Abstract
Background & objectives:
AmpC β-lactamases which are often plasmid mediated hydrolyze all β-lactam antibiotics except cefepime and carbapenems. We evaluated the presence of AmpC β-lactamases among Enterobacteriaceae strains recovered prospectively from patients at five Indian tertiary care centres.
Methods:
The study included 909 consecutive Gram-negative isolates recovered from clinically significant specimens during June 2007 - May 2008 as part of an ICMR-ESBL study. Among the study isolates, 312 were found to be cefoxitin resistant by disc diffusion test (DDT). Minimum inhibitory concentration (MIC) determination by E test was done against amikacin, levofloxacin, impinem, meropenem, ertapenem, tigecycline and piperacillin-tazobactam. Combined DDT using phenyl boronic acid as inhibitor with cefoxitin was used for phenotypic confirmation of AmpC phenotype. The common Amp C genotypes ACC, FOX, MOX, DHA, CIT and EBC were detected by multiplex PCR.
Results:
Plasmid mediated Amp C phenotype was confirmed in 114 of the 312 (36.5%) cefoxitin resistant isolates with 255 (81.7%) showing multidrug resistance. Susceptibility to tigecycline was highest (99%) followed by imipenem, meropenem (97%), ertapenem (89%), amikacin (85%), and piperacillin-tazobactam (74.6%). Levofloxacin resistance was 82 per cent. ESBL co carriage was observed among 92 per cent of Amp C producers. Among 114 Amp C producers, 48 could be assigned a genotype, this included CIT- FOX (n=25), EBC (n=10), FOX (n = 4), CIT (n=3), EBC-ACC (n=2) and one each of DHA, EBC-DHA, FOX -DHA and FOX-EBC-DHA.
Interpretation & Conclusions:
Overall, AmpC phenotypes were found in 12.5 per cent isolates, multidrug resistance and ESBL co-carriage among them was high suggesting plasmid mediated spread. The study results have implications in rational antimicrobial therapy and continued surveillance of mechanisms of resistance among nosocomial pathogens.
Keywords: AmpC β-lactamases, Amp C genotypes, cefoxitin resistance, ESBL, multidrug resistance
AmpC β-lactamases are clinically important cephalosporinases produced by many Enterobacteriaceae strains and mediate resistance to cephalothin, cefazolin, cefoxitin, most penicillins and β-lactam/β-lactam inhibitor combinations1. Plasmid-mediated AmpC genes first reported in 1988, constitute an emerging therapeutic problem2. The plasmid-mediated AmpC genes are derived from inducible chromosomal genes that have become mobilized. Commonly reported genotypes are ACC, FOX, MOX, DHA, CIT and EBC3–5. These enzymes confer a resistance pattern similar to the overproduction of chromosomal AmpC β-lactamases, which may involve all β-lactam antibiotics except for carbapenems and cefepime6.
In many bacteria, AmpC enzymes are inducible and can be expressed at high levels by mutation. Overexpression confers resistance to broad-spectrum cephalosporins including cefotaxime, ceftazidime, and ceftriaxone and is a problem especially in infections due to Enterobacter aerogenes and E. cloacae, where an isolate initially susceptible to these agents may become resistant upon therapy7. Transmissible plasmids have acquired genes for AmpC enzymes, which consequently can now appear in bacteria lacking or poorly expressing a chromosomal AmpC gene, such as Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. Resistance due to plasmid mediated AmpC enzymes is less common than ESBL production in most parts of the world but may be both harder to detect and broader in spectrum7.
Plasmid-mediated AmpC genes are of special interest because their mobility allows them to emerge in one genus or species and spread to different organisms. The prevalence of plasmid mediated AmpC-type resistance at the national level in most countries is unknown because studies have not examined the strains at the molecular level required to elucidate the different mechanisms involved. A 2004 report from the United States documented 7 to 8.5 per cent of the Klebsiella spp. and 4 per cent of the Escherichia coli isolates contained plasmid mediated AmpC type enzymes8. In a 2005 Canadian study, 13.5 per cent of the isolates harboured a gene that correlated with acquired AmpC CMY-2 type resistance, and in all strains the gene was identified as CMY-29. In 2006, a study from US reported the rate of transferable AmpC producer to be 3.3 per cent with FOX-5 genes most predominantly occurring in K. pneumoniae10. Plasmid mediated AmpC was present in 26 per cent of study isolates, with CMY like enzymes detected predominantly in E.coli and DHA like enzymes predominantly in K. pneumoniae in a study from Singapore11.
In India, AmpC prevalence has been reported in Klebsiella spp (24.1%) and E coli (37.5%)12. In another Indian study, 3.3 per cent of isolates produced AmpC β lactamases13. In this report, we present the detailed phenotypic and molecular characterization of prospectively collected cefoxitin-resistant E. coli, Klebsiella spp. and Enterobacter spp. from a multicentric Indian survey. The usefulness of a screening test using phenyl boronic acid as inhibitor of AmpC enzymes was evaluated.
Material & Methods
Study design: In this prospective laboratory based surveillance study (June 2007 to May 2008), 909 non repeat Gram-negative strains [E.coli (n=517), Klebsiella spp. (n=331) and Enterobacter spp. (n=61)] determined to be clinically significant [skin and soft tissue (132), blood (n= 91), and urinary tract infection (89)] were collected from five Indian tertiary care centres All India Institute of Medical Sciences (AIIMS), New Delhi, Amrita Institute of Medical Sciences (AIMS), Kochi, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, Mahatma Gandhi Institute of Medical Sciences (MGIMS), Wardha, and Jawaharlal Institute of Post Graduate Medical Education & Research (JIPMER), Puducherry. This survey was done as a part of Indian Council of Medical Research (ICMR) sponsored initiative on “Clinico-epidemiologic and molecular characterization of extended-spectrum beta-lactamase (ESBL) producing E. coli, Klebsiella spp. and Enterobacter spp. causing nosocomial and community Infections”.
The identities of all strains submitted were reconfirmed by conventional biochemical methods and API (Biomerieux, Craponne, France) system. The isolates were then tested for extended-spectrum β-lactamase (ESBL) production using the screening criteria described by Clinical and Laboratory Standards Institute (CLSI) 200914 at the study central monitoring laboratory (BMPLIII) at CMC, Vellore. From this collection (n=909), 312 cefoxitin resistant isolates [(E. coli (n=152), Klebsiella spp. (n=117) and Enterobacter spp. (n= 43)] were further characterized for AmpC expression.
Antimicrobial susceptibility testing (AST): ESBL-producing isolates were confirmed to be cefoxitin (30 μg) resistant by the Kirby Bauer disc diffusion method as per CLSI 200914. The minimum inhibitory concentrations (MICs) of cephalosporins, with and without clavulanic acid, for ESBL detection and confirmation as well as the MICs of amikacin, levofloxacin, piperacillin-tazobactam, imipenem, meropenem, ertapenem and tigeycycline for all cefoxitin-resistant isolates detected in this study were determined by E test (Biomerieux, Craponne, France) and results interpreted as per manufacturers and CLSI 2009 guidelines14. Multidrug resistance (MDR) was defined as resistance to three or more antimicrobial classes.
Combined disc diffusion test: The differences in inhibition zones for cefoxitin (30 μg) discs alone and in combination with (400 μg) of phenyl boronic acid was determined4. The zone diameters were similar and reproducible when the procedure was repeated. An increase of >5 mm in zone diameter in the presence of phenyl boronic acid compared with cefoxitin tested alone was considered to be positive for the presence of an AmpC β-lactamase production.
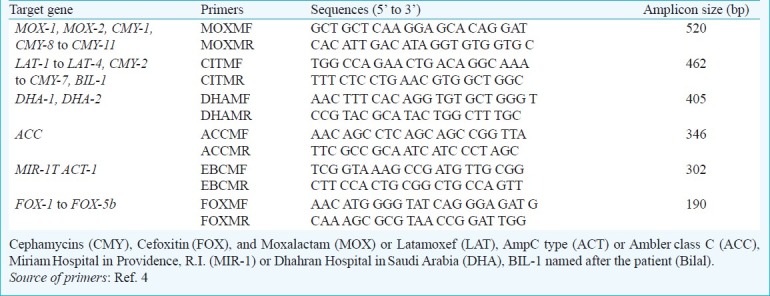
Molecular characterization of Ambler class C (AmpC) resistance determinants: Multiplex PCR was used to detect the most common plasmid mediated AmpC genes ACC, FOX, MOX, DHA, CIT and EBC reported in literature using protocol previously reported4,15.
Preparation of template DNA - A single colony of each organism was inoculated from a Mac Conkey agar plate into 5 ml of nutrient broth (Becton Dickinson, Maryland, USA) and incubated for 16-18 h at 37°C. Cells from 1.5 ml of the overnight culture were harvested by centrifugation at 17,310 g for 5 min. After the supernatant was decanted, the pellet was resuspended in 500 μl of distilled water. The cells were lysed by heating at 95°C for 10 min, and cellular debris was removed by centrifugation at 17,310 g for 5 min. Supernatant (2 μl) was used as the DNA template source for amplification.
PCR was performed with a final volume of 25 μl in 0.2-ml thin-walled tubes. The primers used for PCR amplification are listed in Table I. Each reaction contained 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 0.2 mM each deoxynucleoside triphosphate; 1.5 mM MgCl2; 0.6 mM primers MOXMF, MOXMR, CITMF, CITMR, DHAMF, and DHAMR; 0.5 mM primers ACCMF, ACCMR, EBCMF, and EBCMR; 0.4 mM primers FOXMF and FOXMR4; and 1.25 U of Taq DNA polymerase (Life Technologies, Rockville, USA). Template DNA (2 μl) was added to 23 μl of the master mixture. The PCR programme consisted of an initial denaturation step at 95°C for 2 min, followed by 30 cycles of DNA denaturation at 94°C for 45 sec, primer annealing at 62°C for 45 sec, and primer extension at 72°C for 1 min. After the last cycle, a final extension step at 72°C for 5 min was added. PCR product (15 μl) was analyzed by gel electrophoresis with 2 per cent agarose (USB Corporation, Cleveland, USA.). Gels were stained with ethidium bromide at 5 μg/ml and visualized by UV transillumination. A 100-bp DNA ladder (Fermentas International Inc. Burlington, Canada) was used as a molecular ladder. Negative controls were PCR mix with water in place of template DNA.
Table I.
Primers used for characterization of AmpC β-lactamases
Control strains: Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 27853, and E. coli ATCC 25922 were used to quality check the media, biochemical tests and susceptibility testing. ATCC standard strains were used to quality check E test and test ranges interpreted as per manufacturers, CLSI 2009 guidelines14. Amp C genotype standard strains A7 (ACC), A9 (CMY-2), and PMG252 (FOX-5) (provided by Dr George A Jacoby, Lahey Clinic, Burlington, Massachusetts, USA), and clinical isolate ADB05 (DHA), ADB42 (EBC) from our laboratory were used as PCR control strains.
Results
Overall, among 909 Gram-negative isolates, 312 were deemed cefoxitin resistant by Kirby Bauer disc diffusion test. This included E. coli (n=152), Klebsiella spp. (n=117) and Enterobacter spp. (n=43).
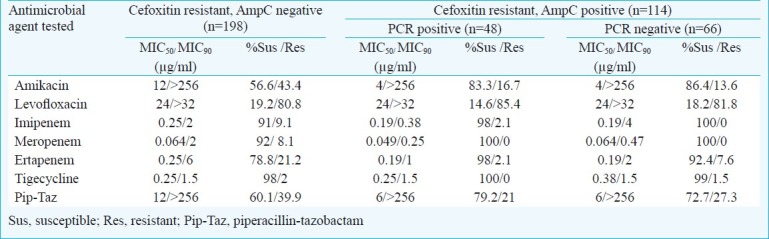
Among 312 cefoxitin resistant isolates, AmpC phenotype was confirmed by the combined disc diffusion testing in 36.5 per cent (n=114), in the remaining (n=198) it was not detectable. The overall occurrence of Amp C in the study was determined to be 12.5 per cent. Among the 312 cefoxitin resistant isolates, highest susceptibility to tigecycline (99%) was seen followed by imipenem, meropenem (97%), ertapenem (89%), amikacin (85%), and piperacillin- tazobactam (74.6%), resistance to levofloxacin was high (82%). There were no significant differences in susceptibility as well as MIC50 and MIC90 values (Table II) among the AmpC producers (n=114) and Amp C negative isolates (n=198). A very high proportion (n=92%) of the AmpC phenotypes was also found to be ESBL producers with a significant number of them (n=81.7%) showing multidrug resistance.
Table II.
Antimicrobial susceptibility profile of 312 cefoxitin resistant study isolates
Among 114 isolates of Amp C producers, 48 could be assigned a genotype, this included CIT- FOX (n=25), EBC (n=10), FOX (n = 4), CIT (n=3), and EBC-ACC (n=2), one each of DHA, EBC-DHA, FOX -DHA and FOX-EBC-DHA.
Isolates with plasmid mediated AmpC were recovered from patients with urinary tract (n=21), skin and soft tissue (n=17) and blood stream infection (n=10). Of these, 79.4 per cent were classified to be nosocomial in origin. The phenylboronic acid-cefoxitin disc tests showed corresponding sensitivity of 72.9 per cent, specificity of 45.4 per cent, positive predict value of 49.2 per cent, and negative predict value of 69.7 per cent when compared with PCR. Overall, presence of common plasmid mediated AmpC genotypes among Gram-negative isolates was low (5.2%). In our study, CIT- FOX (21.9%) and EBC Amp C (8.7%) genotypes were predominant. CIT-FOX like enzymes were common in E.coli (43.7%) and EBC like enzymes in K. pneumoniae (16.6%).
Discussion
The prevalence of plasmid AmpC-mediated resistance in India is not known, due to the limited number of surveillance studies seeking clinical strains producing AmpC β-lactamases and the difficulty that laboratories have in accurately detecting this resistance mechanism. The present study showed plasmid mediated AmpC β lactamases in 12.5 per cent isolates, with commonly reported genotypes seen among 5.2 per cent of them.
Plasmid AmpC β-lactamases have differential activity on β-lactamases inhibitors, E.coli derived enzymes have shown to exhibit resistance to inhibitor combinations with possible exception of piperacillin-tazobactam1. In previous Indian studies, cefoxitin resistant strains were tested for the production of AmpC β lactamases by three dimensional extract methods16,17. A recent Indian study has recommended use of piperacillin and piperacillin-tazobactam discs for AmpC screening18. Cefotetan with phenyl boronic acid has also been used to detect AmpC especially MOX-1, FOX-1, ACT-1 producing isolates19. Detecting plasmid mediated AmpC with co-existing ESBL is very challenging. Given these difficulties in detecting plasmid mediated AmpC β-lactamases, their prevalence is currently being underestimated.
AmpC β-lactamases also have differential activity on substrates. E.coli with ACC–1 can be resistant to ceftazidime but not to cefotaxime or cefotetan while an isolate with DHA-2 may show intermediate resistance to cefoxitin but susceptible to cefotaxime or ceftazidime1. In our study, we could not discern such findings, further analysis of isolates including cephamycin hydrolysis assay is necessary to verify these effects.
In the study, sizeable number of cefoxitin resistant isolates were not positive for AmpC production by the disc potentiation test or PCR, this warrants further investigation into the other mechanisms of resistance and their laboratory detection. Clinical isolates rarely express more than one plasmid-mediated AmpC β-lactamases. Two reasons could explain this observation. First, the inability of current phenotypic tests to accurately detect the type of transferable AmpC β-lactamase does not allow for the differentiation of multiple AmpC enzymes. Second, it is possible that there is a limit to the amount of AmpC β-lactamase that a bacterial cell can accommodate and still be a viable pathogen20. A single type of test will not be able to accurately characterize the resistance mechanisms in these complex organisms. Although automated systems are available for susceptibility testing, the accuracy of these are inadequate for organisms expressing plasmid-mediated AmpC β-lactamases alone or in combinations with ESBLs21–23.
The use of cefoxitin resistance as a screening agent/marker for AmpC production is quite reliable with a good negative predictive value as found in our study. The use of phenylboronic acid in combination with cefoxitin as a phenotypic screening method may be a better tool for laboratory diagnosis and confirmation of AmpC producing Gram-negative bacteria. The disc potentiation test reliably detected AmpC β-lactamase when compared against the PCR in the present study. Clinical laboratories interested in distinguishing AmpC mediated resistance from other β-lactamase resistance mechanisms will need to use combination of phenotypic and molecular identification methods. The multiplex PCR technique described in this study will be an important tool for the detection of plasmid-mediated AmpC β-lactamases genes in Gram-negative bacteria.
In the present study, MDR among AmpC positive study isolates was high suggesting plasmid mediated spread. Current therapeutic options include use of cefepime or carbapenems7, however, the high co-carriage of ESBL and AmpC in this study and the fact that majority of these were nosocomial in origin is a cause for concern. This study findings indicate the necessity for continued surveillance of mechanisms of resistance among nosocomial pathogens and evolving preventive measures aimed at reducing their spread.
Acknowledgment
Authors acknowledge the Indian Council of Medical Research (ICMR), New Delhi, for financial assistance, and Christian Medical College, Vellore Institutional Review Board for approving conduct of this study (R.C.No. 5342 dated 20.04. 2004).
References
- 1.Philippon A, Arlet G, Jacoby GA. Plasmid determined AmpC type β-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swaminathan B, Barrett T, Hunter SB, Tauxe R PulseNet. The molecular subtyping network for foodborne disease surveillance in the United States. Emerg Infect Dis. 2001;7:382–9. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson ND. AmpC β-lactamases: what do we need to know for the future? J Antimicrob Chemother. 2003;52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Pérez FJ, Hanson ND. Detection of plasmid mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papanicolaou GA, Medeiros AA, Jacoby GA. Novel plasmid mediated β-lactamases (MIR-1) conferring resistance to oxyimino and α methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–9. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Por Beatriz Mirelis A, Alba Rivera A, Elisenda Miró B, Raúl J, Mesa A, Ferran Navarro A, Pere Coll P. A simple phenotypic method for differentiation between acquired and chromosomal AmpC β-lactamases in Escherichia coli. Enferm Infect Microbiol Clin. 2006;24:370–2. doi: 10.1157/13089690. [DOI] [PubMed] [Google Scholar]
- 7.Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22:161–82. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez M, Tran JH, Chow N, Jacoby GA. Epidemiology of conjugative plasmid mediated AmpC β-lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–7. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulvey MR, Bryce E, Boyd DA, Ofner-Agostini M, Land AM. Molecular characterization of cefoxitin resistant Escherichia coli from Canadian hospitals. Antimicrob Agents Chemother. 2005;49:358–65. doi: 10.1128/AAC.49.1.358-365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moland ES, Hanson ND, Black JA, Hossain A, Song W, Thomson KS. Prevalence of newer β-lactamases in Gram negative clinical isolates collected in the United States from 2001 to 2002. J Clin Microbiol. 2006;44:3318–24. doi: 10.1128/JCM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan TY, Ng SY, Teo L, Koh Y, Teok CH. Detection of plasmid mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J Clin Pathol. 2008;61:642–4. doi: 10.1136/jcp.2007.053470. [DOI] [PubMed] [Google Scholar]
- 12.Subha A, Renuka Devi V, Ananthan S. AmpC β-lactamases producing multidrug resistant strains of Klebsiella spp. & Escherichia coli isolated from children under five in Chennai India. Indian J Med Res. 2003;117:13–8. [PubMed] [Google Scholar]
- 13.Ratna AK, Menon I, Kapur I, Kulkarni R. Occurrence & detection of Amp C β-lactamases at a referral hospital in Karnataka. Indian J Med Res. 2003;118:29–32. [PubMed] [Google Scholar]
- 14.Performance standards for antimicrobial susceptibility testing, 19th informational supplement, CLSI document M100-S19. 3. Vol. 29. Wayne, PA: CLSI; 2009. Clinical and Laboratory Standards Institute (CLSI) [Google Scholar]
- 15.Thomson KS. Controversies about extended-spectrum and AmpC β-lactamases. Emerg Infect Dis. 2001;7:333–6. doi: 10.3201/eid0702.010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow M, Hall BG. Experimental prediction of the evolution of cefepime resistance from the CMY-2 AmpC β-lactamase. Genetics. 2003;164:23–9. doi: 10.1093/genetics/164.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manchanda V, Singh NP, Shamweel A, Eideh HK, Thukral SS. Molecular epidemiology of clinical isolates of AmpC producing Klebsiella pneumoniae. Indian J Med Microbiol. 2006;24:177–81. [PubMed] [Google Scholar]
- 18.Taneja N, Rao P, Arora J, Dogra A. Occurrence of ESBL and Amp-C beta-lactamases and susceptibility to newer antimicrobial agents in complicated UTI. Indian J Med Res. 2008;127:85–8. [PubMed] [Google Scholar]
- 19.Pitout JD, Le PG, Moore KL, Church DL, Gregson DB. Detection of AmpC beta-lactamases in , Klebsiella spp, Salmonella spp and Proteus mirabilis in a regional clinical microbiology laboratory. Clin Microbiol Infect. 2010;16:165–70. doi: 10.1111/j.1469-0691.2009.02756.x. [DOI] [PubMed] [Google Scholar]
- 20.Morosini MI, Ayala JA, Baquero F, Martinez JL, Blazquez J. Biological cost of AmpC production for Salmonella enterica serotype typhimurium. Antimicrob Agents Chemother. 2000;44:3137–43. doi: 10.1128/aac.44.11.3137-3143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canton R, Perez-Vazquez M, Oliver A, Sanchez Del Saz B, Gutierrez MO, Martinez Ferrer M, et al. Evaluation of the Wider system, a new computer-assisted image-processing device for bacterial identification and susceptibility testing. J Clin Microbiol. 2000;38:1339–46. doi: 10.1128/jcm.38.4.1339-1346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzouvelekis LS, Vatopoulos AC, Katsanis G, Tzelepi E. Rare case of failure by an automated system to detect extended-spectrum beta-lactamase in a cephalosporin resistant Klebsiella pneumoniae isolate. J Clin Microbiol. 1999;37:2388. doi: 10.1128/jcm.37.7.2388-2388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders CC, Peyret M, Moland ES, Cavalieri SJ, Shubert C, Thomson KS, et al. Potential impact of the VITEK 2 system and the Advanced Expert System on the clinical laboratory of a university-based hospital. J Clin Microbiol. 2001;39:2379–85. doi: 10.1128/JCM.39.7.2379-2385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


