Abstract
Background & objectives:
This study was carried out to evaluate the association between the antibiotic susceptibility patterns and the antibiotic resistance genes in staphylococcal isolates obtained from various clinical samples of patients attending a teaching hospital in Hatay, Turkey.
Methods:
A total of 298 staphylococci clinical isolates were subjected to antimicrobial susceptibility testing. The genes implicated in resistance to oxacillin (mecA), gentamicin (aac(6’)/aph(2”), aph(3’-IIIa, ant(4’)-Ia), erythromycin (ermA, ermB, ermC, and msrA), tetracyclin (tetK, tetM), and penicillin (blaZ) were amplified using multiplex PCR method.
Results:
Methicillin resistance rate among 139 Staphlococcus aureus isolates was 16.5 and 25.9 per cent of S. aureus carried mecA gene. Of the 159 CoNS isolates, methicillin resistance rate was 18.9 and 29.6 per cent carried mecA gene. Ninety four isolates identified as gentamicin resistant phenotypically, contained at least one of the gentamicin resistance genes [aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia], 17 gentamicin-susceptible isolates were found as positive in terms of one or more resistance genes [aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia] by multiplex PCR. A total of 165 isolates were resistant to erythromycin, and contained at least one of the erythromycin resistance genes (ermA, ermB, ermC and msrA). Phenotypically, 106 staphylococcal isolates were resistant to tetracycline, 121 isolates carried either tetK or tetM or both resistance genes. The majority of staphylococci tested possessed the blaZ gene (89.9%).
Interpretation & conclusions:
The present results showed that the phenotypic antibiotic susceptibility patterns were not similar to those obtained by genotyping done by multiplex PCR. Rapid and reliable methods for antibiotic susceptibility are important to determine the appropriate therapy decisions. Multiplex PCR can be used for confirmation of the results obtained by conventional phenotypic methods, when needed.
Keywords: Antibiotic resistance genes, antibiotic susceptibility, mecA gene, staphylococci
The resistance to antimicrobial agents is an increasingly global problem worldwide, especially among nosocomial pathogens. Staphylococci have become one of the most common causes of nosocomial infections. Multidrug-resistant staphylococci pose a growing problem for human health. The rise of drug-resistant virulent strains of Staphylococcus aureus, particularly methicillin-resistant S. aureus (MRSA) is a serious problem in the treatment and control of staphylococcal infections1,2
Methicillin-resistant staphylococci (MRS) cause hard-to-treat infections because these are resistant to most of the antibiotics such as beta-lactams, aminoglycosides, and macrolides. The most important mechanism of resistance to penicillin is production of beta-lactamase which inactivates penicillin by hydrolysis of its beta-lactam ring. Another mechanism is associated with penicillin-binding protein 2a (PBP2a), encoded by mecA2. Another gene involved in penicillin resistance in staphylococci is blaZ which encodes β-lactamase2.
Aminoglycoside modifying enzymes (AMEs) are major factors responsible for resistance to aminoglycoside in staphylococci. Until now, three classes of AMEs have been identified: acetyltransferase (AAC), aminoglycoside phosphotransferase (APH), and aminoglycoside nucleotidyltransferase (ANT)3,4. The most important mechanism of aminoglycoside resistance in staphylococci is drug inactivation by AMEs like aminoglycoside nucleotidyltransferases (APHs). AMEs can be plasmid or chromosome encoded. In staphylococcal strains, the most commonly found AME is aac(6’)/aph(2”). The bifunctional enzyme aac(6’)/aph(2”) is encoded by the aac(6’)/aph(2”) gene. In addition, APH(3’)-III is encoded by aph(3’)- IIIa gene and the ANT(4’)-I by ant(4’)-Ia gene, are also found in staphylococcal isolates5–8
The accurate and rapid diagnosis of antibiotic resistance genes in the treatment of staphylococcal infections is extremely important in preventing the spread of infections. PCR-based molecular methods are often preferred for determination of antibiotic resistance genes9. The present study was aimed to investigate the relation between the antibiotic susceptibility patterns and the antibiotic resistance genes (mecA, aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia, erm(A), erm(B), erm(C), tet(K), tet(M), msrA, blaZ) by multiplex PCR assay in staphylococci isolates obtained from clinical specimens of patients at a teaching hospital in the South of Turkey.
Material & Methods
The study was carried out in Mustafa Kemal University, Faculty of Medicine, Department of Medical Microbiology (Hatay, Turkey) between July 2007 and August 2009. A total of 298 clinical isolates of staphylococcai were isolated from blood (29, 9.7%), wounds (145, 48.7%), urine (42, 14.1%), pus (59, 19.8%), and other sources samples (23, 7.7%). Of the 298 staphylococcal isolates, 159 (53.4%) were coagulase-negative staphylococci (CONS) and 139 (46.6%) isolates were identified as the S. aureus.
The samples were placed into modified amines charcoal transport medium (Difco Laboratories, USA), and were sent immediately to the microbiology laboratory. The samples were inoculated onto 5 per cent sheep blood agar plates (Difco Laboratories, USA), and incubated at 37°C for 48 h. The identification of staphylococci was on the basis of colony morphology, Gram staining, biochemical tests such as catalase and coagulase tests10. Isolates were stored at -70°C in Mueller-Hinton Broth (Merck, Germany) supplemented with 40 per cent glycerol (v/v).
Susceptibility testing
Disc diffusion - Antimicrobial susceptibilities of the isolates were tested by the agar disk diffusion method on Mueller-Hinton agar (Tiantan Biotechnology, PR China) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines11. Antibiotic discs (Becton Dickinson, USA) were placed on Mueller-Hinton agar plates, incubated at 37°C for 24 h, and the diameter of each zone was measured in millimeters. The following antibiotic discs were used: penicillin (10 U), erythromycin (15 μg), oxacillin (1 μg), tetracycline (30 μg), gentamicin (10 μg), amoxicillin-clavulanic acid (30 μg), clindamycin (2 μg), vancomycin (30 μg), trimethoprim-sulphamethoxazole (25 μg), and ciprofloxacin (5 μg).
Oxacillin disc diffusion test - Oxacillin disc susceptibility testing was performed on all isolates of S. aureus according to CLSI recommendations using a 1 μg oxacillin disc. Oxacillin disc (Becton Dickinson Microbiology Systems, USA) were placed on Mueller-Hinton agar plates without NaCl supplementation. Subsequently, the plates were incubated 24 h at 37°C. The zone size was interpreted according to the CLSI criteria11. S. aureus ATCC 29213 and S. aureus ATCC (43300) were chosen as the negative and positive control strains, respectively.
Determination of minimum inhibitory concentrations (MIC): MIC values of antibiotics were determined by the broth microdilution test. All isolates were subcultured on blood agar and incubated for 24 h at 37 °C. Then, two-fold serial dilutions of each antibiotic were made in Mueller-Hinton broth to achieve a concentration range from 0 to 256 μg/ml. After incubation at 37° for 24 h, the MIC was defined as the lowest concentration of antibiotics that produced no growth. The broth microdilution tests were performed according to the CLSI guidelines11
DNA isolation: For nucleic acid isolation from staphylococcal isolates, the frozen samples were thawed rapidly, and were cultivated in brain-heart infusion broth (Merck, Germany) at 37°C with shaking overnight. Total DNA was isolated from 5 ml of a broth culture grown overnight12. After incubation, bacterial cells were harvested by centrifugation at 3000 × g for 10 min, the cell pellet was re-suspended in phosphate-buffered saline with 100 μg of lysostaphin (Sigma, USA) per ml, and incubated at 37°C for 30 min. The phenol/chloroform extraction method was used for nucleic acid extraction and DNA was precipitated in 1 ml 70 per cent ethanol. The DNA precipitate was dissolved in 50 μl of TE buffer [10 mM Tris chloride-1 mM EDTA (pH 8.0)], and stored at -20°C until processing.
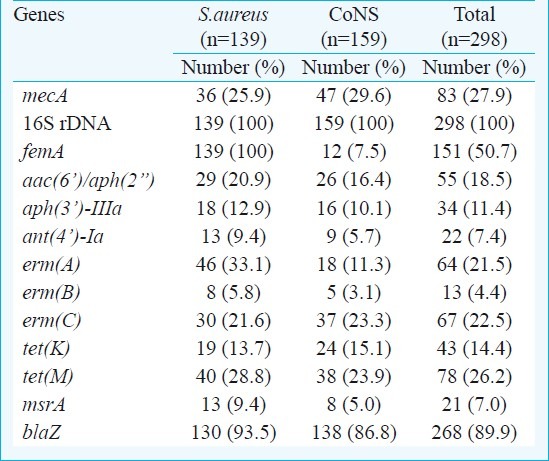
The oligonucleotide primers3 for the mecA, femA, 16S rDNA, aac(6’)/aph(2”), aph(3’)-IIIa and ant(4’)-Ia genes were selected and the primers for erm(A), erm(B), erm(C), tet(K), tet(M), clf A, msrA and blaZ, genes were selected based on earlier studies13,14 (Table I).
Table I.
The primer sequences and predicted sizes used in the multiplex PCRs

PCR method for aac(6’)/aph(2”), aph(3’)-IIIa and ant(4’)-Ia: The PCR amplification was performed in a 25 μl reaction mixture [2.5 ml of 10x reaction buffer without MgCl2 (Promega Corp., USA); 200 μM of each deoxynucleoside triphospate (AB Gene, UK), 2 mM MgCl2; 2.5 pmol of each primer (aac(6’)/aph(2”), aph(3’)-IIIa and ant(4’)-Ia] and approximately 10 ng of template DNA, and brought up to a 25 μl final volume with distilled water. In order to reduce the formation of nonspecific extension products, a “hot-start” protocol was used12. PCR reactions were hot started for 5 min at 95°C and placed on ice, and 1 U of Taq polymerase (Fermentas, USA) was added. Reaction mixtures were subjected to 30 PCR cycles (95°C for 2 min, 1 min at 54°C and 1 min at 72 °C). A final elongation step at 72°C for 7 min was applied in a thermal cycler (Bioder/Thermal Blocks xp Cycler, Tokyo, Japan).
PCR method for 16S rDNA, mecA and femA: Multiplex PCR conditions for 16S rDNA, mecA and femA included the same constituents as the procedure of aminoglycoside resistance genes except for the MgCl2 concentration (3.0 mM) and the primers, which were used at 20 pmol of primers for 16S rDNA, mecA and femA.
DNA amplification was carried out in a thermal cycler and reactions were hot started for 5 min at 94°C and placed on ice, and 1 U of Taq polymerase (Fermentas, USA) was added. Reaction mixtures were subjected to 35 PCR cycles (94°C for 2 min, 2 min at 55°C, and 1 min at 72°C). A final elongation step at 72°C for 7 min was also applied.
PCR method for erm(A), erm(B), erm(C), tet(K), tet(M), msrA, blaZ: The PCR amplification was carried out in a total volume of 25 μl. PCR amplification was achieved as follows: 5 μl of genomic DNA (approximately 50 ng) sample was added to 20 μl of PCR mixture (20 mmol/l Tris-HCl, pH 8.4; 50 mmol/l KCl, 10 mmol/l MgCl2, and 200 μmol/l each of deoxynucleoside triphosphates (dNTPs), 0.6 μmol/l each primers and 1 U Taq DNA polymerase. The amplification process was started with an initial denaturation step (95 °C, 3 min). Each cycle consisted of three steps (denaturation, annealing, and extension). Each PCR reaction consisted of 30 cycles of amplification (denaturation at 95°C for 30 sec, annealing at 54 °C for 30 sec, and DNA chain extension at 72°C for 30 sec). A final extension cycle was performed at 72°C for 4 min. The resistance genes were studied in five different groups: group 1 (mecA and femA); group 2 [(aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia)]; group 3 (16S rDNA, ermA, ermC); group 4 (msrA, 16S rDNA, ermC); and group 5 (erm B).
After amplification of the resistance genes, 10 μl of the PCR products were mixed with 3 μl of loading buffer (10 %, w/v, Ficoll 400; 10 mmol/l Tris-HCl, pH 7.5; 50 mmol/l EDTA; 0.25% bromophenol blue) and then loaded onto a 2 per cent agarose gel and electrophoresis was performed in Tris-borate-EDTA buffer containing 0.5 g of ethidium bromide per ml. The PCR products were analyzed in a 2 per cent (w/v) agarose gel in 1 × TAE buffer (40 mmol/l Tris-acetate, 1 mmol/l EDTA). Ethidium bromide (0.5 μg/ml TAE)-stained DNA amplicons were visualized using a gel imaging system (Wealtec, Dolphin-View, USA).
Statistical analysis: All data were evaluated by χ2 test using Statistical Package for Social Sciences (SPSS1 for Windows V. 11.5, Chicago, USA) software.
Results
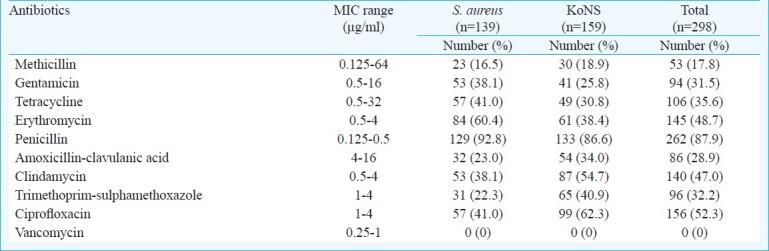
A total of 16.5 per cent (23/139) of the coagulase positive staphylococcal isolates showed resistance to methicillin and 18.9 per cent (30/159) coagulase negative isolates showed methicillin resistance, phenotypically. The highest drug resistance was obtained against penicillin in both S. aureus 92.8 per cent and the CoNS 86.6 per cent. Overall, antibiotics resistance rates for gentamicin, tetracycline and erythromycin were 31.5, 35.6, 48.7 and 87.9 per cent, respectively (Table II). The staphylococcal isolates showed moderate resistance to amoxicillin-clavulanic acid (28.9%), and trimethoprim-sulphamethoxazole (32.2%). Both CoNS and S.aureus strains were vancomycin susceptible Table II.
Table II.
The MIC values and the resistance patterns of staphylococci by the disc diffusion method
The susceptibility results determined by conventional methods were compared with the results of the multiplex PCR assay (Table III). All S. aureus isolates (positive for coagulase test) carried the femA gene. Methicillin resistance rate among 139 S. aureus isolates was 16.5 and 25.9 per cent (36/139) of S. aureus isolates carried mecA gene. Methicillin resistance rate in CoNS was 18.9 and 29.6 per cent of CoNS carried mecA gene. Thirty isolates which were phenotypically methicillin-susceptible, were found to carry the mecA gene. While a total of 139 isolates were identified as S. aureus by phenotypic methods, the presence of the femA gene was determined in 151 (Table III).
Table III.
The rates of antibiotic resistance genes in staphylococci by multiplex PCR method
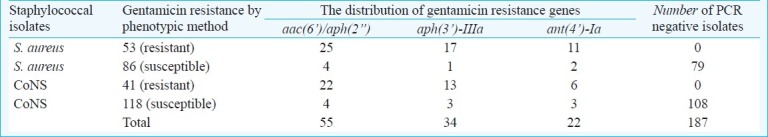
Among AME genes, aac(6’)/aph(2”) was detected in 18.5 per cent, aph(3’)-IIIa and ant(4’)-Ia were found in 11.4 and 7.4 per cent of the isolates, respectively. All 94 isolates identified as gentamicin resistant phenotypically contained at least one of the gentamicin resistance genes [aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia] (Fig. 1). A total of 17 gentamicin-susceptible isolates were found positive in terms of one or more resistance genes [aac(6’)/aph(2”), aph(3”)-IIIa, ant(4’)-Ia] by multiplex PCR (Table IV).
Fig. 1.
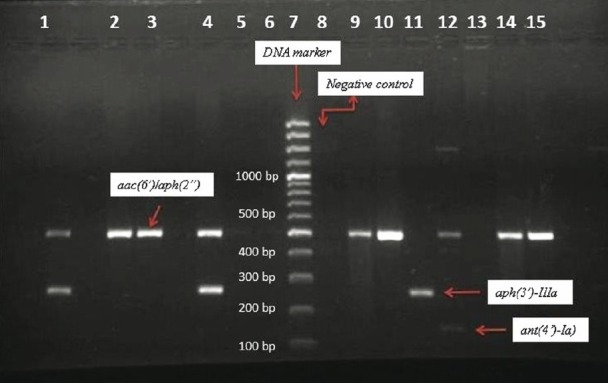
Ethidium bromide-stained multiplex PCR products after gel electrophoresis for the aac(6’)/aph(2”) (491 bp), aph(3’)-IIIa (242 bp) and ant(4’)-Ia (135 bp) genes. DNA molecular size marker (100 bp). Lanes: 1-4, 9, 10, 12, 14 and 15; aac(6’)/aph(2”); Lane 7: DNA marker (molecular size standard); Lane 8: Negative control; Lane 1 and 4: aac(6’)/aph(2”) and aph(3’)-IIIa; Lane 11: aph(3’)-IIIa; Lane 12: aac(6’)/aph(2”) and ant(4’)-Ia.
Table IV.
Relationship between gentamicin resistance and the presence of three resistance genes (aac(6’)/aph(2”), aph(3’)-IIIa, ant(4’)-Ia)
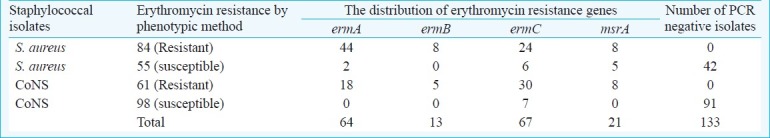
A total of 145 isolates were resistant to erythromycin, and contained at least one of the erythromycin resistance genes (ermA, ermB, ermC and msrA). The ermA and ermC genes were presenting 77; ermB could be detected in 13 isolates (Fig. 2). Eleven isolates carried both ermA and ermB. A total of 21 isolates had msrA gene (Table V).
Fig. 2.
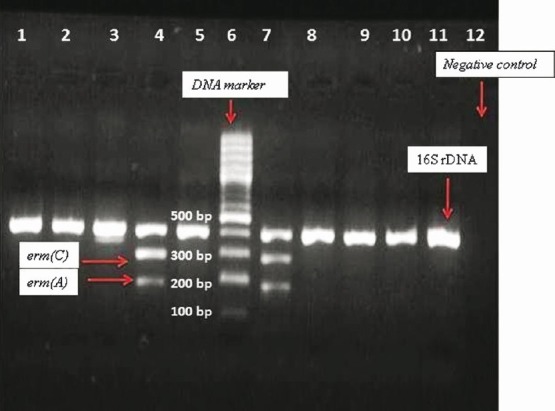
Ethidium bromide-stained multiplex PCR products after gel electrophoresis for the 16S rDNA (420 bp), erm(A) (190 bp) and erm(C) (299 bp) genes. DNA molecular size marker (100 bp). Lanes: 1-5, and 7-11; 16S rDNA; Lane 4 and 7: 16S rDNA, erm(A) and erm(C); Lane 6: DNA marker (molecular size standard); Lane 12: Negative control.
Table V.
Relationship between erythromycin resistance and the presence of four resistance genes (ermA, ermB, ermC, or msrA)
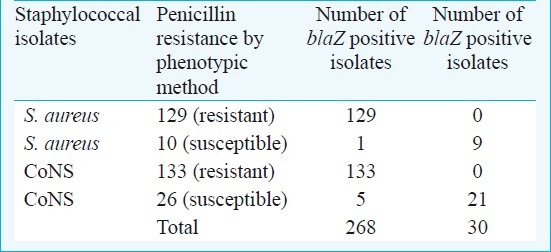
A total of 106 staphylococcal isolates were found resistant to tetracycline, phenotypically, 121 isolates were resistant to tetracycline and carried either tetK or tetM or both resistance genes (Table VI). With regards to blaZ gene, the majority of tested staphylococci possessed the blaZ gene. The 130 (93.5%) S. aureus isolates carrying the blaZ gene expressed the phenotype, and the presence of the blaZ gene was found as 86.8 per cent (138/159) among the coagulase negative staphylococci (Table VII).
Table VI.
Relationship between tetracycline resistance and the presence of the tetK and tetM genes
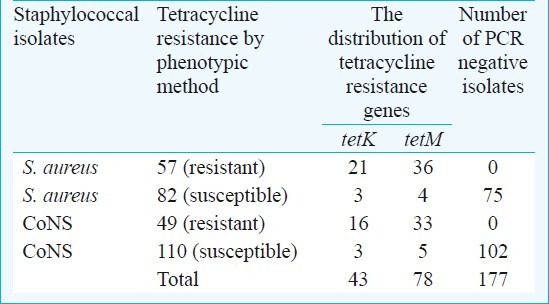
Table VII.
Relationship between penicillin resistance and the presence of the blaZ gene
Discussion
In the present study, the results of antibiotic susceptibility by disk diffusion method were compared with gene analysis results in staphylococcal isolates. The phenotypic expression of antimicrobial resistance has been reported to be influenced by various factors15. The antimicrobial susceptibility test was performed for all isolates of S. aureus and CoNS by disc diffusion and broth microdilution test. Antibiotic susceptibility test results showed a high level of resistance among the staphylococal isolates to most of the commonly used antimicrobials. In our study, all staphylococcal isolates were found as positive by PCR for 16S rDNA gene and, all S. aureus isolates were positive to the species specific femA gene.
Methicillin resistance in staphylococci has been reported to be associated with the presence of penicillin-binding proteins, PBP2’ (PBP2a), encoded by the mecA gene. The mecA gene may be heterogeneously expressed and, therefore, all methicillin-resistant staphylococcal strains may not be detectable with phenotypical methods2–5. However, the detection of mecA gene by PCR techniques is considered the gold standard method. Also, phenotypic methods require at least 24 h for evaluation of the results2,3
Methicillin resistance was observed in 17.8 per cent isolates when tested by methicillin disk diffusion method, whereas 27.9 per cent isolates had mecA gene. Phenotypically methicillin susceptible 30 isolates also carried the mecA gene. Comparison of conventional method and multiplex PCR assay did not show a good agreement. Results about resistance to methicillin found in this study were lower than those reported from other parts of Turkey16,17 , but were in accordance with those in similar studies from Europe (25%) but were lower than those in USA18,19
Our results showed that the blaZ gene was widely spread among both S. aureus and CoNS. In various studies from our country, penicillin resistance has been reported between 61 and 100 per cent20,21. The resistance rate in various countries of the world ranged from 50 to >91 per cent22,23.
The staphylococcal isolates were analyzed for the presence of the aminoglycoside-modifying enzyme genes aac(6’)/aph(2”), aph(3’)-IIIa, and ant(4’)-Ia by multiplex PCR. The presence of at least one of the AmE genes was seen in 111 of 298 isolates. A total 62.8 per cent isolates negative for AME genes were resistant to gentamicin. In accordance with previous studies3,5, the aac(6’)/aph(2”) gene was the most prevalent AME gene in staphylococci. In our study, aac(6’)-aph(2”) gene was the most frequently found gene among MRSA isolated. This result was similar to the studies carried out in Europe4. Ida et al6 from Japan reported that aac(6‘)/aph(2”) gene in MRSA strains was encountered less frequently among clinical isolates6.
Erytromycin resistance in staphylococci is encoded by erm genes24. All isolates found to be resistant to erythromycin by phenotypic methods contained at least 1 erythromycin resistance gene. While resistance to erythromycin in staphylococci was found as 48.7 per cent by phenotypic methods, this ratio was 55.4 per cent by multiplex PCR assay. erm A, erm B and msrA genes in S. aureus isolates were found at higher rates than in CoNS, and among the CoNS, ermC gene was the more prevalent. In 34 isolates, neither ermA nor ermC gene was detected, although these were detected as resistant. Erytromycin resistance might be due to the presence of msrA or ermB gene as previously described in CoNS24.
Lim et al25 reported that the ermA gene was more prevalent than the other erytromycin resistance genes in S. aureus isolates, and ermC gene was found mostly in CoNS. Similarly, in a study performed by Martineau et al14, the ermC gene has been reported to be more prevalent in CoNS. However, ermA has been reported to be the more common gene in CoNS in another study26. Our results were consistent with that of Lim et al25 and Martineau et al14, but were contradictory to that of Thakker-varia et al26.
In the present study, the phenotypic resistance to tetracycline in S. aureus and CoNS was observed as 41 and 30.8 per cent, respectively. Whereas 42.4 per cent S. aureus isolates carried the tetK and tetM genes, 39 per cent of CoNS were found to carry these genes. Tetracycline resistance genes tetK and tetM were found positive by PCR method in 15 isolates which were phenotypically sensitive to tetracycline.
Identification and determination of the susceptibility to antibiotics of staphylococci by conventional methods requires a minimum of two-days period, whereas the detection of antibiotic resistance genes by PCR assay can be done within a few hours. Rapid and reliable methods for antibiotic susceptibility are important to institute appropriate therapy. Multiplex PCR can be used for confirmation of the results obtained by conventional phenotypic methods when needed.
Acknowledgment
This work was supported by the Research Fund of Mustafa Kemal University.
References
- 1.Livermore DM. Antibiotic resistance in staphylococci. Int J Antimicrob Agents. 2000;16:S3–S10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- 2.Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32:361–85. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 3.Choi SM, Kim SH, Kim HJ, Lee DG, Choi JH, Yoo JH, et al. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J Korean Med Sci. 2003;18:631–6. doi: 10.3346/jkms.2003.18.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanhoof R, Godard C, Content J, Nyssen HJ, Hannecart-Pokorni E Belgian Study Group of Hospital Infections (GDEPIH/GOSPIZ) Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J Med Microbiol. 1994;41:282–90. doi: 10.1099/00222615-41-4-282. [DOI] [PubMed] [Google Scholar]
- 5.Yadegar A, Sattari M, Mozafari NA, Goudarzi GR. Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb Drug Resist. 2009;15:109–13. doi: 10.1089/mdr.2009.0897. [DOI] [PubMed] [Google Scholar]
- 6.Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbiol. 2001;39:3115–21. doi: 10.1128/JCM.39.9.3115-3121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HB, Kim T, Lee BB, Kim US, Park SW, Shin JW, et al. Frequency of resistance to aminoglycoside antibiotics in Staphylococcus aureus isolates from tertiary hospitals. Korean J Infect Dis. 2002;34:39–46. [Google Scholar]
- 8.Sekiguchi J, Fujino T, Saruta K, Konosaki H, Nishimura H, Kawana A, et al. Prevalence of erythromycin-, tetracycline-, and aminoglycoside- resistance genes in methicillin-resistant Staphylococcus aureus in hospitals in Tokyo and Kumamoto. Jpn J Infect Dis. 2004;57:74–7. [PubMed] [Google Scholar]
- 9.Woodford N, Sundsfjord A. Molecular detection of antibiotic resistance: when and where? J Antimicrob Chemother. 2005;56:259–61. doi: 10.1093/jac/dki195. [DOI] [PubMed] [Google Scholar]
- 10.Holt JG, Krieg NR, Sneath PHA, Williams ST. Staphylococcus spp. In: Bergey's manual of determinative bacteriology. 9th ed. Baltimore, MD: Williams & Wilkins; 1994. pp. 544–51. [Google Scholar]
- 11.Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute; 2007. Clinical and Laboratory Standards Institute. [Google Scholar]
- 12.Johnson WM, Tyler SD, Ewan EP, Ashton FE, Pollard DR, Rozee KR. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in S. aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–30. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–94. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martineau F, Picard FJ, Lansac N, Menard C, Roy PH, Ouellette M, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic suseptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baddour MM, AbuElKheir MM, Fatani AJ. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2007;55:473–9. doi: 10.1007/s00284-007-9015-6. [DOI] [PubMed] [Google Scholar]
- 16.Kurutepe S, Surucuoglu S, Gazi H, Teker A, Ozbakkaloglu B. Antibiotic resistance rates of methicillin resistant and susceptible Staphylococcus aureus strains. Turk J Infect. 2007;21:187–91. [Google Scholar]
- 17.Dogan O, Cirak MY, Engin D, Turet S. Methicillin resistance and in-vitro susceptibility to various antibiotics of staphylococci isolated from clinical samples. J Ankem. 2005;19:39–42. [Google Scholar]
- 18.Marshall SA, Wilke WW, Pfaller MA, Jones RN. Staphylococcus aureus and coagulase negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn Microbiol Infect Dis. 1998;30:205–14. doi: 10.1016/s0732-8893(97)00212-5. [DOI] [PubMed] [Google Scholar]
- 19.Fluit AC, Weilders CLC, Verhoef J, Schmitz FJ. Epidemiology and susceptibility of 3051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY Study. Clin Microbiol. 2001;39:3727–32. doi: 10.1128/JCM.39.10.3727-3732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirmatel F, Zeyrek F, Erkmen O. Antibiotic resistance in nosocomial Staphylococcus strains with broth microdilution method. J Ankem. 2004;18:200–4. [Google Scholar]
- 21.Savas L, Duran N, Onlen Y, Savas N, Erayman M. Prospective analysis of antibiotic susceptibility patterns of MRSA in a Turkish university hospital. Turk J Med Sci. 2005;35:323–7. [Google Scholar]
- 22.Kantzanou M, Tassios PT, Tseleni-Kotsovili A, Maniatis AN, Vatopoulos AC, Legakis NJ Greek MRSA Study Group. A multi-centre study of nosocomial methicillin-resistant Staphylococcus aureus in Greece. Int J Antimicrob Agents. 1999;12:115–9. doi: 10.1016/s0924-8579(99)00005-9. [DOI] [PubMed] [Google Scholar]
- 23.Zmantar T, Chaieb K, Ben Abdallah F, Ben Kahla-Nakbi A, Ben Hassen A, Mahdouani K, et al. Multiplex PCR detection of the antibiotic resistance genes in Staphylococcus aureus strains isolated from auricular infections. Folia Microbiol. 2008;53:357–62. doi: 10.1007/s12223-008-0055-5. [DOI] [PubMed] [Google Scholar]
- 24.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–85. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim JA, Kwon AR, Kim SK, Chong Y, Lee K, Choi EC. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in Gram-positive cocci isolated in a Korean hospital. J Antimicrob Chemother. 2002;49:489–95. doi: 10.1093/jac/49.3.489. [DOI] [PubMed] [Google Scholar]
- 26.Thakker-Varia S, Jenssen WD, Moon-McDermott L, Weinstein MP, Dubin DT. Molecular epidemiology of macrolides-lincosamides-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:735–43. doi: 10.1128/aac.31.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]


