Protein quality control mechanisms are essential to ensure the proper levels of functional proteins within the cell and to eliminate nonfunctional proteins from the cell. To this end, the cell maintains an array of molecular chaperones to help fold proteins into their native conformations, prevent aggregation, and assist in assembly of multiprotein complexes, along with ATP-dependent proteases to degrade damaged or improperly synthesized proteins (reviewed in refs. 1–4). Also, the intracellular concentrations or activities of a limited number of normal cellular proteins are modulated by these same proteases and chaperones. These natural protein targets are often key regulatory proteins involved in vital processes such as cell division, stress responses, and developmental changes. To avoid potentially lethal damage to inappropriate cellular components, the chaperones and proteases have evolved rather complex energy-dependent mechanisms to assure accurate substrate recognition.
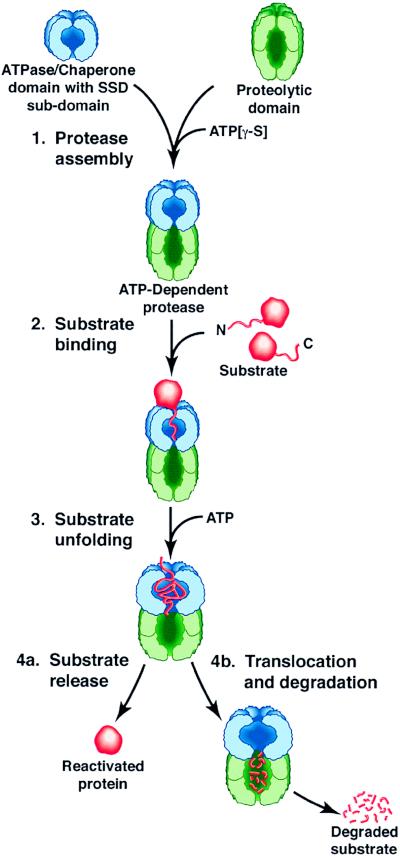
The prokaryotic energy-dependent proteases, as exemplified by the two-component Clp proteases, including ClpAP, ClpXP, and ClpYQ in Escherichia coli, are elaborate complexes that couple the protein unfolding activity of a molecular chaperone with the degradative activity of an endopeptidase. The current model of substrate recognition, unfolding, translocation, and degradation by these proteases is depicted in Fig. 1. The ATPase components, ClpA, ClpX, or ClpY, form ring-like structures composed of six or seven subunits (5–8) and have protein reactivation and remodeling activities characteristic of classical molecular chaperones (refs. 9–12 and reviewed in refs. 1, 13, 14). When in complexes with proteolytic components, the ATPases are positioned at one or both ends of the proteolytic component. They regulate degradation by binding, unfolding, and translocating specific substrates into the proteolytic chamber. The protease active sites of both ClpP and ClpQ are located in an internal cavity generated by two stacked rings of six or seven identical subunits (15). The substrate is indiscriminately degraded in the proteolytic chamber and the products diffuse out. The Lon protease, like the Clp proteases, is another oligomeric ATP-dependent protease with an ATPase domain and a proteolytic domain within one polypeptide chain.
Figure 1.
Working model of substrate recognition and discrimination by the chaperone domains of Clp proteins and Lon protease. The SSD subdomain of the ATPase domain or component binds appropriate substrates by recognizing motifs near the C or N terminus of substrates. Perhaps the strength and fidelity of the binding are increased by the presence of secondary binding sites also located in the ATPase domain. Conformational changes associated with ATP binding and/or hydrolysis promote either protein remodeling and reactivation or protein unfolding and translocation to the proteolytic component.
The most recent advance in defining substrate recognition motifs in the chaperone components of prokaryotic ATP-dependent proteases is reported by C. K. Smith, T. A. Baker, and R. T. Sauer in a recent issue of the Proceedings (16). They searched for the domains of Clp ATPases and Lon protease that interact with substrates and identified domains with similar predicted structures. They refer to these domains as “sensor and substrate-discrimination,” or SSD, domains. Protein fragments corresponding to ClpA, ClpY, and Lon SSD domains formed stably folded structures. The analogous regions from ClpX and ClpB were less stable. Gratifyingly, the isolated ClpA, ClpX, ClpB, ClpY, and Lon SSD domains retain the ability to bind several substrates. The substrates tested included SsrA-tagged Arc, which is degraded by ClpAP and ClpXP in vivo (see below); σ32, which is degraded by another ATP-dependent protease, FtsH; and UmuD, which is degraded by Lon. Binding was measured by an indirect ELISA procedure, and specificity was assessed by comparing the binding of the wild-type substrate with a corresponding variant that was mutated in its recognition tag and not recognized by proteases in vivo. The variants included Arc-SsrA, in which the two terminal amino acids were changed from Ala to Asp; σ32, carrying a mutation changing the two C-terminal amino acids to Asp; and UmuD′, a homodimer that lacks the N-terminal 28 amino acids. Substrate fidelity for a particular SSD domain was good, but not perfect. ClpX and to a lesser extent ClpA specifically bound Arc-SsrA as is seen in vivo. Similarly, Lon specifically bound UmuD. However, several SSD domains crossreacted with substrates that are normally degraded by other proteases. For example, ClpY interacted with Arc-SsrA and ClpA bound σ32.
Substrate recognition requires not only a binding site on the protease but also a recognition motif on the substrate. In earlier work, Sauer and his collaborators discovered that a class of abnormal proteins is tagged with a unique amino acid sequence that targets them for degradation by Clp proteases (17, 18). Proteins translated from truncated messenger RNA are released from the ribosome after addition of an 11-aa extension encoded by a modular template, a 10 S RNA encoded by the ssrA gene. In addition to this C-terminal tagging by SsrA, N-terminal recognition has also been demonstrated. Certain β-galactosidase fusion proteins are rapidly degraded by ClpAP protease only when the fusions have hydrophobic N-terminal amino acids (19). As with C-terminal tagging, there is an enzymatic system for specifically adding hydrophobic amino acids to the amino terminus of some proteins (20), thus demonstrating that modification of the ends of proteins is a physiologically important mechanism for tagging proteins for degradation.
Because the tags identified to date are located near the N or C terminus of polypeptides, there has been close examination of these regions of naturally unstable proteins for possible recognition sites. A striking aspect of the results reported to date is the diversity in the sequences recognized by the different proteases and also by any single protease. One criterion for a recognition tag is functioning in trans on proteins. For MuA, a substrate for ClpX and ClpXP, a recognition motif is located within the C-terminal 10 amino acids (21). This signal can be transferred to an otherwise stable Arc repressor protein, which thereby is able to bind ClpX and be degraded by ClpXP (22). Other studies show, however, that another ClpX substrate, the O protein of phage λ, is targeted to ClpXP via sequences contained within the N terminus (23). The RepA protein, which is specifically recognized by ClpAP, is also targeted by a transferable signal located within the N-terminal 20 amino acids (J. Hoskins and S.W., unpublished work). UmuD, a protein specifically degraded by Lon protease, is recognized by virtue of a transferable motif located within the first 24 amino acids (24), whereas CcdA and SulA, two other Lon substrates, have recognition motifs in the C-terminal region (25, 26). Proteins with multiple recognition motifs can be degraded by more than one protease. For example, when the C terminus of the Muvir repressor, a ClpX substrate, was fused to CcdA, the resulting protein was degraded by both Lon and ClpXP (27). These data argue in favor of a direct interaction between recognition sites on substrates and specific regions on the proteases.
Well known classes of proteins that recognize C-terminal peptide extensions in proteins are the PDZ and PTB proteins of eukaryotic cells. These proteins are involved in a variety of protein transfer reactions needed in signal transduction and in receptor and channel clustering pathways (28). This functional similarity had led the Baker and Sauer labs to look for PDZ-like domains in ClpX and ClpA (22). Although the regions that they initially proposed as interaction sites probably have structures that are quite different from PDZ domains, having significantly more alpha-helical structure than the primarily beta-stranded structures found in bona fide PDZ domains, the sites specifically bound proteins containing the SsrA tag as well as proteins carrying the C terminus of MuA. These motifs overlap but are not identical to the SSD-like domains discussed above.
Recent sequence analysis suggests that the SSD motif is part of a key structural feature, specifically the sensor 2 domain, exhibited by members of a superfamily of ATPases referred to as the AAA family (29). AAA family members are involved in assembly, disassembly, and functioning of a broad class of protein complexes and molecular machines. Sensor 2 domains consist of three or four alpha helices and, in many AAA proteins, contain a critical arginine residue that interacts with the bound nucleotide and is needed for ATPase activity. Binding of protein segments to the sensor domains could affect ATP hydrolysis and provide a means for coupling ATP hydrolysis to unfolding or translocation of substrates. These regions normally occur adjacent to the ATP hydrolysis to unfolding or translocation of substrates. These regions normally occur adjacent to the ATP-binding domain of AAA proteins and are found either at the C terminus of the proteins or in regions bridging the ATPase domain and a specific functional domain. Thus, an intriguing possibility is that the substrate interaction sites for the ATP-dependent proteases may serve parallel roles for all AAA proteins.
Important questions regarding the role of SSD domains in substrate selection remain. Are the SSD domains the only interaction sites for substrates? There are several observations pointing to the possibility that substrate recognition and binding are more complex: first, the deviations in fidelity from the in vivo situation of the SSD domains observed by Smith et al. hint that other as yet unidentified factors may impact on substrate selection (16). Those factors could influence the discriminatory ability of an SSD domain, perhaps by altering its conformation or by affecting its availability. Second, binding affinities between substrates and isolated SSD domains are rather weak, suggesting that binding to SSD domains is not the sole basis for substrate interaction. For example, the binding of ClpA to RepA (30) or ClpX to λO protein (23) is significantly stronger than the interactions between isolated SSD domains and substrates (16). Binding by SSD domains is also weaker than that observed for PDZ-domain fusion proteins interacting with peptide ligands (31). In addition, the affinity of ClpX for one substrate tag, the N-terminal 28-aa peptide of λO, is an order of magnitude lower than for intact λO (J. Rozycki and M.M., unpublished results). Additional binding sites on the ATP component that contribute to binding, perhaps by interacting with different portions of a substrate molecule, could explain some of the above observations and would increase the fidelity of substrate recognition (1, 13, 32).
The physical location of the SSD domains is a matter of intense interest and has important implications regarding the availability of the sites to substrates. If SSD domains were the initial sites of interaction with substrates, they might be expected to be located on the exterior surface of the ATPase rings. In fact, electron microscopic studies indicate that RepA binds on the apical surface of the ClpA rings, away from ClpP (F. Beuron, M. Kessel, D. Belnap, S.W., M.M., and A. Steven, unpublished work). However, in ClpA and ClpX, the SSD domains are located in the C-terminal regions, and biochemical data suggest that the C-terminal domains are near the interface of ClpA and ClpX with ClpP (S. K. Singh and M.M., unpublished data). In Lon, the SSD domain is similarly located between the ATPase domain and the proteolytic domain. Alternatively, the SSD domains could be located within the axial channel or in the cavity formed by the interface of the ATPase and protease components, where they would be accessible to an N- or C-terminal extension of the substrate. In this case, binding to SSD sites within the protease complex might follow initial interaction of substrates at the surface, constituting a second level of substrate discrimination.
What is the nature of the binding interaction between the recognition tags on the substrates and the SSD domains? SSD domains interact with sites near the N and C termini of substrates, suggesting that the binding site should be able to accommodate both terminal COO− and NH3+. The orientation of the bound peptide extension thus presents a puzzle. If binding requires a free end group, either the binding site has to accept peptides in both orientations or there have to be separate subsites for the different termini. A free end group may not be essential, in which case both terminal peptides could bind in the same orientation with their charged groups at opposite ends of a binding site. Such a binding site might also allow interaction with any flexible polypeptide segment carrying a positive charge at one end and a negative charge at the other. In this case, degradation motifs might occur anywhere within protein sequences and not just at the ends, although this has not been observed to date.
The current results hold the promise that, as with other biological interactions of seemingly unmanageable complexity, substrate recognition by chaperone components of ATP-dependent proteases may use a relatively straightforward mechanism—a structurally well conserved site with mutable local elements serves as a docking site for recognition motifs located in accessible extensions near the N or C terminus of target proteins. Whether this simple mechanism will explain substrate selection remains to be seen, but, in any event, identifying the specific residues or subsites within the SSD domains that interact with substrates and help fine tune substrate selection should be a challenging and informative undertaking.
Acknowledgments
We thank Susan Gottesman for comments on the manuscript.
Footnotes
The companion to this commentary begins on page 6678 in issue 12 of volume 96.
References
- 1.Gottesman S, Wickner S, Maurizi M. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 2.Larsen C N, Finley D. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 6.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 7.Kessel M, Wu W, Gottesman S, Kocsis E, Steven A C, Maurizi M R. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 8.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 9.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 10.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 12.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawrzynow A, Banecki B, Zylicz M. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- 14.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 15.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 16.Smith C K, Baker T A, Sauer R T. Proc Natl Acad Sci USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiler K C, Waller P R, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1376. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 20.Shrader T E, Tobias J W, Varshavsky A. J Bacteriol. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 22.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 23.Gonciarz-Swiatek M, Wawrzynow A, Um S-J, Learn B A, McMacken R, Kelley W L, Georgopoulos C, Sliekers O, Zylicz M. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez M, Frank E G, Levine A S, Woodgate R. Genes Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Melderen L, Dao Thi M H, Lecchi P, Gottesman S, Couturier M, Maurizi M R. J Biol Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 26.Sonezaki S, Ishii Y, Okita K, Sugino T, Kondo A, Kato Y. Appl Microbiol Biotechnol. 1995;43:304–309. doi: 10.1007/BF00172829. [DOI] [PubMed] [Google Scholar]
- 27.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Tousaint A. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison S C. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- 29.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 30.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman S, Maurizi M R. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]