Abstract
Background & objectives:
Infections due to seafood associated Salmonella serovars are great risk to public health. Different phenotypic characteristics have been used previously for epidemiological investigation of Salmonella. Beyond the phenotypic characterization, a reliable genetic level discriminatory method is required. Therefore, this study was attempted to use different phenotypic and molecular fingerprinting methods for investigation of genetic diversity among seafood associated nontyphoidal Salmonella serovars.
Methods:
Fifty eight seafood associated Salmonella isolates were included in this study. All isolates were serotyped and epidemiological investigation was carried out using molecular fingerprinting methods, random amplified polymorphic DNA (RAPD) and enterobacterial repetitive intergenic consensus sequence based-PCR (ERIC-PCR) along with whole cell protein profiling using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) in our study.
Results:
Among the 58 Salmonella isolates, S. Weltevreden was observed to be the most predominant serovar. Typing of Salmonella serovars using RAPD and ERIC-PCR suggested the existence of a genetic diversity. Though both PCR based techniques were found to have a good discriminatory index, a better discriminatory ability was observed when the results obtained by the two techniques were combined and taken for composite analysis. Protein profiling of whole cells using SDS-PAGE demonstrated the presence of several bands with two bands of sizes 38 kDa and 46 kDa common among all 58 isolates.
Interpretation & conclusions:
Our study shows that use of protein profiling in combination with established typing methods such as RAPD and ERIC-PCR may provide useful information in typing of non-typhoidal Salmonella isolates associated with seafood and to develop strategies to protect public from Salmonella infections.
Keywords: ERIC-PCR, RAPD-PCR, Salmonella spp., SDS-PAGE, seafood, serotype
Food borne infections due to Salmonella serovars are a major concern worldwide. Many non typhoidal Salmonella (NTS) serovar infections result in diarrhoeal disease, bacteraemia and extraintestinal focal infection in infants and more serious complications among the elderly and immunocompromised adults1. Salmonella spp. associated with gastrointestinal tract of animals, including birds2 reach to aquatic environments through faecal contamination. Presence of NTS serovars in fish, shellfish and other seafood has been reported3,4. Filter feeding organisms such as oysters and clams harvested from contaminated waters are known to concentrate high levels of Salmonella serovars leading to high incidence of this pathogen in seafood5. Though there are several reports from India on the prevalence of Salmonella in seafood4,6, information on molecular characterization and genetic relatedness among the different NTS serovars is limited.
Different phenotypic characteristics used for epidemiological investigation of Salmonella include serotyping, based on the differentiation of O and H antigen7 and though most widely used it lacks the ability to differentiate isolates having same serotype. Fingerprinting techniques have been used to determine the source of infection, transmission of organisms, stage of infection, detection of particularly virulent isolates as well as the host distribution of specific pathogen8. Differentiation of Salmonella serovars has been studied using several DNA based typing methods4,9. A rapid and precise subtyping was achieved by randomly amplified polymorphic DNA (RAPD) and enterobacterial repetitive intergenic consensus (ERIC) fingerprinting8,9. While RAPD uses a single primer of arbitrary nucleotide sequence which targets random segments of genomic DNA to reveal polymorphisms10 , ERIC is a short interspersed repetitive consensus sequence originally found in Escherichia coli and Salmonella and ERIC-PCR uses outward facing primers complementary to each end of the repeat in a PCR11. Whole-cell polypeptides solubilized with sodium dodecyl sulphate (SDS) have also been used for typing Salmonella spp.12. In the present study, the serotyped NTS isolates from seafood prevalent along the southwest coast of India were subjected to DNA based fingerprinting (RAPD and ERIC) and whole cell SDS-PAGE profiling to look at the genetic relatedness/distance by assessing the discriminatory index provided by each of the methods individually and in combination.
Material & Methods
Bacterial isolates: The study was carried out in the Department of Fishery Microbiology, Karnataka Veterinary, Animal and Fisheries Sciences University, College of Fisheries, Mangalore with samples drawn during the period March 2002-April 2007. Fifty eight Salmonella isolates obtained from seafood (20 isolates from oyster, 16 isolates from clam, 16 isolates from fish and 6 isolates from shrimp) were included in this study. Oyster, clam and fish samples were harvested biweekly from two estuaries, Mulki (site 1) and Sasthan (site 2), which are located along the southwest coast of India, 60 km away from Mangalore. Shrimp samples (6) were collected from culture ponds in Kundapur (site 3), which is located about 100 km North of Mangalore on the same coast. Salmonella isolates were isolated during March 2002-April 2007. Salmonella isolates characterized biochemically as per the protocol recommended by FDA Bacteriological Analytical Manual13 were maintained at -80° C in nutrient broth containing 30 per cent glycerol (Sanyo Corporation, Japan). For activation, the isolates were grown overnight at 37° C in tryptone soya broth with continuous aeration in a shaker water bath (150 rev/min). A loopful of the culture was subcultured on tryptone soya agar (TSA). Isolated colonies were picked and maintained in TSA slants for further work. S. Typhimurium, ATCC-14028 was used as a reference strain.
Polymerase chain reaction confirmation: Primer pair specific for hns14 and invA15 genes were used for the confirmation of the Salmonella isolates. The DNA was extracted from the isolates following the protocol described by Ausubel et al16. DNA concentration and purity was determined using Nanodrop spectrophotometer (Nanodrop, USA). PCR was performed in 50 μl volumes containing 5 μl of 10X buffer (Bangalore Genei, Bangalore, India) consisting of 0.1 mol/l Tris-HCl (pH 8.3), 0.02 mol/l MgCl2, 0.5 mol/l KCl and 1 per cent gelatin, 0.2 μ mol/l of each of the 4 deoxyribonucleotide triphosphates (dATP, dGTP, dCTP and dTTP), 0.1 pmol/1 of each primer and 1.25 U of Taq polymerase (Bangalore Genei) using a gradient thermocycler (M.J. Research, USA). The cycling conditions followed were as described by Jones et al14 for hns and Rahn et al15 for invA gene.
All PCR confirmed isolates of Salmonella were sent for serotyping to the reference centre for Salmonella and E. coli at the Central Research Institute, Kasauli, Himachal Pradesh, India.
RAPD-PCR: Ten different RAPD primers which included 6 PM primers (PM1 to PM6)17 and 4 CRA primers (CRA22 to CRA26)18 were initially used in order to select the one that would give reproducible RAPD fingerprints. Among the primers used, PM5 (5’ CGA CGC CCT G 3’) that generated a reproducible and well resolved pattern was used for further analysis. PCR was performed in 50 μl volumes containing 5 μl of 10X buffer [0.1 mol l/1 Tris-HCl (pH 8.3), 0.02 mol l-1MgCl2, 0.5 mol l/1 KCl, 1% gelatin], 0.2 μ mol l/1 of each of the four deoxyribonucleotide triphosphates (dATP, dGTP, dCTP and dTTP), 100 pmol of primer and 1.25 U of Taq polymerase. The PCR conditions included an initial denaturation of 94°C for 5 min, followed by 30 cycles of denaturation 94°C for 50 sec, primer annealing at 36°C for 36 sec, extension at 72°C for 30 sec and a final delay at 72°C for 5 min. The PCR products were resolved on a 1 per cent agarose gel, stained with ethidium bromide (5 ng/ml) and bands observed using a transilluminator (Herolab, Wiesloch, Germany). The PCR was performed in replicates to see the reproducibility of the result.
ERIC-PCR: The ERIC-PCR as described in Millemann et al19 was performed in 50 μl volumes containing 5 μl of 10X buffer (0.1 mol/l Tris-HCl (pH 8.3), 0.02 mol/l MgCl2, 0.5 mol/l KCl, 1% gelatin), 0.2 μ mol/l of each of the four deoxyribonucleotide triphosphates (dATP, dGTP, dCTP and dTTP), 100 pmol of each primer pair and 1.25 U of Taq polymerase. The amplification reaction consisted of initial denaturation at 90° C for 3 min, followed by 30 cycles of denaturation at 90° C for 30 sec, primer annealing at 52° C for 1 min, extension at 72° C for 8 min and a final delay step of 72° C for 16 min. The products were resolved on a 1 per cent agarose gel, stained with ethidium bromide (5 ng/ml) and bands photographed using a transilluminator (Herolab, Wiesloch, Germany). The PCR was performed in replicates to see the reproducibility of the result.
Whole cell protein profiling: Whole cell protein profiles of the 58 Salmonella isolates were obtained by SDS- PAGE, using the discontinuous buffer system of with 5 per cent stacking and 10 per cent resolving gels20. Each sample (20 μl) was loaded and run on a vertical slab electrophoresis unit for 3 h at a constant voltage of 150 V. At the end of electrophoresis the gel was stained with c0 oomassie b0 rilliant b0 lue R-250. PMW-M was used as molecular weight marker (Bangalore Genei, Bangalore). The whole cell protein profiling was performed in replicates to see the reproducibility of the result.
Statistical analysis: The DNA banding patterns generated by RAPD and ERIC-PCR methods were used in generating dendrograms using the software Gelcompare II version 2.5 (Applied Maths, Sint-Martens-Latem, Belgium). The relatedness between the profiles was derived based on unweighted pair group method with arithmetic mean (UPGMA) using the Dice correlation coefficient method and expressed as percentage similarity. The discriminatory index was calculated for RAPD-PCR and ERIC-PCR by using the Simpson's index of diversity21. Average similarity value was calculated on the basis of the similarity matrix according to Nei and Lie22, in order to choose the cut-off to divide the isolates in to different clusters.
Results
The isolates were confirmed as Salmonella as all were positive for hns and invA gene giving amplicons of 152 and 284 bp, respectively (data not shown). On serotyping, the number of positive isolates was as follows: S. enterica serovar Weltevreden (18), S. enterica serovar Newport (10), S. enterica serovar Bareilly and S. enterica serovar Paratyphi C (8 each), S. enterica serovar Oslo (7), S. enterica serovar Infantis (3), S. enterica serovar Anatum (2), S. enterica serovar Virchow and S. enterica serovar Aba (1 each).
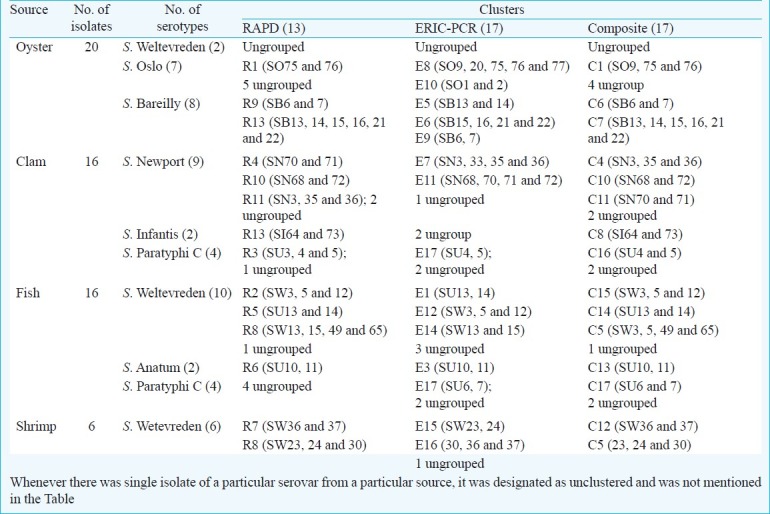
Analysis of serovars by RAPD-PCR: RAPD of the 58 Salmonella serovars yielded different patterns consisting of 5-12 bands ranging approximately from 0.15 to 2.5 kb (Fig. 1). Fig. 2 shows the dendrogram from the RAPD results of the 58 isolates. A common band was found in all 58 isolates at 1 kb. At an average similarity of 54 per cent, 44 of the 58 Salmonella serovars grouped into 13 clusters (R1-R13), while the remaining 14 were unclustered. The unclustered isolates belonged to serovar S. Virchow (SV17), S. Oslo (SO1, SO2, SO9, SO20, and SO77), S. Newport (SN33, SN34 and SN37) and S. Paratyphi C (SU1, SU2, SU6, SU7 and SU12). Heterogeneity was observed within serovars of S. Oslo (6 clusters), S. Weltevreden (5 clusters), S. Newport (6 clusters), S. Paratyphi C (6 clusters) and S. Barielly (2 clusters). S. Weltevreden, the major group with 18 isolates was assigned to RAPD clusters designated R2, R5, R7, R8 and R12. S. Newport isolates grouped in clusters designated R4, R10 and R11. S. Bareilly isolates presented as two groups viz. R9 and R13 of which R13 was further sub-clustered into two where one subgroup belonged to S. Bareilly and the other to S. Infantis at 60 per cent similarity. The single isolate of S. Aba and S. Virchow clustered with S. Infantis and S. Paratyphi C at 72 and 51 per cent, respectively. All the serovars typed in this study were isolated from a particular seafood source. The source of isolates and the serotype results showed commonality. Each serovars was linked to a particular seafood type in most of the isolates. For example, in S. Weltevreden, except for R2 and R5, where the cluster was generated from serovar isolated from fish, the remaining clusters (R7, R8 and R12) were generated from isolates from mixed animal sources of fish, oyster and shrimp.
Fig. 1.
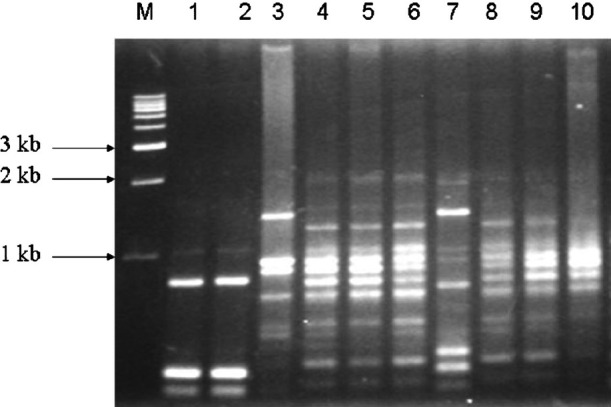
Representative RAPD fingerprint of different Salmonella isolates on 1 per cent agarose gel. M, 1kb DNA marker, Genei, Bangalore; lanes 1-10: SU1, SU2, SI64, SW49, SW30, SW23, SW65, SB6, S132 and SN34.
Fig. 2.
Dendogram showing the percentages of similarity between typable seafood associated Salmonella generated from random amplified polymorphic DNA-PCR (RAPD-PCR) fingerprinting with the band matching coefficient of Dice and the UPGMA clustering method.
Of the seven S. Oslo isolates from oyster, two could be clustered (R1) whereas others presented diversity. Eight S. Bareilly isolated from oysters could be placed in two clusters (R9 and R13). Of 16 clam isolates, nine of S. Newport isolates could be grouped as R4, R10 and R11 with two not clustering while two isolates of S. Infantis and three of the four S. paratyphi C each presented as a single cluster. Sixteen fish isolates included 10 S. Weltevreden, two S. Anatum and for S. Paratyphi C. S. Weltevreden isolates grouped in three clusters (R2, R5 and R8), two S. Anatum isolates presented in a single cluster, whereas the four S. Paratyphi C isolates remained unclusterd. Six S. Weltevreden isolates obtained from shrimp were grouped in two clusters (R7 and R8) (Table). The index of discrimination calculated from Simpson's index of diversity formula for RAPD-PCR was 0.94.
Table I.
RAPD, ERIC-PCR and composite profiles of 58 seafood associated Salmonella isolates
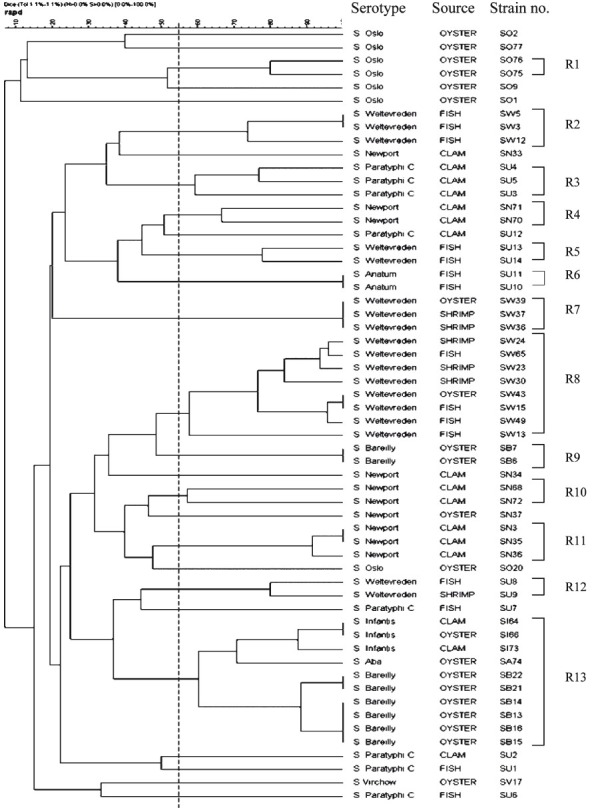
Analysis of serovars by ERIC-PCR: ERIC-PCR fingerprints generated for the 58 isolates comprised 5-15 bands ranging from 0.15 to 4.5 kb (Fig. 3) with a discriminatory index of 0.96. A common band was found in all 58 isolates at 1.5 kb. All the serovars were grouped into 17 clusters (E1-E17) at an average similarity of 51 per cent (Fig. 4). Unlike RAPD where 14 isolates failed to cluster, ERIC analysis resulted in only six which could not be grouped. These included two isolates of S. Paratyphi C and one isolate each of S. Newport, S. Weltevreden, S. Infantis and S. Virchow. It was also observed that except for a few S. Weltevreden isolates which grouped with clusters of other serotypes (S. Oslo in E10, S. Bareilly in E6, and with S. Aba in E1) almost all the isolates within a cluster belonged to the same serotype. Genetic heterogeneity was also observed among the various serovars. Major cluster differentiation was observed for S. Weltevreden which grouped into six clusters designated E1, E2, E12, E14, E15 and E16. Similarly three clusters were observed for S. Bareilly (E5, E6 and E9), two each for S. Paratyphi C (E13 and E17), S. Newport (E7 and E11) and S. Oslo (E8 and E10). Sixteen isolates obtained from clam included nine S. Newport that grouped in two clusters (E7 and E11) and only two of the four S. Paratyphi C grouping as a single cluster (E17). Sixteen fish isolates included 10 S. Weltevreden that grouped in three clusters (E1, E12 and E14), two S. Anatum present in a single cluster and two of the four S. Paratyphi C isolates grouped in one cluster (Table).
Fig. 3.
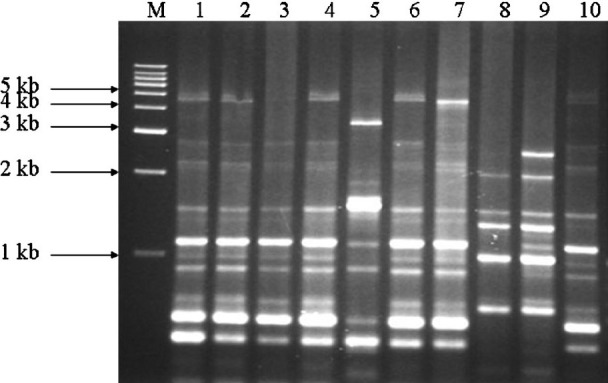
Representative ERIC-PCR fingerprint of different Salmonella isolates on 1 per cent agarose gel. M, 1kb DNA marker; lanes 1-10, SW5, SW13, SW39, SW49, SN34, SW23, SW65, SB6, SO2 and SU1.
Fig. 4.
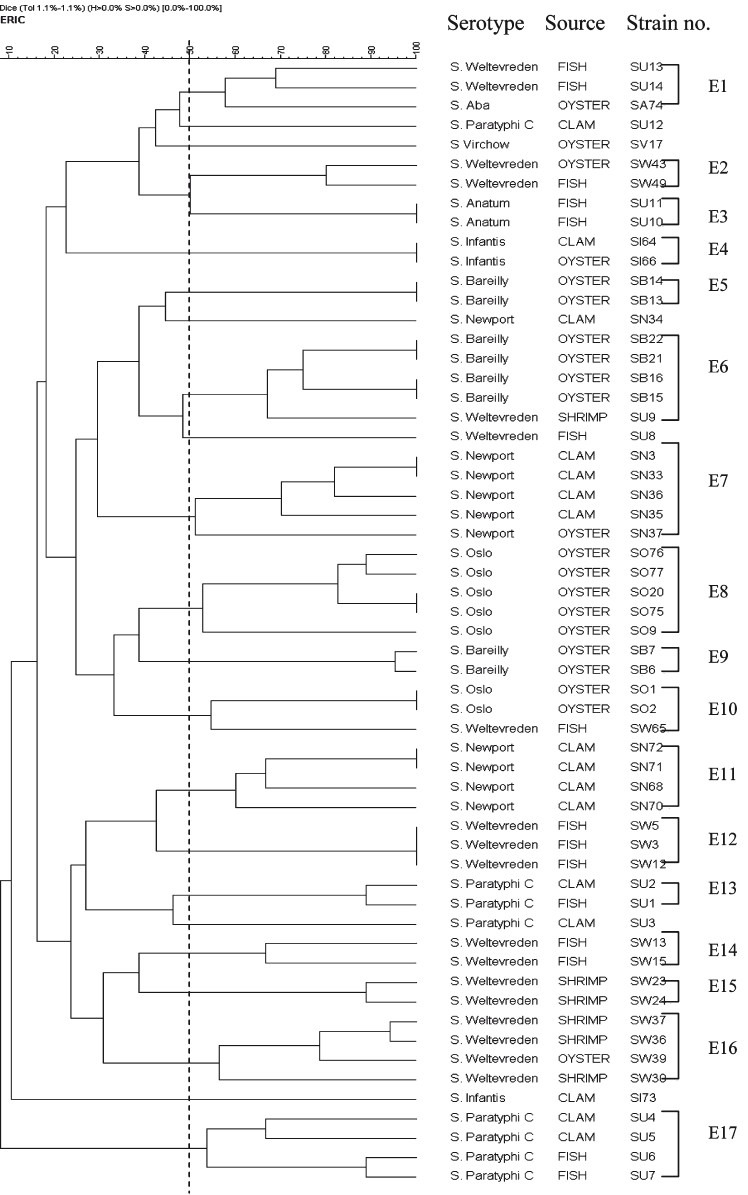
Dendogram showing the percentages of similarity between typable seafood associated Salmonellagenerated from enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) fingerprinting with the band matching coefficient of Dice and the UPGMA clustering method.
Composite analysis of RAPD and ERIC-PCR: Data from the two molecular typing methods were subjected to a composite analysis to determine whether a better clustering of the serovars could be obtained. Clustering based on fragment profiles grouped the serovars into 17 clusters (C1-C17) at an average similarity of 51 per cent (Fig. 5). The 18 isolates of S. Weltevreden were grouped into five clusters (C5, C9, C12, C14, and C15). Majority of the isolates (8 of 18) belonged to the C5 cluster. S. Newport also grouped into four clusters (C2, C4, C10 and C11). Isolate SN34 was observed to be distinct from all others, S. Bareilly was differentiated into two clusters (C6 and C7) and S. Paratyphi C into three clusters (C3, C16 and C17). Two S. Paratyphi isolates (SU12 and SU3) were distinct. Four of seven of S. Oslo isolates were distinct from each other while three isolates (SO76, SO75 and SO9) clustered together. Eight S. Bareilly isolates were grouped in two clusters. Of 16 clam isolates nine were S. Newport and they grouped in three clusters. Two S. Infantis grouped in a single cluster and only two of the four S. Paratyphi C isolates grouped together as a cluster. The Fish isolates (16) included 10 S. Weltevreden that grouped in three clusters, two S. Anatum in a single cluster and two of four S. Paratyphi C as one cluster (Table). The discriminatory index (DI) calculated using composite analysis was 0.95.
Fig. 5.
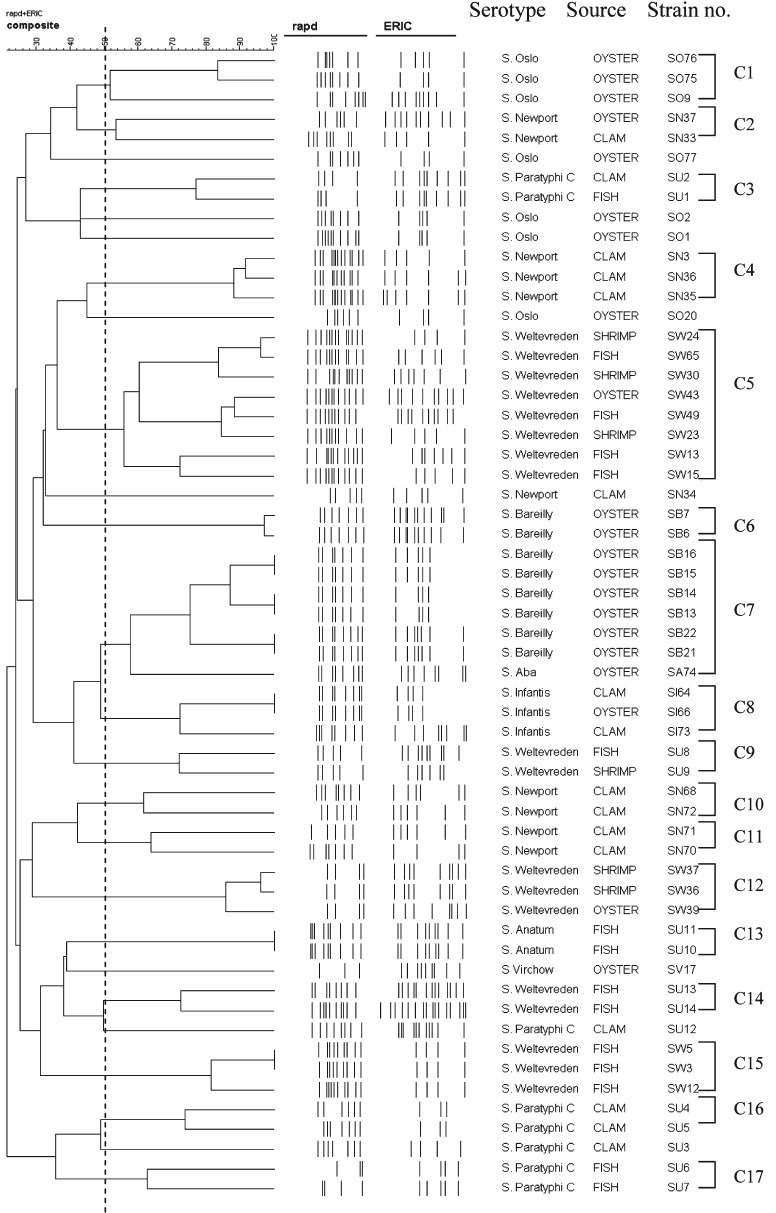
Dendogram showing the percentages of similarity between typable seafood associated Salmonella generated from composite fingerprinting with the band matching coefficient of Dice and the UPGMA clustering method.
Whole cell protein analysis: The whole cell protein profile revealed much commonality among the 58 Salmonella isolates (Fig. 6). The results showed common major protein bands of 38 kDa and 46 kDa among all the isolates of different serovars except two S. Newport isolates (SN37 and SN68) which showed 36 kDa (SN37) and 40 kDa (SN68) protein. There was a high degree of variation in the region between 29-70 kDa. A total of 30-36 minor bands ranging from 10 kDa to 100 kDa were common to all isolates. Additional protein bands were found in some of the isolates which included 42 kDa band in two S. Weltevreden isolates, one from shrimp (SW37) and another one from oyster (SW39). A heavy protein band of 68 kDa was found in S. Infantis (SI73) isolated from clam.
Fig. 6.
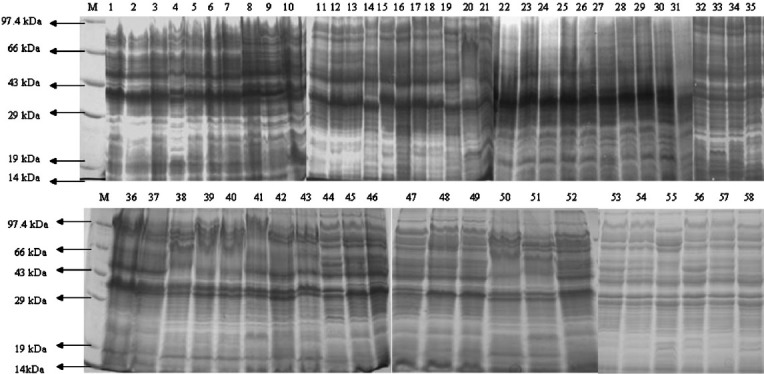
SDS-PAGE analysis of different Salmonella isolates. M, PMW-M protein marker (Bangalore genei, Bangalore). Lane 1, SW3; lane 2, SW5; lane 3, SW12; lane 4, SW13; lane 5, SW15; lane 6, SW24; lane 7, SW36; lane 8, SW37; lane 9, SW39; lane 10, SW43; lane 11, SW30; lane 12, SN35; lane 13, SN36; lane 14, SN37; lane 15, SN68; lane 16, SN70; lane 17, SN71; lane 18, SN72; lane 19, SB6; lane 20, SB7; lane 21, SB13; lane 22, SW65; lane 23, SO1; lane 24, SO2; lane 25, SO9; lane 26, SO20; lane 27, SO75; lane 28, SO76; lane 29, SO77; lane 30, SN3; lane 31, SN33; lane 32, SB2; lane 33, SB22; lane 34, SI64; lane 35, SO22; lane 36, SU1; lane 37, SU2; lane 38, SU3; lane 39, SU4; lane 40, SU5; lane 41, SU6; lane 42, SU7; lane 43, SU8; lane 44, SU9; lane 45, SU10; lane 46, SU1; lane 47, SU14; lane 48, SU13; lane 49, SU12; lane 50, SB14; lane 51, SB15; lane 52, SB16; lane 53, SW23; lane 54, SA74; lane 55, SI73; lane 56, SW49; lane 57, SN34; lane 58, SV17.
Discussion
In the present study, 58 Salmonella isolates obtained from seafood were investigated using phenotypic methods (serotyping) and molecular fingerprinting methods, RAPD and ERIC-PCR along with whole cell protein profiling (SDS-PAGE). Serotyping is the most widely used phenotyping method for epidemiological investigation of Salmonella7. Serotyping results showed that 36 per cent of Salmonella isolates belonged to the serotype S. Weltevreden. S. Weltevreden is being increasingly recorded as the most common nontyphoidal serotype in seafood throughout the world6,23. This serovar has also been reported to be commonly associated with human infections from Malaysia and Thailand24,25. In India, an outbreak of foodborne illness due to S. Weltevreden was recorded as early as in 198526. Since then, except for a few3,27 there are no reports implicating a particular serotype with human infections. Recently, an outbreak of gastroenteritidis among 34 female nursing students due to S. Weltevreden has been reported in Mangalore, India28. S. enterica Paratyphi C was also isolated from a few (14%) of the seafood samples tested and this serovar is known to be associated with enteric fever in humans.
Serotyping lacks the ability to differentiate isolates from the same serotype. PFGE is a well established method for fingerprinting Salmonella spp. with a high discriminatory index. However, the lack of equipment and facility can hamper the use of this useful DNA based technique. RAPD and ERIC-PCR on the other hand are relatively simple typing methods. In our study combination of the two methods presented a high discriminatory index and generated larger number of DNA fingerprints as reported earlier10,11. Our results elucidated the genetic difference within Salmonella serovars associated with seafood. The DI value obtained by RAPD and ERIC-PCR was 0.94 and 0.96, respectively (>0.90) which is the acceptable confidence value for interpreting the level of discrimination8. RAPD with 13 and ERIC with 17 different patterns could differentiate the isolates indicating the presence of diverse Salmonella serotypes in seafood in this region. The different clusters generated by RAPD for various Salmonella serotypes indicated genetic heterogeneity. Our result was in agreement with that of others workers29 who reported that RAPD is one of the most reliable techniques for discriminating different serotypes of Salmonella. An earlier investigation9 compared four molecular typing methods for differentiation of Salmonella spp. and observed ERIC-PCR to be most efficient. Outbreaks and sporadic cases of S. Panama were fingerprinted by ERIC-PCR for epidemiological analysis30. We combined the results of RAPD and ERIC and expressed the diversity as a composite analysis. Results of the combined analysis were highly discriminatory (DI=0.95) and thus considered more efficient. Eriksson et al31 highlighted that S. Mbandaka and S. Livingstone could be well typed using a combination of different typing methods. S. Weltevreden from tropical seafood was typed by ERIC-PCR and RAPD methods6.
Along with RAPD and ERIC-PCR, whole cell protein profiles of 58 Salmonella isolates on SDS-PAGE were also carried out. Although SDS-PAGE profiles of porins, OmpF, OmpC and OmpD of S. Typhimurium have been reported38, there is not much information on whole cell protein profiles of nontyphoidal Salmonella serovars. Begum et al12 found a common 37.81 kDa protein band common to 54 different serovars of Salmonella studied. Our study also showed major bands of ~ 38 kDa and ~ 46 kDa among all the isolates. In addition, all isolated showed several bands in the range ~ 29-70 kDa with a high degree of variation. These variations could be useful for typing of Salmonella. Results of protein profiling suggest that these variations do not allow well defined clustering but could provide information on presence of new proteins which may play a role in pathogenesis, virulence associated mechanisms, biofilm formation and other traits.
Nontyphoidal salmonellae have become a concern in seafood and have been reported from several counties in Asia and South-East Asia. Molecular characterization of these serovars would be useful for tracking the Salmonella serovars involved in outbreaks and those associated with food/water. Composite analysis using RAPD and ERIC-PCR would allow better inter- and intra-serovar strain discrimination. Although protein profiling had less discriminatory power, its use in combination with the other established typing methods may generate useful information on virulence associated traits.
Acknowledgment
This work was carried out as a part of the project funded by Indian Council of Medical Research, New Delhi.
References
- 1.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–22. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Pelzer KD. Salmonellosis. J Am Vet Med Assoc. 1989;195:456–63. [PubMed] [Google Scholar]
- 3.Aissa RB, Al-Gallas N, Troudi H, Belhadj N, Belhadj A. Trends in Salmonella enterica serotypes isolated from human, food, animal and environment in Tunisia, 1994-2004. J Infect. 2007;55:324–39. doi: 10.1016/j.jinf.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Kumar Y, Sharma A, Sehgal R, Kumar S. Distribution trends of Salmonella serovars in India (2001-2005) Trans R Soc Trop Med Hyg. 2008;103:390–4. doi: 10.1016/j.trstmh.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Urtaza J, Saco M, Hernandez-Cordova G, Lozano A, Garcia-Martin O, Espinosa J. Identification of Salmonella serovars isolated from live molluscan shellfish and their significance in the marine environment. J Food Pro. 2003;66:226–32. doi: 10.4315/0362-028x-66.2.226. [DOI] [PubMed] [Google Scholar]
- 6.Shabarinath S, Sanath KH, Khushiramani R, Karunasagar I, Karunasagar I. Detection and characterization of Salmonella associated with tropical seafood. Int J Food Microbiol. 2007;114:227–33. doi: 10.1016/j.ijfoodmicro.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Le Monor L, Rhode R. In: Bergey's manual of determinative bacteriology. 8th ed. Buchanan RE, Gibbons NE, editors. Baltimore: The Williams & Wilkins Co.; 1974. p. 299. [Google Scholar]
- 8.Kumar R, Surendran PK, Thampuran N. Molecular fingerprinting of Salmonella enterica subsp. enterica Typhimurium and Salmonella enterica subsp. enterica Derby isolated from tropical seafood in South India. Mol Biotechnol. 2008;40:95–100. doi: 10.1007/s12033-008-9067-2. [DOI] [PubMed] [Google Scholar]
- 9.Lim H, Lee KH, Hong C-H, Bahk G-J, Choi WS. Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol. 2005;105:411–8. doi: 10.1016/j.ijfoodmicro.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphism amplified arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:531–5. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Versalovic J, Koeuth T, Le Lupsbi JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begum F, Adachi Y, Khan MSR. Characterization of Salmonella serovars in comparison with some enterobacteria by SDS-PAGE analysis. Bangladesh J Vet Med. 2008;6:169–74. [Google Scholar]
- 13.Bacteriological analytical manual. 8th ed. Arlington, VA, USA: Association of Official Analytical Chemists; 2001. Food and Drug Administration. [Google Scholar]
- 14.Jones DD, Law R, Bej AK. Detection of Salmonella spp. in oysters using polymerase chain reaction (PCR) and gene probes. J Food Sci. 1993;8:1191–7. [Google Scholar]
- 15.Rahn K, De-Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probe. 1992;6:271–9. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 16.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Current protocol molecular biology. New York: John Wiley and Sons; 1995. [Google Scholar]
- 17.Tsasanakajon A, Pongsomboon S, Rimphanitchayakit V, Jarayabhand P, Boonsaeng V. Random amplified polymorphic DNA (RAPD) markers for determination of genetic variation in wild population of the black tiger prawn (Penaeus monodon) in Thailand. Mol Mar Biol Biotechnol. 1997;6:110–5. [PubMed] [Google Scholar]
- 18.Neilan BA. Identification and phylogenetic analysis of toxigenic cyanobacteria by multiplex randomly amplified polymorphic DNA PCR. Appl Environ Microbiol. 1995;61:2286–91. doi: 10.1128/aem.61.6.2286-2291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millemann Y, Lesage-Descauses M-C, Lafont J-P, Chaslus-Dancla E. Comparison of random amplified polymorphic DNA analysis and enterobacterial repetitive intergenic consensus PCR for epidemiological studies of Salmonella. FEMS Immunol Med Microbiol. 1996;14:129–34. doi: 10.1111/j.1574-695X.1996.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei M, Lei WH. Mathematical model for studying genetic variation in terms of vostriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–73. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponce E, Khan AA, Chengc M-C, Summage-Westa C, Cernigliaa CE. Prevalence and characterization of Salmonella enterica serovar Weltevreden from imported seafood. Food Microbiol. 2008;25:29–35. doi: 10.1016/j.fm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Padungtod P, Kaneene JB. Salmonella in food animals and humans in northern Thailand. Int J Food Microbiol. 2006;108:346–54. doi: 10.1016/j.ijfoodmicro.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Thong KL, Goh YL, Radu S, Noorzaleha S, Yasin R, Koh YT, et al. Genetic diversity of clinical and environmental strains of Salmonella enterica serotype Weltevreden isolated in Malaysia. J Clin Microbiol. 2002;40:2498–503. doi: 10.1128/JCM.40.7.2498-2503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal P, Singh SM, Bhattacharya MM. An outbreak of food poisoning in a family due to Salmonella Weltevreden at Delhi. J Diarrhoeal Dis Res. 1985;3:224–5. [PubMed] [Google Scholar]
- 27.Patil AB, Krishna BVS, Chandrasekhar MR. Neonatal sepsis caused by Salmonella enterica serovar Weltevreden. Southeast Asian J Trop Med Public Health. 2006;7:1175–8. [PubMed] [Google Scholar]
- 28.Antony B, Dias M, Shetty AK, Rekha B. Food poisoning due to Salmonella enterica serotype Weltevreden in Mangalore. Indian J Med Microbiol. 2009;27:257–8. doi: 10.4103/0255-0857.53211. [DOI] [PubMed] [Google Scholar]
- 29.Learn-Han L, Yoke-Kqueen C, Salleh NA, Sukardi S, Jiun-Horng S, Chai-Hoon K, et al. Analysis of Salmonella Agona and Salmonella Weltevreden in Malaysia by PCR fingerprinting and antibiotic resistance profiling. Antonie Van Leeuwenhock. 2008;94:377–87. doi: 10.1007/s10482-008-9254-y. [DOI] [PubMed] [Google Scholar]
- 30.Soto SM, Guerra B, Del Cerro A, González-Hevia MA, Mendoza MC. Outbreaks and sporadic cases of Salmonella serovar Panama studied by DNA fingerprinting and antimicrobial resistance. Int J Food Microbiol. 2001;71:35–43. doi: 10.1016/s0168-1605(01)00553-0. [DOI] [PubMed] [Google Scholar]
- 31.Eriksson J, Löftrom C, Aspán A, Gunnarsson A, Karlsson I, Borch E, et al. Comparison of genotyping methods by application to Salmonella Livingstone strains associated with an outbreak of human salmonellosis. Int J Food Microbiol. 2005;104:93–103. doi: 10.1016/j.ijfoodmicro.2005.01.011. [DOI] [PubMed] [Google Scholar]


