 100ng/ml. Plates were photographed after 24h and 48h at the identical location of the initial image. All experiments were performed at least three times.
Invasion assay
Matrigel invasion assay was performed by using a Matrigel invasion chamber from BD Biocoat Cellware (San Jose, CA). Cells were seeded at a density of 1×105 cells per insert and cultured overnight. After 16 hours of serum starvation, the culture media were changed to serum free media containing treatments as indicated. Triplicate wells were used for each treatment. Cells were treated with leptin as indicated. After 24 hours of incubation, cells remaining above the insert membrane were removed by gentle scraping with a sterile cotton swab. Cells that had invaded through the Matrigel to the bottom of the insert were fixed in methanol for 10 minutes. After washing with PBS, the cells were stained with hematoxylineosin. The insert was subsequently washed with PBS and briefly air-dried and mounted. The slides were coded to prevent counting bias, and the number of invaded cells on representative sections of each membrane were counted under light microscope. The number of invaded cells for each experimental sample represents the average of triplicate wells. All experiments were performed at least three times.
RNA isolation and RT-PCR
RT-PCR analysis was performed following previously published protocol (Sharma, et al. 2006a), using specific primers for survivin, actin, Notch1, Notch4 and Hey1. Total cellular RNA was extracted using the TRIZOL Reagent kit (Life Technologies, Inc., Rockville, MD) and quantified by UV absorption. RT-PCR was carried out using specific sense and antisense PCR primers for amplification. PCR products were resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primers used were survivin-wt sense, 5’-GGA CCA CCG CAT CTC TAC AT-3’, antisense, 5’-GAC AGA AAG GAA AGC GCA AC-3’; β-actin sense, 5’-ACC ATG GAT GAT GAT ATC GC-3’ and antisense, 5’-ACA TGG CTG GGG TGT TGA AG-3’; Notch 1 sense, 5’-GCA ACA GCT CCT TCC ACT TC-3’, antisense, 5’-GCC TCA GAC ACT TTG AAG CC-3’; Notch 4 sense, 5’-GAA GCC AAA GGC AGA AGT TG -3’, antisense, 5’-TTC CAT CTC AGA TTC CTG GG-3’; Hey1 sense, 5’-AGC TCC TCG GAC AGC GAG CTG-3’, antisense, 5’-TAC CAG CCT TCT CAG CTC AGA CA-3’.
Western Blot
Whole cell lysate was prepared following previously described method (Sharma et al. 2006b). Whole cell lysates were prepared by scraping cells in 250μl of ice cold modified RIPA buffer [50mM Tris-Cl (pH 7.4), 150mM NaCl, 1mM EDTA, 1% NP-40, 0.25% Na-deoxycholate, 1mM PMSF, 10 μg/ml aprotinin, 10μg/ml leupeptin, 1mM Na3VO4 and 1mM NaF]. The lysate was rotated 360° for 1h at 4°C followed by centrifugation at 12,000g for 10min at 4°C to clear the cellular debris. Proteins were quantified using the Bradford protein assay kit (Biorad, Hercules, CA). Equal amounts of proteins were resolved on SDS-polyacrylamide gels and transferred to nitrocellulose membranes, and western blot analyses were performed using the previously described antibodies. Immuno-detection was performed by blocking the membranes for 1h in TBS buffer [20mM Tris-Cl (pH 7.5), 137mM NaCl, 0.05% Tween-20] containing 5% powdered nonfat milk followed by addition of the primary antibody (as indicated) in TBS for 2h at room temperature. Specifically bound primary antibodies were detected with peroxidase-coupled secondary antibodies and developed by enhanced chemiluminescence (ECL system, Amersham Pharmacia Biotech Inc., Arlington Heights, IL) according to manufacturer's instructions. All experiments were performed at least three-five times using independent biological replicates.
Immunohistochemistry
These studies are approved by Institutional Review Board. Archived specimens of paraffin-embedded normal breast tissue (n=4), ductal carcinoma in situ (DCIS) (n=26) and associated invasion ductal carcinoma (n=26) were obtained from the Emory University Hospital Systems Breast Tumor Bank. Sections were deparaffinized, subjected to antigen retrieval, and incubated with primary antibody. The survivin antibody was from Cell Signaling Technology. The leptin antibody (Ob, A-20) was from Santa Cruz Biotechnology, Inc. Immunocomplexes were detected by the labeled polymer method (EnVision Dual-Link-System-HRP, DAKO, Carpentaria, CA). Sections were counterstained with hematoxylin.
Chromatin immunoprecipitation (ChIP)
Chromatin samples were sonicated on ice three times for 10 seconds each (i.e., until the average length of sheared genomic DNA was 1 to 1.5 kb) followed by centrifugation for 10 minutes. The immunoprecipitated DNA was ethanol precipitated and resuspended in 25 μl of H2O. Total input samples were resuspended in 100 μl of H2O and diluted 1:100 before PCR analysis. Initially, PCR was performed with different numbers of cycles and/or dilutions of input DNA to determine the linear range of amplification; all results shown fall within this range. Following 28-30 cycles of amplification, PCR products were run on 1% agarose gel and analyzed by ethidium bromide staining. All ChIP assays were performed at least thrice with similar results.
Immunofluorescence and confocal imaging
Breast cancer cells (5×105 cells/well) were plated in 4-well chamber slides (Nunc, Rochester, NY) followed by treatment with 100ng/ml human recombinant full-length leptin for 2h. Cells were washed three times with 1X PBS and fixed using freshly prepared fixative containing 3.7% formaldehyde, 0.05% glutaraldehyde, and 0.4% Triton-X-100 in PHEMO buffer [0.068mol/L PIPES, 0.025mol/L HEPES, 0.015mol/L EGTA, 0.003mol/L MgCl2 and 10%v/v DMSO] for 10 minutes at room temperature. Primary antibodies (as indicated) were used at 1:500 with an overnight incubation at 4°C followed by an anti-rabbit IgG Alexa Flour 488 secondary antibody used at 1:500 for 1h at room temperature. The nucleus was stained with 4’,6-diamidino-2-phenyl-indole (Sigma-Aldrich) using 300nmol/L at room temperature for 10 minutes before mounting in Gel Mount mounting medium (Biomeda). Fixed and immunofluorescently stained cells were imaged using a Zeiss LSM510 Meta (Zeiss) laser scanning confocal system sonfigured to a Zeiss Axioplan 2 upright microscope with a 63XO (NA 1.4) plan-apochromat objective. All experiments were performed multiple times using independent biological replicates.
Immunoprecipitation of Epidermal Growth Factor Receptor (EGFR)
Whole cell lysates from breast cancer cells were incubated with specific antibodies for EGFR and the immunoprecipitation was performed following previously described method (Saxena et al. 2008). Whole cell lysate from breast cancer cells was incubated with specific antibodies for EGFR and the mixture was rotated slowly at 4°C for 16h. A total of 20μl packed protein A/G agarose beads was added and mixture was incubated at 4°C for 1h with rotation. The beads were collected by gentle centrifugation and washed twice with 1.5ml ice-cold buffer [50mM Tris-Cl (pH 7.4), 150mM NaCl, 1mM EDTA, 1% NP-40, 0.25% Na-deoxycholate, 1mM PMSF, 10μg/ml aprotinin, 10μg/ml leupeptin]. After the final wash, the precipitated protein-beads complexes were resuspended in SDS-sample loading buffer, fractionated by SDS-PAGE, and transferred to nitrocellulose membrane. Immuno-detection was performed by blocking the membranes for 1h in TBS buffer [20mM Tris-Cl (pH 7.5), 137mM NaCl, 0.05% Tween-20] containing 5% powdered nonfat milk followed by addition of the primary antibody (as indicated) in TBS for 2h at room temperature. Specifically bound primary antibodies were detected with peroxidase-coupled secondary antibodies and developed by enhanced chemiluminescence (ECL system, Amersham Pharmacia Biotech Inc., Arlington Heights, IL) according to manufacturer's instructions.
Transfections
Breast cancer cells were transiently transfected with survivin-wild type or psilencer-survivin plasmid or EGFR siRNA (Santa Cruz). Breast cancer cells were transiently transfected with 1.0μg of survivin-wild type or psilencer-survivin plasmid using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Forty-eight hours post-transfection, the cells were treated with 100ng/ml human recombinant full-length leptin for the indicated durations. Cells were harvested and total protein lysates were subjected to Western blot analysis using specific antibodies as described. The experiments were performed in triplicate, and similar results were obtained from at least three independent experiments. Cells transfected with survivin-wild type or psilencer-survivin were used to perform migration assay as described above. Cells were transfected with EGFR-siRNA (Santa Cruz) using Lipofectamine 2000 according to manufacturer's instructions. Cells were subjected to western blot analysis as described above.
Electric cell-substrate impedance sensing (ECIS) Wound-healing assays
Wound-healing assays were performed using the ECIS (Applied BioPhysics) (Saxena et al. 2008). For wound-healing assays, cells were grown to confluence on ECIS plates and serum starved for 16h. The ECIS plates were submitted to an elevated voltage pulse of 40 kHz frequency, 3.5-V amplitude, and 30-s duration, which led to the death and detachment of cells present on the small active electrode resulting in a wound normally healed by cells surrounding the small active electrode that have not been submitted to the elevated voltage pulse. Cells were immediately treated with leptin at 100ng/ml. Wound healing was then assessed by continuous resistance measurements for ~ 24 h. All experiments were performed at least three times in triplicates.
Breast tumorigenesis assay
MDA-MB-231 (5×106) cells in 0.1ml of HBSS were injected subcutaneously into the right gluteal region of 4-6-week-old female athymic nude mice. Two weeks after initial implantation, animals placed into two experimental groups. Mice were treated with intraperitoneal injections of 1) saline or 2) recombinant leptin (dosage of 5 mg/kg), five days a week for the duration of the experiment. Tumors were measured using vernier calipers, with tumor volume calculated using the formula (V= a/2 × b2), where V is the tumor volume in mm3, a and b are the largest and smallest diameters in mm, respectively. All animals were sacrificed after 6 weeks of treatment. Tumors were collected; weighed, fixed in 10% neutral-buffered formalin; and subjected to further analysis by immunohistochemistry. All animal studies were conducted in accordance with the guidelines of University IACUC.
Statistical Analysis
All experiments were independently performed three times in triplicate. Statistical analysis was done using Microsoft Excel software. Significant differences were analyzed using student's t test and two-tailed distribution. Data were considered to be statistically significant if P<0.05. Data are expressed as means ± SE between triplicate experiments.
Results
Leptin treatment increases growth and migration potential of breast cancer cells along with increased survivin expression
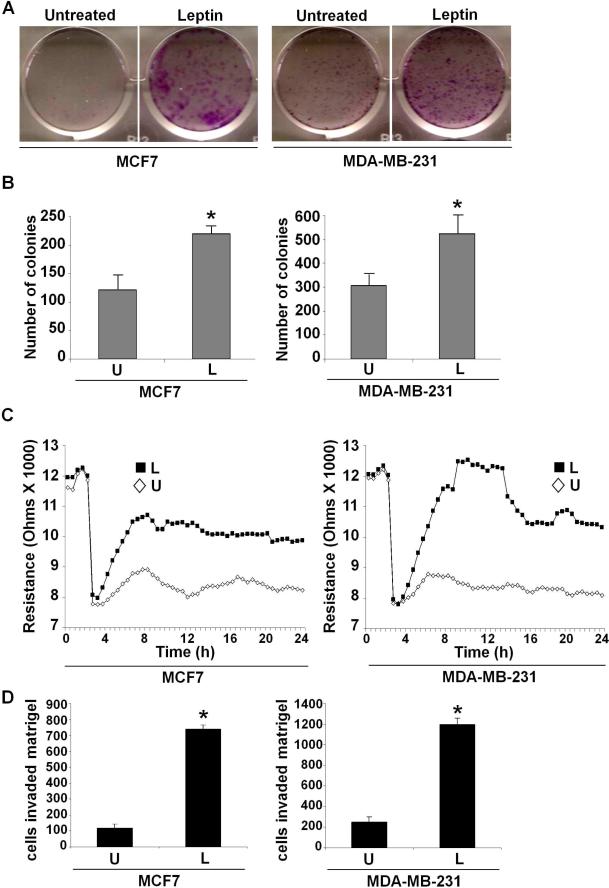
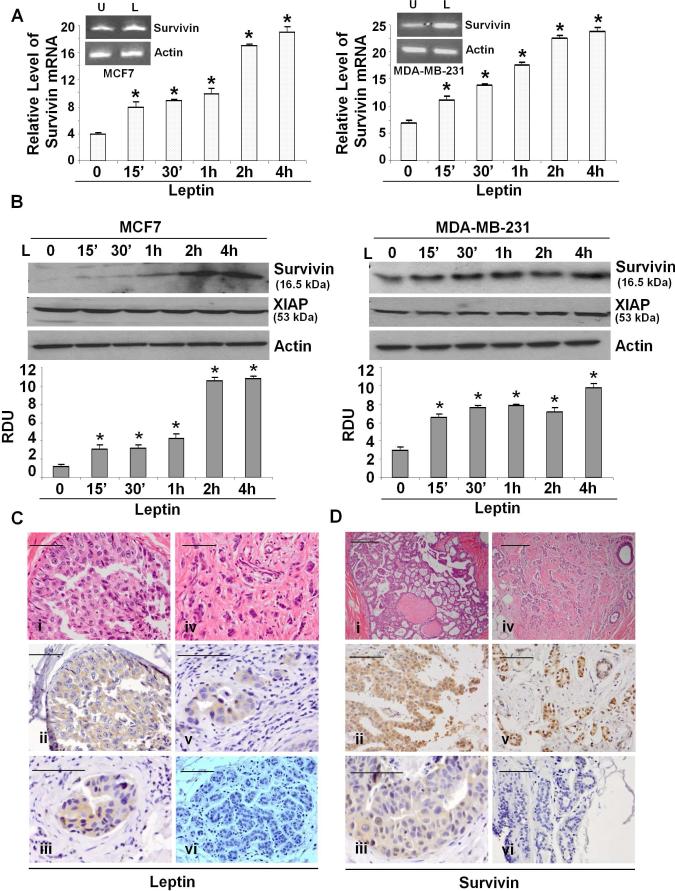
Epidemiological studies have shown that high leptin levels are significantly associated with an increased breast tumor growth and metastasis (Berclaz, et al. 2004; Chen, et al. 2006). Metastatic process is a complex cascade of events involving the ability of cancer cells to migrate and form colonies in an anchorage-independent manner (Gupta and Massague 2006). We examined the effect of leptin on breast cancer cell growth and migration potential. Breast cancer cells were treated with various concentrations of leptin and subjected to growth and clonogenicity assays. Substantial stimulation was observed after treatment of cells at 100 ng/ml leptin, whereas higher concentrations were not particularly more stimulatory (Supplementary Figures 1, 2). Leptin treatment resulted in increased clonogenicity (Figure 1A) in comparison to untreated cells. Next, we performed a soft-agar assay to assess changes in anchorage-independent growth of breast cancer cells in response to leptin treatment. Untreated cells formed very few colonies in soft-agar whereas leptin-treatment resulted in significantly higher number of colonies (Figure 1B). Leptin treatment increased the migration potential of breast cancer cells in a quantitative Electric-cell-substrate-impedance sensing (ECIS) based migration assay (Figure 1C). Leptin increased invasion of breast cancer cells in a matrigel invasion assay (Figure 1D). Uncontrolled leptin-signaling can induce various genes involved in proliferation, adhesion, angiogenesis thereby contributing to various aspects of tumorigenesis and metastasis (Cirillo, et al. 2008). While analyzing the effect of leptin on some key target genes involved in cancer growth regulation, we found that leptin treatment increased survivin mRNA expression in a real-time RT-PCR analysis within 15 minutes (Figure 2A). Leptin increased survivin protein expression within 15 minutes which remained elevated for the duration of the experiment (Figure 2B). Leptin increased survivin expression in a dose-dependent manner. Statistically significant increase in survivin expression was observed at 100 ng/ml leptin, while the higher concentrations were not particularly more stimulatory (Supplementary Figure 3).
Figure 1. Leptin promotes clonogenicity, anchorage-independent growth and migration of breast cancer cells.
A) Breast cancer cells were subjected to colony-formation assay in the presence or absence of leptin. Colonies containing >50 normal-appearing cells were counted. Leptin increases clonogenicity. B) Breast cancer cells were subjected to soft-agar colony-formation assay in the presence (L) or absence (U) of leptin for three weeks. Results are expressed as average number of colonies counted (in six micro-fields). *, P<0.005, compared with untreated controls. Leptin augments anchorage-independent growth. C) Breast cancer cells were subjected to ECIS-migration assay in the absence (U) or the presence of leptin (L) as indicated. Cells were allowed to grow to confluence and initial resistance was measured for few hours. Cells were subjected to high voltage to initiate wound at 3 hours resulting in the drop of resistance. Remaining cells were allowed to migrate in the presence or absence of leptin and resistance changes were measured for ~24 h after the creation of wound. Cells migrate faster in the presence of leptin. D) Breast cancer cells were subjected to Matrigel invasion assay in the presence (L) or absence (U) of leptin for 24 hours. The number of cells that invaded through the Matrigel was counted in six different regions. The slides were blinded to remove counting bias. The results show mean of three independent experiments performed in triplicates. *, P<0.001, compared with untreated controls.
Figure 2. Evidence of leptin-induced survivin upregulation in breast cancer cells and survivin and leptin overexpression in primary DCIS and invasive ductal carcinoma.
A) Breast cancer cells were treated with leptin (L) for indicated time intervals followed by real-time PCR analysis using specific primer sets for survivin. *, P<0.001, compared with untreated controls. Insert shows conventional RT-PCR analysis of survivin in leptin treated cells. Leptin induces survivin mRNA. B) Breast cancer cells were treated with leptin for indicated time intervals, lysates were probed with indicated antibodies. The histogram is the mean of densitometric analysis showing relative density units (RDU) of the western-blot signal for survivin normalized to actin in three separate experiments. *, P<0.005, compared with untreated controls. Leptin increases survivin protein expression. C). Immunohistochemistry was performed on formalin-fixed, paraffin-embedded ER, PR positive breast tumor tissue samples. Leptin expression in DCIS, 40X magnification (ii) and 60X magnification (iii). Leptin expression in IDC, 60X magnification (v). Note increased cytoplasmic staining of leptin in lobular epithelial cells of DCIS and IDC in comparison to normal breast lobular epithelium (vi). D) Survivin expression in DCIS, 40X magnification (ii) and 60X magnification (iii). Survivin expression in IDC, 40X magnification (v). Survivin is overexpressed in epithelial cells in IDC and DCIS as compared to normal breast lobular epithelial (vi). Both survivin and leptin were overexpressed in DCIS and IDC tumor samples. The data represents the results of all DCIS and IDC samples analyzed with IHC. (scale bar, 100μm)
Overexpression of survivin and leptin is associated with tumor progression in breast tumors
The above results prompted us to investigate the pattern of survivin and leptin expression in a panel of normal breast tissue and ER, PR positive tumors. Immunohistochemical studies on normal breast tissue, ductal carcinoma in situ (DCIS) (n=26) and invasive ductal carcinoma (IDC) (n=26) tumor samples showed increased survivin expression in 60% of DCIS and 85% of IDC samples compared to normal breast tissue. Survivin expression was enhanced in the cytoplasmic region in breast cancer cells filling the ducts. In agreement with our studies, leptin overexpression was observed in DCIS and IDC samples exhibiting overexpression of survivin whereas normal breast epithelial cells showed minimal staining (Figure 2C and D). Adipocytes and stromal cells surrounding tumoral region also showed increased leptin expression (data not shown). The overexpression of survivin and leptin in DCIS and IDC as well as overexpression of leptin in the surrounding adipocytes indicates their involvement in the progression of breast cancer.
Notch1 activation plays an important role in leptin-induced survivin overexpression in breast cancer cells
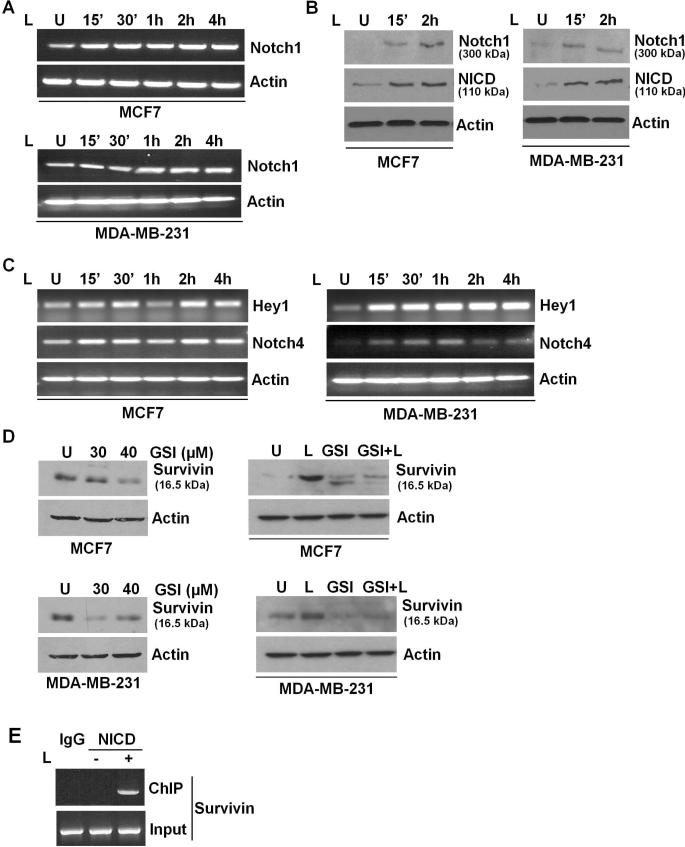
We next sought to determine the biological mechanism by which survivin is upregulated in the context of oncogenic effects of leptin. Recent studies have suggested that activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway results in the upregulation of survivin expression (Wang and Greene 2005). We found that inhibition of Akt or ERK activation using specific inhibitors did not affect leptin-induced activation of survivin (data not shown). Interactions between various growth factor signaling pathways such as crosstalk between IGF-1R and EGFR (Higashiyama, et al. 2008), and EGFR and Notch1 (Wu, et al. 2007), have been observed in response to specific stimuli. In particular, Notch1 signaling has emerged as a pivotal target in breast cancer with Notch1 receptors acting as survival factors promoting breast cancer. Mammalian Notch1 receptors are type-1 membrane proteins (Wu et al. 2007). Intriguingly, we found that leptin treatment increased Notch1 mRNA expression within 15 minutes of treatment (Figure 3A). Leptin also increased Notch1 protein expression within 15 minutes post-treatment. Notch1 receptors are heterodimeric proteins consisting of extracellular Notch (NEC) and transmembrane Notch (Nt). Activation of Notch1 receptors involves cleavage of Notch1 receptor by γ-secretase to release transcriptionally active intracellular Notch (NotchIC or NICD) (Callahan and Raafat 2001; Miele 2008). Leptin treatment resulted in release of NICD confirming Notch1 activation (Figure 3B). To unequivocally determine the activity of Notch1 in response to leptin, we also examined the expression of endogenous Notch1 targets, Hey1 and Notch4 (Weijzen, et al. 2002). Leptin increased expression of Hey1 and Notch4 (Figure 3C) showing elevated Notch1 activity.
Figure 3. Leptin-induced Notch1 expression and activity plays an important role in regulating survivin expression in the presence of leptin.
A) Breast cancer cells were treated with leptin for indicated time intervals. Untreated cells are denoted with U. Change in Notch1 mRNA expression was analyzed by RT-PCR analysis using specific primer sets. B) Notch1 protein and NICD levels were detected by western blot analysis using specific antibodies. Leptin treatment increases Notch1 expression and NICD levels. C) Breast cancer cells were treated with leptin for indicated time intervals. Total RNA was isolated and subjected to RT-PCR analysis using specific primer sets for Notch1 targets, Hey1 and Notch4. Leptin induces transcriptional activation of Hey1 and Notch4. D) Breast cancer cells were treated with γ-secretase inhibitor (GSI) and leptin alone or in combination. Untreated cells are denoted with U. Lysates were probed with survivin and actin antibodies. GSI treatment inhibits survivin expression alone and even in the presence of leptin. E) Breast cancer cells were untreated or treated with leptin and subjected to ChIP analysis using NICD antibodies. Immunoprecipitated chromatin was analyzed using specific primer sets encompassing CSL binding site in survivin promoter. IgG was used as a control. NICD gets recruited to survivin promoter in the presence of leptin.
To examine the direct role of Notch1 activation in survivin expression, we treated breast cancer cells with a specific GSI (γ-secretase) inhibitor (LY411,575), and found that inhibition of Notch1 activation inhibits survivin expression (Figure 3D). Importantly, GSI treatment was able to inhibit leptin-induced increase in survivin expression in a co-treatment experiment (Figure 3D). Intracellular domain of Notch1, NICD translocates to the nucleus and interacts with CSL resulting in transcriptional activation of Notch1 targets. In order to define the mechanism of regulation of survivin mediated by Notch1, we searched for possible regulatory elements related to the Notch1 signaling pathway in the survivin promoter. We identified a putative CSL binding site located at position -1218 in the human survivin promoter. The sequence of the putative CSL-binding site is GCTGAGAT; this sequence is different from the consensus sequence for the high-affinity CSL-binding site (GTGGGAA) previously described (Ronchini and Capobianco 2001). However, similar sequences have been identified in the human cyclin D1 promoter and human β-globin locus control region, which have been shown to be authentic CSL-binding sites (Ronchini and Capobianco 2001). We examined the ability of leptin-induced NICD to bind the identified site by chromatin-immunoprecipitation assay. Importantly, we found the recruitment of NICD to survivin promoter in the presence of leptin (Figure 3E) whereas no binding was observed in the absence of leptin. Collectively, these results showed that leptin induces expression and activation of Notch1 and leptin-activated Notch1 binds to survivin promoter hence Notch1 plays an important role in the regulation of survivin expression in breast cancer cells.
Crosstalk between leptin and Notch1 signaling involves EGFR
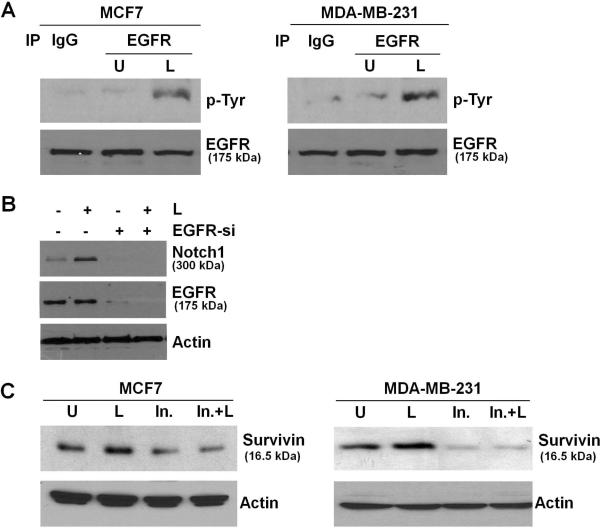
EGFR transactivation observed in response to IGF-1, E-cadherin, integrins and activation of G-protein-coupled receptors, impacts various physiological processes (Higashiyama et al. 2008). We recently showed that co-treatment with leptin and IGF-1 results in increased phosphorylation of EGFR (Saxena et al. 2008). Treatment of MCF7 and MDA-MB-231 cells with leptin resulted in increased phosphorylation of EGFR, as evident by immunoprecipitation of EGFR followed by immunoblotting with an anti-phosphotyrosine antibody (Figure 4A). Phosphorylated-tyrosine bands corresponded to the expected size band for EGFR. We next investigated the role of EGFR in leptin-induced Notch1 activation in breast cancer cells by silencing EGFR using EGFR-siRNA. EGFR-siRNA efficiently inhibited EGFR expression (Figure 4B). Interestingly, leptin-induced Notch1 expression was inhibited in the presence of EGFR-siRNA showing the requirement of EGFR in leptin-induced Notch1 expression (Figure 4B). Activation of the EGFR signaling pathway leads to the up-regulation of survivin gene expression (Asanuma, et al. 2005). We next examined the importance of EGFR activation in leptin-induced increased expression of survivin. We found that pre-incubation of breast cancer cells with EGFR inhibitor erlotinib significantly inhibited the stimulatory effect of leptin on survivin expression (Figure 4C) showing that EGFR activation is required for leptin-induced survivin upregulation. Together, these data showed that transactivation of EGFR is upstream of the activation of Notch1 in the context of leptin, revealing the hierarchy of these events and involvement of leptin-EGFR-Notch1 axis in survivin upregulation.
Figure 4. Involvement of EGFR transactivation in Notch1 and survivin induction in the presence of leptin.
A) Breast cancer cells were untreated (U) or treated with leptin (L). Lysates were immunoprecipitated with EGFR antibody and probed with anti-Phospho-tyrosine antibody. The membranes were re-blotted using anti-EGFR antibody as control. Leptin induces phosphorylation of EGFR. B) Breast cancer cells were transfected with EGFR-siRNA and treated with or without leptin (L). EGFR-siRNA resulted in EGFR silencing. Leptin treatment does not induce Notch1 expression when EGFR is silenced. C) Breast cancer cells were pretreated with EGFR inhibitor followed by leptin treatment as indicated (In.+L). Cells were also treated with leptin alone (L) or inhibitor alone (In.). Untreated cells are denoted with U. Lysates were probed with indicated antibodies. EGFR inhibition by erlotinib inhibits leptin-induced survivin expression.
Inhibition of survivin disrupts leptin-induced migration of breast cancer cells
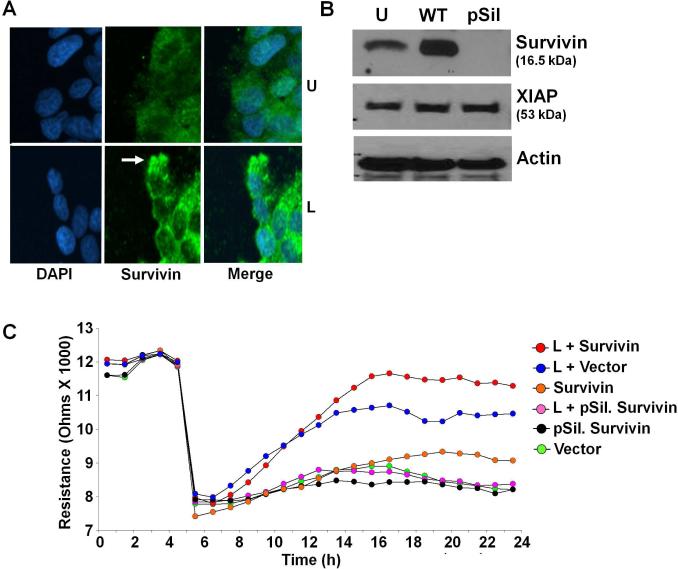
While cancer cell metastasis is a complex process involving various intermediary steps from alteration of microenvironment to colonization in distant organ sites, migration is one of the most important early events (Gupta and Massague 2006). After examining the molecular mechanisms involved in leptin-induced survivin upregulation, we next examined the role of survivin in biological functions of leptin. Migratory cells typically undergo morphological changes as individual cells show podia formation, irregular membrane ruffling and the formation of protrusions at the leading edge of these podias (Insall and Machesky 2009). Confocal immunofluorescence microscopy of breast cancer cells forced to be motile in the presence of leptin revealed that survivin protein was highly concentrated at the tips of podia of migrating cells (Figure 5A). Based on the above morphologic observations, we speculated that leptin-mediated induction of survivin might increase cell motility. To test this hypothesis, we modulated survivin expression in breast cancer cells by transfecting full-length wild-type-survivin and p-silencer-survivin plasmids (Figure 5B). These breast cancer cells were subjected to scratch-migration assay in the presence or absence of leptin. Leptin induced migration of breast cancer cells within 6h of treatment. Survivin overexpression further increased leptin-induced migration whereas silencing survivin abrogated leptin-induced migration (Data not shown). Next, we performed a quantitative real-time impedance assay using an ECIS-based technique to follow migration of breast cancer cells. Leptin-treated vector-transfected cells displayed an increase in resistance, showing increased migration in comparison with untreated vector-transfected cells. Cells overexpressing survivin migrated at a higher rate than cells transfected with either vector or p-silencer-survivin. Cells overexpressing survivin rapidly migrated to reach the resistance values of the nonwounded cells at the start of the experiment in response to leptin. Whereas cells transfected with p-silencer-survivin showed no significant change in migration even in the presence of leptin (Figure 5C).
Figure 5. Survivin plays an integral role in leptin-induced breast cancer cell migration.
A) Breast cancer cells were scratched and forced to migrate in the absence (U) or presence of leptin (L). Scratched edges were immunostained as indicated and analyzed with confocal imaging and photographed. Arrow indicates increased accumulation of survivin at the leading edge of migrating cells in the presence of leptin (similar accumulation was observed in various micro-fields examined). B) Breast cancer cells were transfected with plasmids expressing WT-survivin and p-silencer-survivin. Vector transfected cells are denoted with U. Lysates were probed with the indicated antibodies. C) Breast cancer cells were transfected with vector, WT-survivin or pSilencer-survivin and subjected to quantitative ECIS-migration assay in the presence of leptin (L). Cells were allowed to grow to confluence and initial resistance was measured for few hours. Cells were subjected to high voltage to initiate wound at 5 hours resulting in the drop of resistance. Remaining cells were allowed to migrate in the presence or absence of leptin and resistance changes were measured for ~24 h after the creation of wound. Cells migrate faster in the presence of leptin and survivin. Survivin silencing abrogates leptin-induced migration.
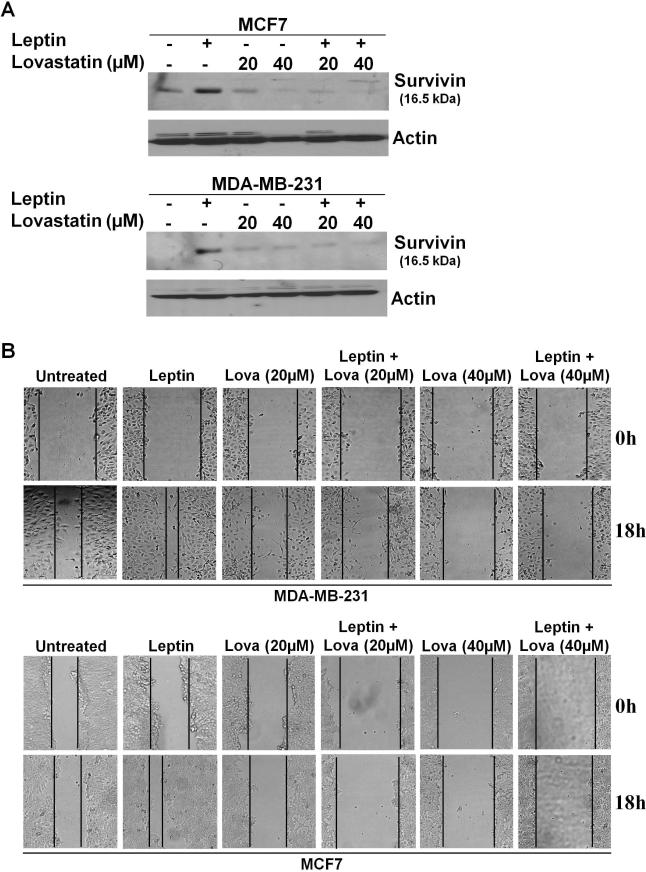
We next examined the requirement of survivin upregulation in leptin-mediated migration of breast cancer cells. HRIs, 3-hydroxy-3-methylglutaryl-coenzyme-A-reductase inhibitors, which are widely used to reduce the serum cholesterol levels in hypercholesterolemia patients, decrease survivin expression in cancer cells (Kaneko, et al. 2007). As shown in figure 6A, leptin increased survivin expression in breast cancer cells whereas treatment with HRI lovastatin inhibited survivin expression. Pre-treatment with HRI lovastatin significantly inhibited leptin-induced survivin expression (Figure 6A). Next, we performed scratch-migration assay in the presence of leptin and lovastatin treatments. As expected, leptin increased migration whereas lovastatin treatment inhibited leptin-induced migration (Figure 6B). These results collectively showed that leptin-induced survivin upregulation is required for leptin-induced migration of breast cancer cells, and that disruption of survivin expression using lovastatin might be a valid therapeutic approach to counter the effects of leptin on breast cancer cells.
Figure 6. Pharmacological inhibition of Survivin expression inhibits leptin-induced increased migration.
A) Breast cancer cells were treated with lovastatin in the presence or absence of leptin. Lysates were probed with the indicated antibodies. Lovastatin inhibits leptin-induced survivin expression. B) Breast cancer cells were subjected to scratch-migration assay in the presence of various treatments as indicated. The plates were photographed at the identical location of the initial image at 18h post-treatment. Lovastatin inhibits leptin-induced migration.
Leptin treatment induces breast tumor progression in athymic nude mice
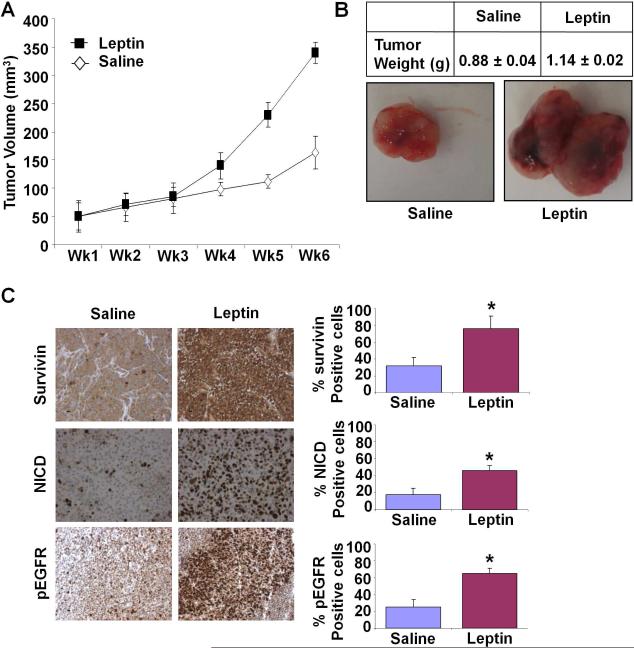
We investigated the physiological relevance of our in vitro findings by evaluating tumor promoting effects of leptin on development of breast cancer in vivo. Leptin treatment significantly increased tumor growth as compared to the saline-treated group (Figure 7A, B). We further confirmed our in vitro findings regarding important signaling molecules. We used tumor sections from this study to determine the effect of leptin administration on the expression of survivin, NICD and pEGFR proteins by immunohistochemistry. Leptin-treated tumors revealed elevated survivin, NICD and pEGFR levels in comparison to saline-treated controls (Figure 7C). Collectively, these in vitro and in vivo studies demonstrated a cogent tumor-promoting effect of leptin and the involvement of Notch-EGFR-Survivin.
Figure 7. Leptin treatment increases breast tumor growth in nude mice.
MDA-MB-231 cells derived tumors were developed in nude mice and treated with saline or leptin. A) Tumor growth was monitored by measuring the tumor volume for 6 weeks. (n = 8 mice per group). B) At the end of six weeks, tumors were collected, measured, weighed and photographed. Leptin treatment increased tumor size as compare to saline treatment. Average tumor weight and representative tumor images are shown here. C) Tumor samples were subjected to immunohistochemical analysis using survivin, NICD and pEGFR-Tyr 992 antibodies. Leptin treatment induced the expression of survivin, NICD and pEGFR as compared to saline treatment. Bar diagrams show quantitation of protein expression in tumors from saline and leptin-treated mice. Columes, mean (n =8); bar, SD. * significantly different (P< 0.05) compared with control.
Discussion
In this study, we have revealed a novel mechanism of regulation of leptin-induced migration of breast cancer cells. The following novel findings are described in this study: (a) leptin-mediated upregulation of survivin requires Notch1 activation; (b) leptin induces recruitment of NICD to survivin promoter; (c) leptin regulates Notch1 via EGFR transactivation; (d) survivin-overexpression further increases leptin-induced migration of breast cancer cells whereas survivin-inhibition abrogates leptin-induced migration; (e) pharmacological inhibition of survivin effectively inhibits leptin-induced migration of breast cancer cells; (f) both survivin and leptin are overexpressed in DCIS and IDC tumors as compared to normal breast tissue. These results show that leptin treatment significantly increases survivin expression via EGFR-Notch1 axis. We also show a novel integral role of anti-apoptotic protein survivin in leptin-mediated migration of breast cancer cells. Thus, pharmacological inhibition of survivin may be a suitable strategy for treating metastatic breast cancer progressing in the presence of high leptin levels.
Survivin is a structurally unique anti-apoptotic protein with known dual function in apoptosis and control of mitosis (Altieri 2004). Survivin acts as caspase inhibitor thus blocking caspase activation and inhibiting biological activities of activated caspases (Altieri 2004). Our current studies demonstrate that leptin induces survivin expression at both mRNA and protein levels in breast cancer cells. Interaction between various growth factor signaling pathways have been previously described. Our previous studies showed that bidirectional crosstalk between leptin and IGF-1 leads to transactivation of EGFR (Saxena et al. 2008). Recent studies have shown that activation of EGFR with EGF treatment upregulates the expression of survivin (Peng, et al. 2006). Thus, leptin-mediated EGFR activation may be responsible for leptin-mediated survivin upregulation. Our studies show for the first time that leptin-mediated survivin upregulation involves EGFR activation. Importantly, inhibition of EGFR activity inhibits leptin-induced survivin upregulation. Our studies support the concept that high levels of leptin and EGF associated with obesity can act in concert to increase survivin expression and increase the negative influence of obesity on breast carcinogenesis.
At present, it is unclear how EGFR signaling leads to overexpression of survivin. EGFR mediated activation of PI3K pathway may lead to survivin upregulation (Wang and Greene 2005). Also, involvement of EGFR-HIF1-α crosstalk in regulation of survivin expression (Peng et al. 2006) has been shown. Our studies demonstrate a novel regulatory mechanism involving EGFR-Notch1 axis for leptin-induced survivin upregulation. Both leptin and Notch1 have been previously linked with poor prognosis in cancer in various epidemiological, clinical and preclinical studies; however, no studies have shown a direct link between the two pro-oncogenic signals. We found that leptin treatment leads to upregulation of Notch1 in breast cancer cells. Notch1 receptors are cell-fate regulatory proteins that also act as survival proteins in cancer. Activation of Notch1 not only cause mammary tumors in mice but it is also associated with poor prognosis for breast cancer development (Dievart, et al. 1999; Stylianou, et al. 2006). Our studies show that the oncogenic potential of leptin might be due in part to activation of Notch1 receptors. In a microarray analysis, Notch1 inhibition has been shown to inhibit EGFR expression in glioma and colon cancer cells (Purow, et al. 2008) thus presenting EGFR as a downstream target of Notch1 signaling. Interestingly, we found crosstalk between Notch1 and EGFR in breast cancer cells where EGFR silencing inhibits leptin-induced Notch1 expression thus showing direct upstream involvement of EGFR in leptin-mediated Notch1 upregulation. We further found that inhibiting Notch1 using γ-secretase inhibitor inhibits survivin expression in breast cancer cells. Our study reveals the direct involvement of leptin-EGFR-Notch1 axis in regulation of survivin expression in breast cancer cells.
The most remarkable feature of survivin is its differential expression in cancer versus normal tissues. Survivin plays an important role in development; it is strongly and diffusely expressed in embryonic and fetal organs while most terminally differentiated normal tissues do not express survivin (Altieri 2003). Notably, overexpression of survivin has been observed in various tumors including lung, colon, stomach, esophagus, pancreas, liver, uterus, ovaries and melanoma in contrast to no survivin expression in corresponding normal tissues (Altieri 2003). Survivin has been shown to be involved with mitotic progression and apoptosis (Altieri 2004). Our study showed yet another important function of survivin in leptin-induced migration of breast cancer cells. Survivin overexpression increases leptin-induced migration whereas silencing of survivin abrogates leptin-induced migration of breast cancer cells. Migration being one of the important early events in metastatic process, our study showed a direct role of survivin in the metastatic process. This becomes an important finding in light of the previous epidemiological and clinical studies linking survivin with increased metastasis and poor prognosis. At present, it is not clear how survivin exerts its promigratory role in the presence of leptin. A recent study linked survivin expression in human melanocytes and melanoma cells with enhanced capacity for migration and showed that survivin knockdown impaired melanoma cell migration (McKenzie, et al. 2010). It has been suggested that survivin-mediated migration promotion may involve Akt-dependent upregulation of α5 integrin (McKenzie, et al. 2010). Integrins promote cell migration through interactions with extracellular matrix proteins that enhance adhesion and transmit intracellular signals (Hood, et al., 2002). Signaling molecules known to be associated with deregulated cancer cell migration include activation of focal adhesion kinase (Sieg, et al., 1999), Src (Klekotka, et al., 2001), MAPK (Zheng, et al. 2000) and Akt (Zhang, et al., 2009). Further analysis of molecular mechanisms underlying leptin-induced migration and contribution of survivin will provide novel insight into breast cancer metastasis.
Because of important roles of survivin in mitosis, apoptosis and now in migration, a survivin-based therapy becomes an attractive option. Also, it would be expected to have limited toxicity towards normal tissues owing to the differential expression of survivin. Amongst various translational approaches targeting survivin, generation of antigen-specific cytolytic T cells against survivin peptides has been considered for vaccination with the advantage of minimal risk of autoimmune effects (Andersen and thor 2002; Kanwar, et al. 2001). Utilizing molecular antagonists to interfere with the survivin pathway is another approach that uses systemic administration of antisense oligonucleotides, ribozymes, or local administration of mutant survivin adenoviruses to inhibit endogenous survivin expression (Chen, et al. 2000; Kanwar et al. 2001). Taking a pharmacological approach, we used HRI (3-hydroxy-3-methylglutaryl-coenzyme-A reductase inhibitor) lovastatin to inhibit leptin-induced survivin expression. Lovastatin treatment also inhibited leptin-mediated survivin-dependent migration of breast cancer cells. Our studies showed that leptin augments survivin expression and migration of breast cancer cells via involvement of the leptin-EGFR-Notch1 axis thus providing mechanistic insights for the development of novel translational applicability of the survivin pathway in cancer therapy.
Supplementary Material
Acknowledgments
Funding: This work was supported by grants NIDDK NIH (K01DK076742 and R03DK089130 to NKS), NCI NIH (R01CA131294 to DS), CDMRP BCRP (BC030963 to DS), The Susan G. Komen for the Cure (BCTR0503526 to DS), and Mary K Ash Foundation (DS).
Footnotes
Declaration of interest: Disclosure Statement: There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported (BBK, GMO, AN, LY, CC, NKS and DS).
References
- Adida C, Haioun C, Gaulard P, Lepage E, Morel P, Briere J, Dombret H, Reyes F, Diebold J, Gisselbrecht C, et al. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood. 2000;96:1921–1925. [PubMed] [Google Scholar]
- Ahima RS, Osei SY. Leptin signaling. Physiol Behav. 2004;81:223–241. doi: 10.1016/j.physbeh.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Molecular circuits of apoptosis regulation and cell division control: the survivin paradigm. J Cell Biochem. 2004;92:656–663. doi: 10.1002/jcb.20140. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Andersen MH, thor SP. Survivin--a universal tumor antigen. Histol Histopathol. 2002;17:669–675. doi: 10.14670/HH-17.669. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- Asanuma H, Torigoe T, Kamiguchi K, Hirohashi Y, Ohmura T, Hirata K, Sato M, Sato N. Survivin expression is regulated by coexpression of human epidermal growth factor receptor 2 and epidermal growth factor receptor via phosphatidylinositol 3-kinase/AKT signaling pathway in breast cancer cells. Cancer Res. 2005;65:11018–11025. doi: 10.1158/0008-5472.CAN-05-0491. [DOI] [PubMed] [Google Scholar]
- Berclaz G, Li S, Price KN, Coates AS, Castiglione-Gertsch M, Rudenstam CM, Holmberg SB, Lindtner J, Erien D, Collins J, et al. Body mass index as a prognostic feature in operable breast cancer: the International Breast Cancer Study Group experience. Ann Oncol. 2004;15:875–884. doi: 10.1093/annonc/mdh222. [DOI] [PubMed] [Google Scholar]
- Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- Chen DC, Chung YF, Yeh YT, Chaung HC, Kuo FC, Fu OY, Chen HY, Hou MF, Yuan SS. Serum adiponectin and leptin levels in Taiwanese breast cancer patients. Cancer Lett. 2006;237:109–114. doi: 10.1016/j.canlet.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu W, Tahir SK, Kroeger PE, Rosenberg SH, Cowsert LM, Bennett F, Krajewski S, Krajewska M, Welsh K, et al. Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia. 2000;2:235–241. doi: 10.1038/sj.neo.7900091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo D, Rachiglio AM, la Montagna R, Giordano A, Normanno N. Leptin signaling in breast cancer: an overview. J Cell Biochem. 2008;105:956–964. doi: 10.1002/jcb.21911. [DOI] [PubMed] [Google Scholar]
- Dan HC, Jiang K, Coppola D, Hamilton A, Nicosia SV, Sebti SM, Cheng JQ. Phosphatidylinositol-3-OH kinase/AKT and survivin pathways as critical targets for geranylgeranyltransferase I inhibitor-induced apoptosis. Oncogene. 2004;23:706–715. doi: 10.1038/sj.onc.1207171. [DOI] [PubMed] [Google Scholar]
- Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- Garofalo C, Koda M, Cascio S, Sulkowska M, Kanczuga-Koda L, Golaszewska J, Russo A, Sulkowski S, Surmacz E. Increased expression of leptin and the leptin receptor as a marker of breast cancer progression: Possible role of obesity-related stimuli. Clin Cancer Res. 2006;12:1447–1453. doi: 10.1158/1078-0432.CCR-05-1913. [DOI] [PubMed] [Google Scholar]
- Gonzalez RR, Cherfils S, Escobar M, Yoo JH, Carino C, Styer AK, Sullivan BT, Sakamoto H, Olawaiye A, Serikawa T, et al. Leptin signaling promotes the growth of mammary tumors and increases the expression of vascular endothelial growth factor (VEGF) and its receptor type two (VEGF-R2). J Biol Chem. 2006;281:26320–26328. doi: 10.1074/jbc.M601991200. [DOI] [PubMed] [Google Scholar]
- Gonzalez RR, Leavis P. Leptin upregulates beta3-integrin expression and interleukin-1beta, upregulates leptin and leptin receptor expression in human endometrial epithelial cell cultures. Endocrine. 2001;16:21–28. doi: 10.1385/ENDO:16:1:21. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Higashiyama S, Iwabuki H, Morimoto C, Hieda M, Inoue H, Matsushita N. Membrane-anchored growth factors, the epidermal growth factor family: beyond receptor ligands. Cancer Sci. 2008;99:214–220. doi: 10.1111/j.1349-7006.2007.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Hood JD, Cheresh JA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Housa D, Housova J, Vernerova Z, Haluzik M. Adipocytokines and cancer. Physiol Res. 2006;55:233–244. doi: 10.33549/physiolres.930848. [DOI] [PubMed] [Google Scholar]
- Huang L, Li C. Leptin: a multifunctional hormone. Cell Res. 2000;10:81–92. doi: 10.1038/sj.cr.7290038. [DOI] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Kitayama J, Nagawa H. Enhanced expression of leptin and leptin receptor (OB-R) in human breast cancer. Clin Cancer Res. 2004;10:4325–4331. doi: 10.1158/1078-0432.CCR-03-0749. [DOI] [PubMed] [Google Scholar]
- Kaneko R, Tsuji N, Asanuma K, Tanabe H, Kobayashi D, Watanabe N. Survivin down-regulation plays a crucial role in 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor-induced apoptosis in cancer. J Biol Chem. 2007;282:19273–19281. doi: 10.1074/jbc.M610350200. [DOI] [PubMed] [Google Scholar]
- Kanwar JR, Shen WP, Kanwar RK, Berg RW, Krissansen GW. Effects of survivin antagonists on growth of established tumors and B7-1 immunogene therapy. J Natl Cancer Inst. 2001;93:1541–1552. doi: 10.1093/jnci/93.20.1541. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071–5074. [PubMed] [Google Scholar]
- Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–209. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- Klekotka PA, Santoro SA, Wang H, Zutter MM. Specific residues within the α 2 integrin subunit cytoplasmic domain regulate migration and cell cycle progression via distinct MAPK pathways. J Biol Chem. 2001;276:32353–32361. doi: 10.1074/jbc.M101921200. [DOI] [PubMed] [Google Scholar]
- Mauro L, Catalano S, Bossi G, Pellegrino M, Barone I, Morales S, Giordano C, Bartella V, Casaburi I, Ando S. Evidences that leptin up-regulates E-cadherin expression in breast cancer: effects on tumor growth and progression. Cancer Res. 2007;67:3412–3421. doi: 10.1158/0008-5472.CAN-06-2890. [DOI] [PubMed] [Google Scholar]
- Miele L. Rational targeting of Notch signaling in breast cancer. Expert Rev Anticancer Ther. 2008;8:1197–1202. doi: 10.1586/14737140.8.8.1197. [DOI] [PubMed] [Google Scholar]
- Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- O'Connor DS, Schechner JS, Adida C, Mesri M, Rothermel AL, Li F, Nath AK, Pober JS, Altieri DC. Control of apoptosis during angiogenesis by survivin expression in endothelial cells. Am J Pathol. 2000;156:393–398. doi: 10.1016/S0002-9440(10)64742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem. 2006;281:25903–25914. doi: 10.1074/jbc.M603414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinteaux E, Inoue W, Schmidt L, Molina-Holgado F, Rothwell NJ, Luheshi GN. Leptin induces interleukin-1beta release from rat microglial cells through a caspase 1 independent mechanism. J Neurochem. 2007;102:826–833. doi: 10.1111/j.1471-4159.2007.04559.x. [DOI] [PubMed] [Google Scholar]
- Purow BW, Sundaresan TK, Burdick MJ, Kefas BA, Comeau LD, Hawkinson MP, Su Q, Kotliarov Y, Lee J, Zhang W, et al. Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis. 2008;29:918–925. doi: 10.1093/carcin/bgn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Kaneko K, Hirota M, Masamune A, Satoh A, Shimosegawa T. Expression of survivin is correlated with cancer cell apoptosis and is involved in the development of human pancreatic duct cell tumors. Cancer. 2001;92:271–278. doi: 10.1002/1097-0142(20010715)92:2<271::aid-cncr1319>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, Anania FA. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007a;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena NK, Taliaferro-Smith L, Knight BB, Merlin D, Anania FA, O'Regan RM, Sharma D. Bidirectional crosstalk between leptin and insulin-like growth factor-I signaling promotes invasion and migration of breast cancer cells via transactivation of epidermal growth factor receptor. Cancer Res. 2008;68:9712–9722. doi: 10.1158/0008-5472.CAN-08-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena NK, Vertino PM, Anania FA, Sharma D. leptin-induced growth stimulation of breast cancer cells involves recruitment of histone acetyltransferases and mediator complex to CYCLIN D1 promoter via activation of Stat3. J Biol Chem. 2007b;282:13316–13325. doi: 10.1074/jbc.M609798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006a;66:6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Saxena NK, Vertino PM, Anania FA. Leptin promotes the proliferative response and invasiveness in human endometrial cancer cells by activating multiple signal-transduction pathways. Endocr Relat Cancer. 2006b;13:629–640. doi: 10.1677/erc.1.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112:2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- Sommer KW, Schamberger CJ, Schmidt GE, Sasgary S, Cerni C. Inhibitor of apoptosis protein (IAP) survivin is upregulated by oncogenic c-H-Ras. Oncogene. 2003;22:4266–4280. doi: 10.1038/sj.onc.1206509. [DOI] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- Taliaferro-Smith L, Nagalingam A, Zhong D, Zhou W, Saxena NK, Sharma D. LKB1 is required for adiponectin-mediated modulation of AMPK-S6K axis and inhibition of migration and invasion of breast cancer cells. Oncogene. 2009;28:2621–2633. doi: 10.1038/onc.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessitore L, Vizio B, Jenkins O, De Stefano I, Ritossa C, Argiles JM, Benedetto C, Mussa A. Leptin expression in colorectal and breast cancer patients. Int J of Mol Med. 2000;5:421–426. doi: 10.3892/ijmm.5.4.421. [DOI] [PubMed] [Google Scholar]
- Wang Q, Greene MI. EGFR enhances Survivin expression through the phosphoinositide 3 (PI-3) kinase signaling pathway. Exp Mol Pathol. 2005;79:100–107. doi: 10.1016/j.yexmp.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Wu F, Stutzman A, Mo YY. Notch signaling and its role in breast cancer. Front Biosci. 2007;12:4370–4383. doi: 10.2741/2394. [DOI] [PubMed] [Google Scholar]
- Xu ZX, Zhao RX, Ding T, Tran TT, Zhang W, Pandolfi PP, Chang KS. Promyelocytic leukemia protein 4 induces apoptosis by inhibition of survivin expression. J Biol Chem. 2004;279:1838–1844. doi: 10.1074/jbc.M310987200. [DOI] [PubMed] [Google Scholar]
- Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]
- Zhang B, Gu F, She C, Guo H, Li W, Niu R, Fu L, Zhang N, Ma Y. Reduction of Akt2 inhibits migration and invasion of glioma cells. Int J Cancer. 2009;125:585–595. doi: 10.1002/ijc.24314. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Woodard AS, Tallini G, Languino LR. Substrate specificity of α(v)β(3) integrin-mediated cell migration and phosphatidylinositol 3-kinase/AKT pathway activation. J Biol Chem. 2000;275:24565–24574. doi: 10.1074/jbc.M002646200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.