Genetics allows the elucidation of gene function through the analysis of gene malfunction. Modern genetics and genomics require ways for in situ modification of genes, by means of point mutations, deletions, and additions. The availability of sequence information of many organisms dictates rapid development of reverse genetics procedures. Until recently, targeting of genes with the help of introduced homologous sequences, here referred to as homologous recombination-dependent gene targeting (hrdGT), was the method of choice, at least for mammalian systems. However, in higher eukaryotic organisms such as mammals and plants, exogenously introduced DNA preferably integrates in random positions in the genome, by the process of illegitimate recombination, and only infrequently can targeted integration events be detected. Recently an alternative strategy became available for precise reverse genetics. Specific chimeric oligonucleotides, COs, consisting of DNA and RNA stretches, were found to induce point mutations in several mammalian genes tested (see below). This technique, here referred to as chimeric oligonucleotide-dependent mismatch repair, cdMMR, has now been used for plants: this issue of the Proceedings includes two reports describing stable changes in the genomes of tobacco and maize after treatment with chimeric oligonucleotides (1, 2).
hrdGT
In the mouse system hrdGT is (almost) routine. Since early pioneering work (3, 4), gene targeting in embryonic stem cells led to the generation of several thousands of “knock-out” mice. Gene targeting frequencies of 10−2 greatly facilitated the work (5) and led to the establishment of extremely interesting loss-of-function phenotypes and animal models for human diseases. In contrast, despite an urgent need for academia and agriculture, hrdGT in plants suffers from a deplorable inefficiency, which has not significantly changed since it was first accomplished (6). Homologous recombination in plants was studied in classical work as well as by using molecular markers (reviewed in refs. 7 and 8). Strategies to improve its frequency by extending the length of homology or applying negative selection to enrich for targeted events had little effect (9, 10). Although the highest reported frequencies range in the order of one in 750 (11) and one in 2,580 events (12), most experiments yielded 10−4, 10−5, or fewer targeted transformants (9, 10). The reasons for these differences are not obvious, but the targeting device (direct DNA transfer or Agrobacterium-mediated DNA transfer), using target sites embedded in regions of active chromatin, target tissue, and experimental plant may contribute. Strategies to improve these frequencies include regimes to activate the recombination target or to enforce the recombination potential by supplying enzymes derived from Escherichia coli. For instance, by transient expression of the restriction enzyme I-SceI, double-strand breaks were introduced in the target gene in the tobacco genome, resulting in hrdGT frequency of up to 10−2, at this site (13). However, only predetermined sites can be targeted, thereby reducing the general applicability of this process. Overexpression of prokaryotic enzymes involved in different aspects of homologous recombination resulted in improvement of extra- and intrachromosomal recombination efficiencies, by one order of magnitude and above (14, 15). Only further work will tell whether this increase in recombination also applies to gene targeting.
An exception in the plant world is the predominantly haploid moss Physcomitrella patens. This organism resembles yeast in that the prevailing mode of insertion of foreign DNA is by means of homology (16). Moss, therefore, may be an excellent model in which to study basic cellular processes, as exemplified by the successful knockout of a gene involved in organelle division (17).
cdMMR in Mammalian Systems
On the basis of analysis of pairing of oligonucleotides to single- and double-stranded DNA (18, 19), specific oligonucleotides were synthesized that combine DNA and RNA nucleotides in complementary configuration (Fig. 1). A “mutator” region of 5 nucleotides complementary to the target except for the mutation is flanked by 2′-O-methyl-RNA bridges of 10 nucleotides each, also complementary to the target locus. The O-methylation protects the RNA from degradation within cells. The loops of 4 T nucleotides are connected to a DNA-based sequence that is complementary to the chimeric strand. A strand break in the lower strand allows topological interwinding of the chimera into the target DNA, whereas the “GC clamp” seems to protect the 3′ terminus from degradation. Such COs were first used to correct a point mutation in cotransfected plasmids coding for an alkaline phosphatase cDNA in Chinese hamster ovary cells (20). In addition, a chromosomal sequence, a mutant β-globin gene causing sickle cell anemia, was found to be corrected with an astonishingly high frequency after delivery of a mutator CO (21) (although alternative explanations have been proposed in refs. 22 and 23). Injection of COs complexed with lactosylated polyethylenimine into tail veins of rats led to reduced factor IX gene activity in liver cells caused by mutation in the targeted gene with high efficiency and precision (24). This result indeed represents gene repair in animal organs. Genetic changes induced by cdMMR have also been shown to be genetically stable in several cell generations, as shown by restoration of pigmentation due to targeted reversion of a tyrosinase gene (25). In all reported cases, mutation was efficient (with exceptions; see note added in proof in ref. 21), and precise—i.e., only targeted sequences seemed to have been changed. In addition, the chimeric nature of the mutagenic oligonucleotide was found to be essential. Targeted gene repair in mammalian systems thus is entering the scene (26), although it is by no means routine.
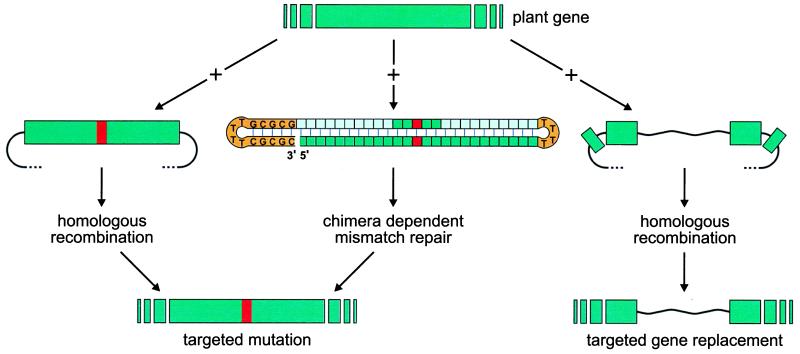
Figure 1.
Two techniques that can be applied for the targeted change of genomic sequences, homologous recombination-dependent gene targeting (hrdGT) and chimeric oligonucleotide-dependent mismatch repair (cdMMR). Both techniques can be used for introducing single base pair changes into genomes (left and center). hrdGT can also be used to introduce larger changes in the genome (right). Color code of the chimeric oligonucleotide: green, DNA sequence representing target sequence; light green, O-methylated RNA sequence representing target sequence; red, mismatch; orange, loops consisting of T residues, and “GC clamp.”
cdMMR in Plants
In contrast to human gene therapy approaches, where somatic tissue is the target, generation of mouse mutants and plant mutants (the latter useful for functional gene analysis and eventually production) requires change in every cell of the organism. Plant transformation mostly is accomplished by stably changing the genetic content of somatic cells, and then regenerating whole plants. In the two reports included in this issue of the Proceedings, use is made of established transformation, screening/selection and, in part, regeneration procedures to test the applicability of cdMMR. A tobacco tissue culture line (1) and cultured maize lines, as well as immature embryos (2), were used as target tissue. In both studies the gene coding for the first enzyme in the biosynthetic pathway of branched amino acids was chosen as the target, which in tobacco is referred to as acetolactate synthase (ALS) and in maize as acetohydroxy acid synthase (AHAS). Mutations of particular amino acids of this protein have been shown to result in plants resistant to the action of the herbicides imidazoline and sulfonylurea (27). COs representing such mutated versions were synthesized. Delivery of COs to plant cells is considerably more difficult than delivery to animal cells because in addition to the cell membrane the relatively rigid plant cell wall has to be passed by the mutagenic oligonucleotide. Therefore microparticle bombardment was used for introduction of COs. The efficiencies of recovery of chlorsulfurone-resistant tobacco clones was 10 to 20 times above background and about 4 and 15 times above background for imazethapyr- and chlorsulfurone-resistant maize clones, respectively. This efficiency was calculated for maize to be in the range of several times 10−7 per cell used for bombardment, or roughly 10−4 per cell receiving an oligonucleotide. Such frequencies are acceptable for selectable targeted mutations in plants, but they seem to be much lower than those reported for mammalian cells.
However, what a surprise when the sequences were analyzed! In the tobacco ALS gene the use of two different COs was expected to mutate CCA, coding for proline, to CAA, coding for glutamine, or to CTA, coding for leucine. Yet, in all analyzed cases in plant cells treated with the first CO the CCA was converted to ACA and in the experiment with the second CO TCA was found, coding for again a different amino acid. Repair thus was apparently shifted from the expected second position in the codon to the first. Yet it appears that the observed sequence changes are a consequence of bombardment with the two specific COs because several control treatments, including the use of an unspecific oligonucleotide or a specific DNA-only oligonucleotide, resulted in only a background resistance level. Analysis of the maize clones reveals a brighter picture: In one of the target positions in the AHAS gene 34 of 40 analyzed resistant clones indeed contained the expected alteration, while in 6 clones other alterations changing the targeted codon as well as the codon 5′ to it (in respect to the coding strand) were found. In the other target position only 2/12 events were precise, whereas in the other described cases the targeted nucleotide was changed differently.
Both studies also included experiments to target a transgenic but nonfunctional green fluorescent protein (GFP) gene (1, 2). A frameshift mutation in this gene in tobacco was found to be reverted, at least phenotypically, but neither sequences nor frequencies were presented (1). In maize, the frequency of detection of GFP-expressing cells, recovered after bombardment of maize cells carrying a GFP gene lacking a termination codon, was estimated to be from 2 × 10−7 to 1.6 × 10−6 per input cell or 1.5 × 10−4 to 1.1 × 10−3 per cell receiving an oligonucleotide (2). GFP-positive plants could be regenerated. One of them was raised to maturity, and it produced offspring with Mendelian segregation of the corrected GFP gene. Repair was at the expected position but not to the expected base. The events described are still rare, visible only through the power of a potent marker gene. However, the beauty and relative ease of the GFP assay system, in connection with the established mutant GFP tobacco and maize lines, will aid in further improvement of the technique.
Mechanistically, chimeric oligonucleotide-mediated gene mutation could resemble a gene conversion event. Recent work involving analysis in human cell extracts demonstrated, however, that the mismatch repair protein Msh2 is involved; extracts of a cell line lacking this protein or depletion of a wild-type extract of Msh2 by a specific antibody had only reduced cdMMR activity (28); cdMMR thus seems to be a true repair process, at least in mammalian systems; hence its name. In line with this argument is the finding that in mammalian systems base substitutions in the target locus occur more frequently than deletions or insertions of one or two bases (26). As Msh2, a key player in mismatch repair, is also found in plants (29), and as mismatch repair could be demonstrated to occur in a plant virus containing an artificial mismatch (30), involvement of the same actors in cdMMR in plants seem likely. It is less clear, however, how the nontargeted mutations arose. Recruitment of mismatch or other repair machinery to a desired site of action seems to allow other DNA- or RNA-targeted activities.
The new developments in gene therapy in plants have to be compared with similar approaches in animal systems. From the published data of cdMMR in mammals (reviewed in ref. 26), compared with the new information on repair in plant cells, the clear message can be derived that efficiency but also precision of gene correction in plants will have to be improved. Any of a number of individual steps culminating in cdMMR may be critical to achieve this: delivery of COs, their alignment to chromosomal sequences, mismatch recognition, and mismatch elimination. Packaging COs in various coats may secure express delivery and nuclease resistance; and providing an extra supply of mismatch repair proteins for the target cells at the time of delivery may increase efficiency and precision of repair. In addition, comparison of cdMMR and hrdGT is interesting in several respects (Fig. 1). If efficient and specific, cdMMR would be the method of choice to introduce a specific change to a specific gene. Efficiencies, as reported here, do not yet allow recovery of targeted events in genes that cannot be selected for. For hrdGT the efficiency of targeting was reported to be in the order of 10−3 to 10−5 targeted versus nontargeted events. cdMMR, as shown here, yielded an efficiency of 10−4 corrected events per cell receiving a repair-promoting oligonucleotide. These numbers are not directly comparable to efficiency of homologous recombination-mediated processes, since in the latter case not every cell receiving a targeting DNA unit gives rise to an integrated event, but possibly every hundredth cell. With efficiencies known to date, both procedures require time-consuming screens. cdMMR is not likely to be able to induce changes larger than single, or possibly several, nucleotides, as judged from animal work. More drastic changes, including gene replacements yielding knock-out plants, remain specialties of hrdGT, at least for the time being (Fig. 1).
For recovery of disrupted genes in plants, we do not depend on homologous recombination. Randomly integrating elements such as the transposable (and to some extent subsequently disposable) elements and T-DNA will serve their duties in years to come to inactivate and sometimes to activate genes in a dependable fashion (31). Less dependable, yet equally, if not more powerful, is the serendipitous or deliberate inactivation of plant genes on the epigenetic level (32). Procedures for introduction of stable and heritable changes on the genetic level will have to be optimized to replace epigenetic changes. The results published in this issue represent a first, difficult, but important step in this direction. As proposed, “at the very least, our data indicate that chimeric RNA/DNA oligonucleotides could be used to assay the poorly characterized mismatch repair pathways in plants” (2). There is this, but there is more.
Acknowledgments
We acknowledge the careful analysis of this commentary by J. Lucht and T. Hohn.
Footnotes
References
- 1.Beetham P R, Kipp P B, Sawycky X L, Arntzen C J, May G D. Proc Natl Acad Sci USA. 1999;96:8774–8778. doi: 10.1073/pnas.96.15.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu T, Peterson D J, Tagliani L, St. Clair G, Baszczynski C, Bowen B. Proc Natl Acad Sci USA. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doetschman T, Gregg R G, Maeda N, Hooper M L, Melton D W, Thompson S, Smithies O. Nature (London) 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 4.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 5.Jasin M, Moynahan M E, Richardson C. Proc Natl Acad Sci USA. 1996;93:8804–8808. doi: 10.1073/pnas.93.17.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paszkowski J, Baur M, Bogucki A, Potrykus I. EMBO J. 1988;7:4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puchta H, Hohn B. Trends Plant Sci. 1996;1:340–348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 8.Vergunst A C, Hooykaas P J J. Crit Rev Plant Sci. 1999;18:1–31. [Google Scholar]
- 9.Thykjaer T, Finnemann J, Schauser L, Christensen L, Poulsen C, Stougaard J. Plant Mol Biol. 1997;35:523–530. doi: 10.1023/a:1005865600319. [DOI] [PubMed] [Google Scholar]
- 10.Gallego M E, Sirand-Pugnet P, White C I. Plant Mol Biol. 1999;39:83–93. doi: 10.1023/a:1006192225464. [DOI] [PubMed] [Google Scholar]
- 11.Kempin S A, Liljegren S J, Block L M, Rounsley S D, Yanofsky M F, Lam E. Nature (London) 1997;389:802–803. doi: 10.1038/39770. [DOI] [PubMed] [Google Scholar]
- 12.Miao Z-H, Lam E. Plant J. 1995;7:359–365. doi: 10.1046/j.1365-313x.1995.7020359.x. [DOI] [PubMed] [Google Scholar]
- 13.Puchta H, Dujon B, Hohn B. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiss B, Klemm M, Kosak H, Schell J. Proc Natl Acad Sci USA. 1996;93:3094–3098. doi: 10.1073/pnas.93.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalev G, Sitrit Y, Avivi-Ragolski N, Lichtenstein C, Levy A A. Proc Natl Acad Sci USA. 1999;96:7398–7402. doi: 10.1073/pnas.96.13.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer D G, Zrÿd J P. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 17.Strepp R, Scholz S, Kruse S, Speth V, Reski R. Proc Natl Acad Sci USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotani H, Kmiec E B. Mol Cell Biol. 1994;14:7163–7172. doi: 10.1128/mcb.14.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havre P A, Kmiec E B. Mol Gen Genet. 1998;258:580–586. doi: 10.1007/s004380050771. [DOI] [PubMed] [Google Scholar]
- 20.Yoon K, Cole-Strauss A, Kmiec E B. Proc Natl Acad Sci USA. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole-Strauss A, Yoon K, Yiang Y, Byrne B C, Rice M C, Gryn J, Holloman W K, Kmiec E B. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 22.Thomas K R, Capecchi M R. Science. 1997;275:1404–1405. doi: 10.1126/science.275.5305.1401e. [DOI] [PubMed] [Google Scholar]
- 23.Stasiak A, West S C, Egelman E H. Science. 1997;277:460–462. doi: 10.1126/science.277.5325.459c. [DOI] [PubMed] [Google Scholar]
- 24.Kren B T, Bandyopadhyay P, Steer C J. Nat Med. 1998;4:1–6. doi: 10.1038/nm0398-285. [DOI] [PubMed] [Google Scholar]
- 25.Alexeev V, Yoon K. Nat Biotechnol. 1998;13:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 26.Ye S, Cole-Strauss A C, Frank B, Kmiec E B. Mol Med Today. 1998;4:431–437. doi: 10.1016/s1357-4310(98)01344-6. [DOI] [PubMed] [Google Scholar]
- 27.Lee K Y, Townsend J, Tapperman J, Chui C F, Mazur B, Dunsmuir P, Bedbrook J. EMBO J. 1988;7:1241–1248. doi: 10.1002/j.1460-2075.1988.tb02937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole-Strauss A, Gamper H, Holloman W K, Munoz M, Cheng N, Kmiec E B. Nucleic Acids Res. 1999;27:1323–1330. doi: 10.1093/nar/27.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Culligan K M, Hays J B. Plant Physiol. 1997;115:833–839. doi: 10.1104/pp.115.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riederer M A, Grimsley N H, Hohn B, Jiricny J. J Gen Virol. 1992;73:1449–1456. doi: 10.1099/0022-1317-73-6-1449. [DOI] [PubMed] [Google Scholar]
- 31.Azpiroz-Leeham R, Feldmann K A. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- 32.Matzke A J, Matzke M A. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]