Abstract
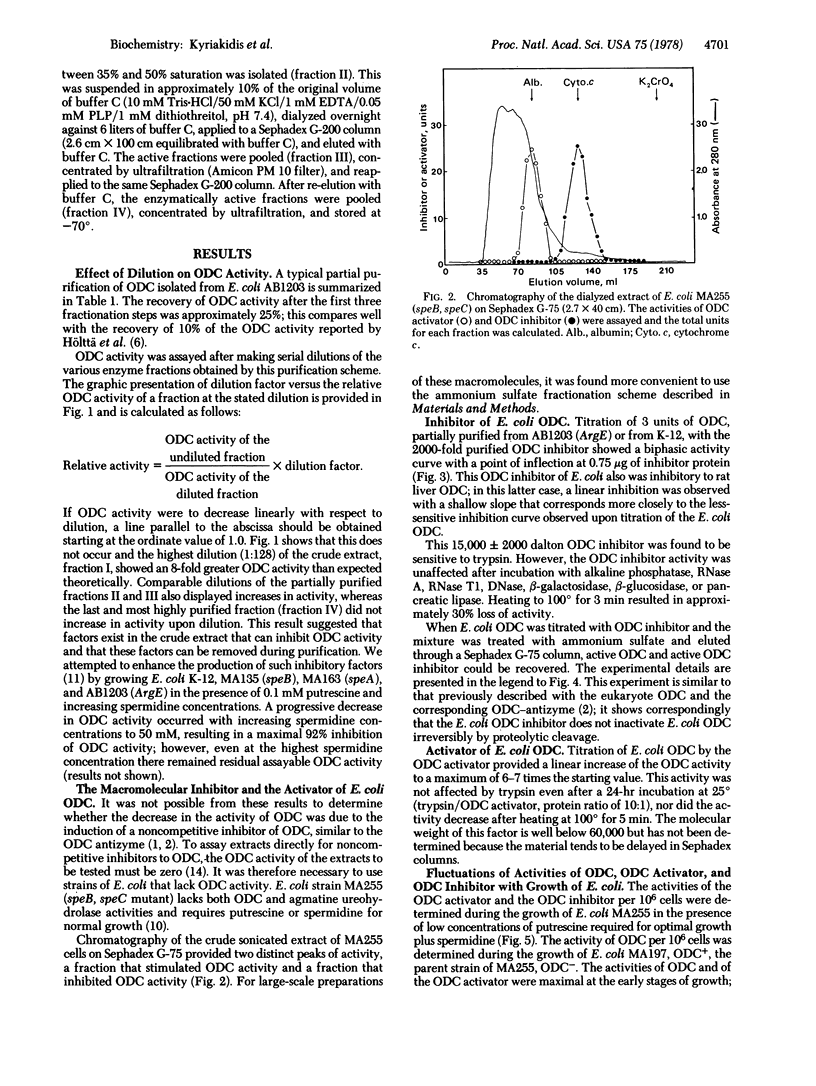
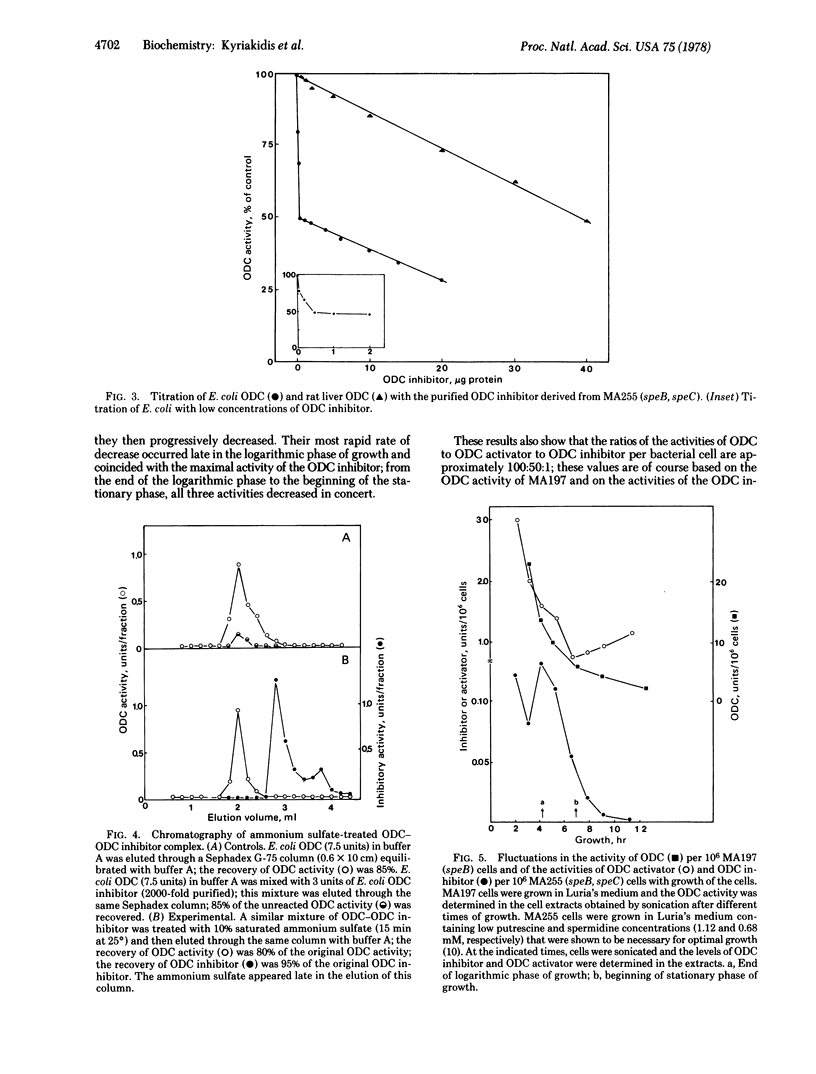
Two effectors of ornithine decarboxylase (ODC; L-ornithine carboxy-lyase, EC 4.1.1.17) have been extracted from an ODC- (speC-) mutant, Escherichia coli MA 255. One of these is an ODC inhibitor (Mr 15,000 +/- 2000) that is labile to trypsin; its activity increases 20-fold in response to increased polyamine levels in the growth medium. It has additional characteristics similar to those of the ODC antizyme of eukaryote cells: it is a noncompetitive inhibitor of ODC; the complex formed between ODC and the ODC inhibitor can be dissociated with salt to provide active ODC and active ODC inhibitor; furthermore, this E. coli ODC inhibitor is inhibitory to eukaryote ODC. A thermostable nondialyzable factor that activates ODC in vitro has also been extracted from MA255; increased polyamine levels in the growth medium caused a 1.6-fold increase in the activity of this ODC activator. Effectors with comparable activities have also been identified in the parent ODC+ (speC+) strain MA197. The fluctuations of the intracellular levels of these two ODC effectors during the growth of E. coli MA255 have been related to the temporal changes of the activity of ODC in the parent ODC+ MA197 strain. The mode of interaction of these three macromolecules, as reflected in the changes of the activity of ODC, appears to be complex. The results suggest that ODC activity may be controlled post-translationally by macromolecules that act as positive and negative effectors and whose levels fluctuate in response to the concentration of the end products of the reaction.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMES B. N., GARRY B., HERZENBERG L. A. The genetic control of the enzymes of histidine biosynthesis in Salmonella typhimurium. J Gen Microbiol. 1960 Apr;22:369–378. doi: 10.1099/00221287-22-2-369. [DOI] [PubMed] [Google Scholar]
- Applebaum D., Sabo D. L., Fischer E. H., Morris D. R. Biodegradative ornithine decarboxylase of Escherichia coli. Purification, properties, and pyridoxal 5'-phosphate binding site. Biochemistry. 1975 Aug 12;14(16):3675–3681. doi: 10.1021/bi00687a025. [DOI] [PubMed] [Google Scholar]
- Cunningham-Rundles S., Maas W. K. Isolation, characterization, and mapping of Escherichia coli mutants blocked in the synthesis of ornithine decarboxylase. J Bacteriol. 1975 Nov;124(2):791–799. doi: 10.1128/jb.124.2.791-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong W. F., Heller J. S., Canellakis E. S. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim Biophys Acta. 1976 Apr 23;428(2):456–465. doi: 10.1016/0304-4165(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Hafner E. W., Tabor C. W., Tabor H. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J Bacteriol. 1977 Dec;132(3):832–840. doi: 10.1128/jb.132.3.832-840.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J. S., Chen K. Y., Kyriakidis D. A., Fong W. F., Canellakis E. S. The modulation of the induction of ornithine decarboxylase by spermine, spermidine and diamines. J Cell Physiol. 1978 Aug;96(2):225–234. doi: 10.1002/jcp.1040960211. [DOI] [PubMed] [Google Scholar]
- Heller J. S., Fong W. F., Canellakis E. S. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J. S., Kyriakidis D., Fong W. F., Canellakis E. S. Ornithine decarboxylase antizyme is a normal component of uninduced H-35 cells and rat liver. Eur J Biochem. 1977 Dec;81(3):545–550. doi: 10.1111/j.1432-1033.1977.tb11980.x. [DOI] [PubMed] [Google Scholar]
- Hölttä E., Jänne J., Pispa J. Ornithine decarboxylase from Escherichia coli: stimulation of the enzyme activity by nucleotides. Biochem Biophys Res Commun. 1972 Jun 9;47(5):1165–1171. doi: 10.1016/0006-291x(72)90957-6. [DOI] [PubMed] [Google Scholar]
- Hölttä E., Jänne J., Pispa J. The regulation of polyamine synthesis during the stringent control in Escherichia coli. Biochem Biophys Res Commun. 1974 Aug 5;59(3):1104–1111. doi: 10.1016/s0006-291x(74)80092-6. [DOI] [PubMed] [Google Scholar]
- Morris D. R., Pardee A. B. Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem. 1966 Jul 10;241(13):3129–3135. [PubMed] [Google Scholar]
- Sakai T. T., Cohen S. S. Regulation of ornithine decarboxylase activity by guanine nucleotides: in vivo test in potassium-depleted Escherichia coli. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3502–3505. doi: 10.1073/pnas.73.10.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


