Abstract
The Na+-glucose cotransporter hSGLT1 is a member of a class of membrane proteins that harness Na+ electrochemical gradients to drive uphill solute transport. Although hSGLT1 belongs to one gene family (SLC5), recent structural studies of bacterial Na+ cotransporters have shown that Na+ transporters in different gene families have the same structural fold. We have constructed homology models of hSGLT1 in two conformations, the inward-facing occluded (based on vSGLT) and the outward open conformations (based on Mhp1), mutated in turn each of the conserved gates and ligand binding residues, expressed the SGLT1 mutants in Xenopus oocytes, and determined the functional consequences using biophysical and biochemical assays. The results establish that mutating the ligand binding residues produces profound changes in the ligand affinity (the half-saturation concentration, K0.5); e.g., mutating sugar binding residues increases the glucose K0.5 by up to three orders of magnitude. Mutation of the external gate residues increases the Na+ to sugar transport stoichiometry, demonstrating that these residues are critical for efficient cotransport. The changes in phlorizin inhibition constant (Ki) are proportional to the changes in sugar K0.5, except in the case of F101C, where phlorizin Ki increases by orders of magnitude without a change in glucose K0.5. We conclude that glucose and phlorizin occupy the same binding site and that F101 is involved in binding to the phloretin group of the inhibitor. Substituted-cysteine accessibility methods show that the cysteine residues at the position of the gates and sugar binding site are largely accessible only to external hydrophilic methanethiosulfonate reagents in the presence of external Na+, demonstrating that the external sugar (and phlorizin) binding vestibule is opened by the presence of external Na+ and closes after the binding of sugar and phlorizin. Overall, the present results provide a bridge between kinetics and structural studies of cotransporters.
Keywords: human SGLT1, vSGLT, stoichiometry, alternating access, kinetics, human SGLT1 homology models
sglt1 was the first member of the SLC5 gene family to be cloned and is one of the most thoroughly studied of all the Na+ cotransporters (47, 48). Integration of detailed biochemical and biophysical studies has resulted in a quantitative kinetic model of SGLT1 function. The crystal structure of a bacterial homolog, vSGLT, has now been solved, and to date, four different gene families of Na+ cotransporters are known to share this structure, the LeuT structural fold (2, 6). These structures have been solved in different conformations representing steps in the transport cycle, i.e., in the outward-facing open (Mhp1), outward-facing substrate-bound (LeuT1 and Mhp1), inward-facing substrate-bound occluded (vSGLT), substrate-bound intermediate between outward- and inward-facing (BetP), and inward-facing substrate-free open conformation (vSGLT and Mhp1) (40, 43, 45, 46, 50). A distinctive feature of these structures is that the organic substrate occupies a binding site in the middle of the protein where it is occluded from both sides of the plasma membrane, suggesting that transport involves a coordinated opening and closing of external and internal gates (2, 18). Our goal is to integrate this structural information with the kinetics of SGLT1.
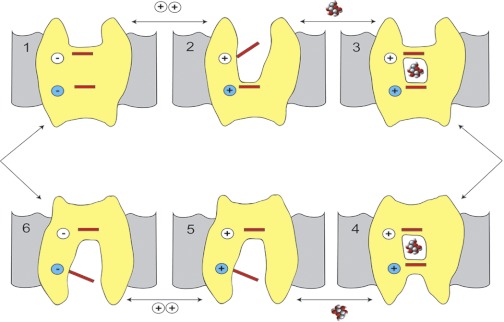
The SGLT1 kinetic model is based on an alternating access mechanism with six conformations (Fig. 1). The probability of each conformation is highly dependent on the membrane potential and the Na+ and glucose concentrations on each side of the membrane (27, 37). In the absence of ligands the transporter exists in two conformations, outward- and inward-facing ligand-free conformations [C1] and [C6], where the distribution between them is governed by voltage. In saturating external Na+, two cations bind to [C1] to form the outward-facing open conformation [C2Na2]. The protein is now poised to bind external glucose (or the nontransported competitive inhibitor phlorizin), and binding triggers the closing of the external gate to form an outward-facing occluded conformation [C3Na2S] (or [C3Na2Pz]). [C3Na2S] rapidly isomerizes to [C4Na2S], the inward-facing occluded conformation. Na+ and sugar are then released into the cytoplasm via an internal vestibule to give the inward open conformation [C6]. The kinetic model predicts that sugar exits first, whereas molecular dynamics simulations of vSGLT suggest that Na+ exit triggers sugar release (45). The final step in the transport cycle is the isomerization of [C6] back to [C1]. Of these six kinetic states, [C4Na2S] likely corresponds to the inward-facing occluded vSGLT crystal structure (6), [C6] to the inward open conformation of vSGLT (45), and [C2Na2] to the outward-facing open conformation of Mhp1 (46).
Fig. 1.
A 6-state model of Na+-glucose cotransporters (SGLTs) to integrate the kinetic and structural data. Na+ binds first to the outside (C1) to open the outside gate [C2Na2] (C2), permitting outside sugar to bind and be trapped in the binding site. This is followed by a conformational change from an outward occluded state [C3Na2S] (C3) to an inward occluded state [C4Na2S] (C4) . On opening of the inward gate (C5→C6); the Na+ ions and sugar are released into the cell interior. There is a paucity of data addressing the order of the ligand dissociation at the cytosolic surface. The transport cycle is completed by the change in conformation from the inward-facing ligand-free state (C6) to the outward-facing ligand-free state (C1). At 22°C the turnover number for the transporter is ∼50 s−1. The phlorizin-bound state [C7Na2Pz], not shown, is reasoned to be similar to [C3Na2S], but this is a dead-end state. The structural states [C3N2S] and [C4Na2S] described here are different from the states C3 and C4 identified in previous kinetic studies (37). The structural states are occluded, whereas kinetic states C3 and C4 are open to the outer and inner membrane surfaces, respectively. The occluded states are intermediates: C3 ⇋ [C3Na2S] ⇋ [C4Na2S] ⇋ C4. For simplicity, we have used C3 and C4 to represent [C3Na2S] and [C4Na2S]. [Modified from Abramson and Wright (2) with permission from Elsevier.]
Our approach to integrating kinetics with the structure was to first generate homology models of hSGLT1 in two conformations, the outward-facing open and the inward-facing occluded conformations. We then mutated each of the conserved gates and ligand binding residues of hSGLT1 and used biophysical and biochemical methods to determine the functional consequences. Transport kinetics of each mutant expressed in Xenopus laevis oocytes were determined using a two-electrode voltage-clamp method. We chose to replace each conserved residue with cysteine because this also enabled us to explore the ligand-induced conformational changes by monitoring their accessibility to cysteine reagents (9, 17). The results show that the effects of mutagenesis are consistent with the homology models and furthermore allow us to speculate on the structural changes that follow the binding of external ligands (Na+, glucose, and phlorizin).
MATERIALS AND METHODS
Homology Modeling
An inward-facing occluded model for hSGLT1 was constructed by using a sequence alignment between hSGLT1 and vSGLT (ClustalW: 32% sequence identity, 60% similarity) and threaded onto the inward-facing conformation of vSGLT (PDB code 3DH4) using the program Modeller (5). The threaded model was manually inspected and further energy-minimized with Refmac (1). The model was validated in Coot (4), showing that 98.2% of the residues reside in the most favorable regions of the Ramachandran plot.
An outward-facing open model was generated using the outward open conformation of Mhp1 as a template (PDB code 2JLN). This structure was chosen over others based on the conservation of two helix-breaking residues in TM10 (P436 and G437 in vSGLT) that are conserved between Mhp1 and the SGLT family. A structure-based sequence alignment between vSGLT and Mhp1 was created using the web server 3Dcoffee (www.tcoffee.org) and then manually optimized by comparing the two structures where gaps were introduced in loop segments to conserve transmembrane helices (Fig. 2). The hSGLT1 model was generated from the inward-facing vSGLT model by threading the hSGLT1 amino acid sequence onto the vSGLT inward-facing model as before. Inverted repeats are colored in blue and red (Fig. 2). Transmembrane helices observed in the structure are depicted on top (vSGLT) and bottom (Mhp1) of the alignment (Fig. 2). The models in shown in Figs. 3 and 8 were prepared in Pymol (http://www.pymol.org/).
Fig. 2.
Sequence alignment of vSGLT and Mhp1 based on structure comparison. A sequence alignment between vSGLT in the inward-facing conformation (PDB code 3DH4) and Mhp1 in the outward open conformation (PDB code 2JLN) was generated using 3-dimensional information via the web server 3Dcoffee (www.tcoffee.org). The sequence alignment was then manually optimized by comparing structural alignments of the 2 structures using Secondary Structure Matching. Gaps were introduced in loops to conserve transmembrane helices. Inverted repeats are colored in blue and red. Transmembrane helices (TM) observed on the structure are depicted on top (vSGLT) and bottom (Mhp1) of the alignment.
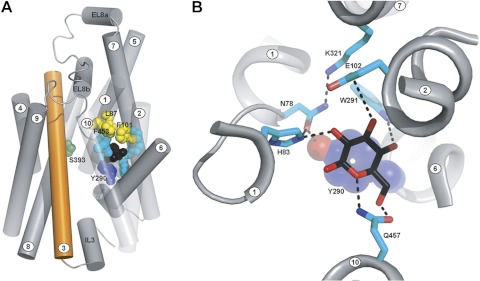
Fig. 3.
A: human SGLT1 (hSGLT1) inward-facing conformation: homology model of hSGLT1 based on the structure of vSGLT, seen from the membrane plane. Helices are represented as tubes with helix 3 colored orange for reference. For clarity, helices −1 and 11–14 have been removed and helices 1, 2, and 10 are depicted as transparent. Black spheres represent the position occupied by galactose in the vSGLT structure. Residues are shown as spheres, colored by function: Y290, lower gate, blue; L87, F101, and F453, upper gates, yellow; N78, H83, E102, W291, K321, and Q457, sugar binding residues, cyan; and S392 and S393, sodium binding residues, dark green. Note that the TMs are numbered according to the LeuT convention (4). B: hSGLT1 glucose binding site: homology model of hSGLT1 based on the structure of vSGLT, zoomed on the sugar binding site. The glucose molecule (black sticks with oxygen in red) was modeled in the position occupied by galactose in the vSGLT structure. Hydrogen bonds are depicted as black dashed lines.
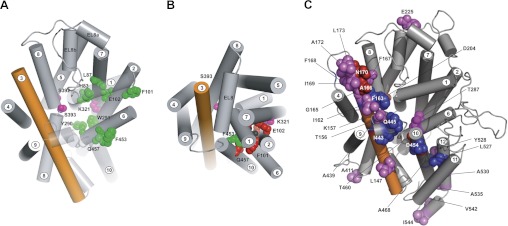
Fig. 8.
A: accessibility of external MTS reagents to hSGLT1 mutants in outward-facing conformation [C2Na2]. The model was generated by threading the human sequence on a model of the inward-facing conformation of vSGLT, made by comparison with the outward-facing conformation of Mhp1 and using the sequence alignment shown in Fig. 2. The model is shown from the external side of the membrane. The residues highlighted in green are cysteine mutants of the gate and ligand binding residues (Fig. 3) that were accessible to polar MTS reagents in the external solution in the [C2Na2] conformation. The mutants of the Na+ site residues S392C and S393C (red) were slightly accessible to methyl methanethiosulfonate (MeMTS). The access of K321C (pink) could not be determined due to low expression. Helices 1 and 10 are transparent for clarity. Note that the TMs are numbered according to the LeuT convention (2). B: accessibility of external MTS reagents to hSGLT1 mutants in inward-facing conformation [C4Na2S]. Accessibility to the residues indicated in A was changed in the inward occluded conformation. Only F453C (green) was accessible to polar cysteine reagents, whereas residues N78C, H83C, L87C, F101C, E102C, Y290, W291, and Q457 (red) were not accessible. S392C and S393C were only accessible (poorly) to MeMTS, and this was independent of conformation. The transparency of helices 1 and 10 (A) has been removed to show the steric hindrance created by these structures. The view of the protein model (Fig. 3) is shown from the external face of the membrane. C: summary of other SGLT1 mutations. Shown is the location of previously reported SGLT1 cysteine mutations that 1) do not significantly alter transport kinetics (violet and blue) or 2) are inhibited (or accessible) to external polar MTS reagents (blue). Those highlighted in red show partial sensitivity to MTS. (See Refs. 3, 12, 14, 15, 19–21, 30, 31, 34, 35, 39; Sala-Rabanal M, Martin M, and Wright EM, unpublished observations; and Jiang X, Hirayama BA, Loo DDF, and Wright EM, unpublished observations.)
Materials
Tetramethylrhodamine-6-maleimide (TMR6M) was purchased from Invitrogen-Molecular Probes (Carlsbad, CA), and sodium (2-sulfonatoethyl) methanethiosulfonate (MTSES−), 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET+), methyl methanethiosulfonate (MeMTS0), 2-aminoethyl methanethiosulfonate (MTSEA+), and tetramethylrhodamine methanethiosulfonate (TMR-MTS) were obtained from Toronto Research Chemicals (North York, ON, Canada). Capped cRNA was synthesized using kits from Ambion (Austin, TX), and [14C]α-methyl-d-glucopyranoside ([14C]αMDG) was obtained from Amersham (GE Healthcare, Piscataway, NJ). X. laevis were purchased from either Xenopus 1 (Dexter, MI) or Nasco (Modesto, CA). Enzymes were obtained from New England Biolabs (Ipswich, MA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Molecular Biology
Cysteine and alanine mutants were made using wild-type (WT) hSGLT1 cDNA in pBluescript (10) and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The integrity of all constructs was verified by full sequencing. Capped cRNA for WT and mutant hSGLTs was synthesized using the T3 mMessageMachine kit from Ambion.
Functional Expression of hSGLT1 in Oocytes
All animal protocols followed guidelines approved by the University of California Chancellor's Committee on Animal Research. Mature female X. laevis were anesthetized with 0.1% tricaine (Sigma-Aldrich) buffered with 0.1% NaHCO3 to surgically remove a portion of the ovary. Stage V–VI oocytes were selected and maintained at 18°C in modified Barth's solution, supplemented with 50 mg/l gentamicin (Sigma), 5.75 mg/l ciprofloxacin (Bayer), and 100 mg/l streptomycin sulfate-100,000 U/l penicillin G sodium (Invitrogen, Carlsbad, CA). One day after isolation, oocytes were injected with 50 ng of cRNA coding for WT hSGLT1 or mutants and incubated at 18°C for 4–7 days. Experiments were performed at 20–22°C. Noninjected oocytes served as controls.
α-Methyl-d-glucopyranoside Uptake Assays
Na+-dependent uptake of α-methyl-d-glucopyranoside (αMDG), a specific nonmetabolized SGLT substrate, was measured as described previously (16). Briefly, oocytes were incubated for 60 min in the presence of 50 μM αMDG (5 μM [14C]αMDG) in a buffer containing (in mM) 100 NaCl, 2 KCl, 1 MgCl2, 1 CaCl2, and 10 HEPES-Tris (pH 7.5), in the absence or presence of 500 μM phlorizin. After incubation, oocytes were rinsed thoroughly with ice-cold buffer containing (in mM) 100 choline-Cl, 2 KCl, 1 MgCl2, 1 CaCl2, and 10 HEPES-Tris (pH 7.5), individually lysed with 5% sodium dodecyl sulfate, and assayed for radioactivity.
Electrophysiology
Two-electrode voltage-clamp studies in oocytes expressing WT or mutant hSGLT1 were carried out as described previously (25, 36). Oocytes were normally bathed in 100 mM NaCl buffer with membrane potential held at −50 mV (Vh). Total current across the oocyte membrane consists of the endogenous background current, the Na+-uniporter current (SGLT1 leak current), which can be blocked by the inhibitor phlorizin, and the sugar-coupled cotransporter current, which is the difference in current recorded in the presence of αMDG and Na+ and in the absence of the sugar.
Current-voltage (I-V) relations of the sugar-induced current (see Fig. 6) were obtained using a standard pulse protocol where membrane potential was stepped from Vh to various test values (from +50 to −150 mV in 20-mV decrements) for 100 ms before returning to Vh. Steady-state sugar-induced current was measured at 100 ms. All experiments were performed at room temperature (20–23°C).
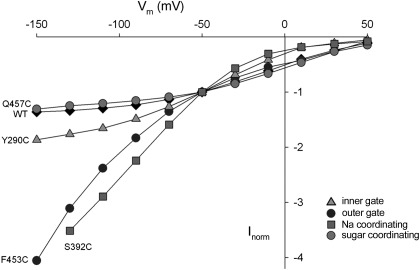
Fig. 6.
Current-voltage (I-V) curves of the WT hSGLT1 and selected mutants (data from Table 2). The maximal sugar-induced current (Imax) for WT and Q457C saturate with voltages more negative than −100 mV. Although the current for Y290C also saturated at large negative voltages, those for F453 and S292C did not. The WT, Q457C, and S392C currents are Imax, but those for Y290C and S392C at 100 mM sugar are not, since for these mutants K0.5αMDG values are >>100 mM. Therefore, the maximum rates of transport by Y290C and S392C are indeterminate.
SGLT1 exhibits a presteady-state current (charge movement) with step jumps in membrane voltage in NaCl buffer in the absence of sugar (21, 22). The presteady-state current was isolated by applying the pulse protocol and fitting the total membrane current (Itot) to
where Icmexp(t/τcm) is the membrane bilayer capacitance transient with initial value Icm and time constant τcm, Ipssexp(−t/τ) is the SGLT1 presteady-state current with initial value Ipss and time constant τ, and Iss is the steady-state current. The presteady-state current was obtained by subtracting the bilayer capacitive and steady-state components from Itot. Charge movement (Q) was obtained from the integral of the presteady-state current (22). Charge vs. voltage (Q-V) relations were fitted with a single Boltzmann function: (Q − Qhyp)/Qmax = 1/{1 + exp[zδ(V − V0.5)F/RT]}, where Qmax = (Qdep − Qhyp) is the maximal charge, Qdep and Qhyp are the Q (absolute value) at depolarizing and hyperpolarizing limits, V is membrane potential, F is Faraday's constant, R is gas constant, T is absolute temperature, V0.5 is midpoint voltage (voltage at 50% Qmax), z is the apparent valence of the moveable charge, and δ is the fraction of the membrane electric field traversed by the charge.
The half-saturation concentration (or apparent affinity constant) for αMDG (K0.5αMDG) was estimated in 100 mM NaCl buffer (at −50 mV) by fitting the sugar-induced current (IαMDG) vs. external αMDG concentration ([αMDG]o) data to a hyperbolic relation: IαMDG = Imax[αMDG]o/(K0.5αMDG + [αMDG]o), where Imax is the maximal sugar-induced current. For mutants Y290C, H83C, E102C, W291C, and S392A/C, the IαMDG vs. [αMDG]o curves did not saturate at 100 mM αMDG, and the estimated K0.5αMDG values were denoted as >100 mM. In these experiments we did not compensate for changes in osmolarity due to the addition of sugar (up to 200 mM), because the exposure was brief (<1 min) and studies showed that the addition of up to 200 mM mannitol did not influence the SGLT1 sugar currents.
Half-saturation concentration for Na+ (K0.5Na) was estimated in the presence of a saturating concentration of αMDG, i.e., 10 mM for WT hSGLT1; 50 mM for L87C, F101C, F453C, N78A/C, and S393A/C; and 100 mM for Q457C (see Table 2). The αMDG-induced current was determined as [Na+]o was varied, and the data were fitted to IαMDG = Imax([Na+]o)n/[(K0.5αMDG)n + ([Na+]o)n], where n is the Hill coefficient. For mutants Y290C, H83C, E102C, W291C, and S392A/C with K0.5αMDG >100 mM, the K0.5Na values were estimated from the dependence of the presteady-state currents on [Na+]o. In brief, there is a shift of the midpoint voltage (V0.5) to more negative values as [Na+]o is reduced in the absence of sugar (see Fig. 22, D and E in Ref. 47). Using the V0.5 shift, simulations were performed on the six-state kinetic model (Fig. 1) to estimate the K0.5Na under saturating sugar concentrations (Loo DDF, Hirayama BA, and Wright EM, unpublished observations). As internal control, the simulated and measured K0.5Na values were in agreement for WT hSGLT1 and mutants where K0.5Na could be determined, i.e., sugar-induced current saturated at the highest sugar concentration used.
Table 2.
Summary of mutant hSGLT1 kinetics
| K0.5αMDG, mM | K0.5Na, mM | Turnover (Imax/Qmax), s−1 | KiPz, μM | Qmax, nC | Imax, nA | |
|---|---|---|---|---|---|---|
| hSGLT1 | 0.3–0.49 | 20–40 | 57 | 0.22 | ≈10 | 500–1,000 |
| Sugar binding | ||||||
| H83C | >100 | 66* | >>> 70 | 4 ± 1 | 6 ± 3 | 165 ± 21 |
| E102C | >100 | nm | nm | nm | nm | 37 ± 11 |
| W291C | >100 | 46* | >>> 22 ± 5 | 68 ± 15 | 8 ± 2.0 | 177 ± 57 |
| K321C | 43 ± 10† | 76 ± 14‡ | nm | 850 ± 50‡ | nm | nm |
| Q457C | 13 ± 2 | 34 ± 2 | 49 ± 5 | 6 ± 1 | 6 ± 1 | 378 ± 60 |
| N78A/C | 6 ± 1 | 18 ± 1 | >56 ± 7 | 4 ± 2 | 0.9 ± 0.2 | 68 ± 17 |
| Na+ binding | ||||||
| S392A/C(3) | >100 | 66* | >>>81 ± 6 | 4 ± 1 | 8 ± 1 | 264 ± 39 |
| S393A/C(3) | 4 ± 1 | 32 ± 3 | 56 ± 10 | 0.5 ± 0.2 | 2 ± 1 | 83 ± 15 |
| Outer gates | ||||||
| L87C | 6 ± 1 | 23 ± 1 | >75 ± 8 | 1.2 ± 0.1 | 2 ± 1 | 165 ± 21 |
| F101C | 1.1 ± 0.2 | 9 ± 1 | >>>107 ± 14 | 37 ± 12 | 7 ± 2 | 1,067 ± 350 |
| F453C | 1.8 ± 0.2 | 21 ± 1 | >>>127 ± 16 | 1.0 ± 0.3 | 6 ± 1 | 690 ± 40 |
| Inner gate | ||||||
| Y290C | >100 | 46* | >>100 | 36 ± 6 | 11 ± 1 | 565 ± 226 |
Kinetics are means ± SE of 3–9 oocytes from at least 2 donor frogs. Kinetic parameters [K0.5αMDG, half-saturation concentration of α-methyl-d-glucopyranoside (αMDG); K0.5Na, half-saturation concentration of Na+; KiPz, inhibitory constant for phlorizin) were determined at a membrane potential of −50 mV.
K0.5Na values estimated from Na+ dependence of charge movement. Note that for the other mutants, estimates are similar for all 3 assays of K0.5Na (see methods).
Rabbit K321C at −150 mV.
Rabbit K321C at −50 mV(34). §Mean of Ala and Cys mutants for S392 and S393.
Imax, maximal sugar-induced current; Qmax, maximal charge movement; nm, not measurable.
KiPz, the inhibitory constant for phlorizin, was estimated by 1) reduction of maximal charge Qmax by phlorizin (H83C, W291C, and S393A/C) (11); the phlorizin-inhibited charge (QPz) vs. [Pz]o fitted a hyperbolic relation with half-maximal concentration KiPz; 2) inhibition of the uniporter (leak) currents in the absence of glucose (F101C, F453C, Q457C, H83C, and Y290C) (26); or 3) phlorizin blockade of the sugar-induced current at the K0.5αMDG concentration for the mutant (N78C, S392A/C). Under these latter conditions, the concentration of phlorizin that inhibited the sugar current 50% was twice the KiPz (11).
Transport protein expression.
The number of WT and mutant SGLT1 proteins in the oocyte plasma membrane was estimated by measuring the maximum hSGLT1 charge movement, Qmax (23). We have previously described that Qmax is proportional to the number of functional SGLT transporters in the plasma membrane; for example, a Qmax of 10 nC is equivalent to 1 × 1010 SGLT1 proteins in the oocyte membrane (23, 51).
Imax/Qmax.
The ratio of the maximal αMDG-induced current Imax (at −150 mV) and Qmax provides a measure of the cotransporter turnover rate, i.e., the number of times the transporter cycles per second, if the sugar-induced current is strictly coupled to sugar transport (23). The ratio provides a clue to uncoupling between Na+ and sugar. For example, if there is uncoupling between the ligands, Imax/Qmax could be greater than that of WT hSGLT1. When the I-V curves were nonsaturating at the largest hyperpolarizing voltages, the Imax at −150 mV was underestimated and Imax/Qmax was designated as “>”. Likewise, when Imax was not able to be determined for mutants with K0.5αMDG > 100 mM, their Imax/Qmax ratio was also designated as “>”.
Stoichiometry.
The Na+-to-sugar transport stoichiometry was obtained for SGLT1 mutants by simultaneously measuring sugar-stimulated Na+ and [14C]αMDG uptakes in the same cell (29, 39). Briefly, voltage-clamped oocytes (at −90 mV) were superfused with 0.2–1 mM [14C]αMDG in 100 mM NaCl buffer for 2–10 min while the sugar-induced inward Na+ current (obtained by subtracting the total current in the presence of sugar from the background current in the absence of sugar) was continuously recorded. At the end of the experiment, the oocyte was assayed for [14C]αMDG uptake (moles) and inward charge, which was determined by integrating the inward Na+ current induced by αMDG and converted to molar equivalent using the Faraday constant. These direct measurements of stoichiometry were not performed on the mutants with K0.5αMDG > 100 mM because the high sugar concentrations required to produce current dramatically reduce the specific activity of [14C]αMDG and compromise the accuracy of sugar uptakes. Transporter stoichiometry is the ratio of the inward charge to the inward sugar flux. Current is a valid measure of Na+ uptake because the sugar-induced current is Na+ dependent, and we have previously shown with 22Na+ uptakes under voltage clamp that the current is carried by Na+ (25). As internal controls we measured the stoichiometry for WT SGLT1, the mutant G507C hSGLT1 [a mutant that does not change the Na+-sugar transport kinetics (25)], and control oocytes not expressing SGLTs.
MTS effects.
The substituted-cysteine accessibility method (SCAM) (17) was used to determine how hSGLT1 conformation affects exposure of specific amino acid positions of the protein (24). The method requires creation of cysteine mutants and an assay for the modification by cysteine reagents, such as inhibition of transport. Experiments were done with the oocytes mounted in a continuous flow chamber on the two-electrode voltage-clamp setup; the bath electrode was equipped with a 3 M KCl-agarose bridge to protect the Ag-AgCl electrodes from the reactive cysteine reagents. Steady-state transporter currents were measured at a Vh of −50 mV. After a stable current was observed in Na+ buffer, we added 100 mM αMDG, which induced an inward current due to the cotransport of 2 Na+ ions with each sugar. We then removed the substrates by superfusing the oocyte with choline buffer. This procedure was repeated after each exposure to the cysteine reagent in the presence or absence of Na+, 100 mM glucose, or 500 μM phlorizin.
Reaction rates in Na+ were estimated from inhibition following the method of Pascual and Karlin (17) using the conditions of the labeling protocol in Na+. The second-order rate constant (κ) was estimated as
where [MTS] is the concentration of the reactive methanethiosulfonate agent and t is time of exposure in seconds.
The conformation of hSGLT1 was manipulated during the experiment through combinations of substrate, inhibitor, and membrane potential (see for example, Refs. 24 and 25). In the presence of Na+ buffer alone with Vh = −100 mV, [C2Na2], the Na+-bound outside conformation, is favored (occupancy probability, Po ∼ 0.9); in the absence of Na+ at Vh = 0 mV, [C6], the empty inside conformation, is most likely (Po ∼ 0.9); Na+ with 100 mM αMDG favors [C5Na2] (Po ∼ 0.6); and in 500 μM phlorizin, the protein is bound to phlorizin ([C3Na2Pz]) (Po ∼ 0.7) (see Fig. 3 and Ref. 25).
RESULTS
Homology Modeling
The initial step of the study was to construct a homology model of hSGLT1 based on the inward occluded structure of vSGLT (6). The ligand binding and gating residues identified in vSGLT (3, 25) are largely conserved in the six members of the SGLT gene family (47). Table 1 shows the conservation between vSGLT and hSGLT1. A side view of the model in the plasma membrane is shown in Fig. 3A with glucose (black spheres) trapped in the center of the protein between the outer gates (L87 on TM1, F101 on TM2, and F453 on TM10; yellow) and the inner gate (Y290 on TM6; blue). Also highlighted are the sugar binding residues (cyan) and the Na+ coordinating residues (green) of the putative Na2 site, which is conserved in the Na+ cotransporters in the LeuT structural family (2, 33, 50). The other Na+ binding site (Na1) of hSGLT1 remains to be identified (unlike vSGLT, 2 Na+ ions are cotransported with glucose in SGLT1). Note that numbering of the transmembrane (TM) helices follows the convention adopted for the LeuT structural fold (2).
Table 1.
Corresponding residues between vSGLT and hSGLT1
| vSGLT | hSGLT1 |
|---|---|
| Sugar binding | |
| Q69 | H83 |
| E88 | E102 |
| S91 | A105 |
| K294 | K321 |
| N260 | T287 |
| W264 | W291 |
| Q428 | Q457 |
| N64 | N78 |
| Na+ binding | |
| A62 | A76 |
| I65 | I79 |
| A361 | S389 |
| S364 | S392 |
| S365 | S393 |
| Outer gates | |
| M73 | L87 |
| Y87 | F101 |
| F424 | F453 |
| Inner gate | |
| Y263 | Y290 |
The human SGLT1 (hSGLT1) residues at the conserved ligand binding sites and gates between vSGLT and hSGLT1 were identified from the sequence alignment (Fig. 2).
The residues that coordinate with glucose in the model are shown in more detail in Fig. 3B: H83 and N78 (TM1), E102 (TM2), Y290 and W291 (TM6), K321 (TM7) and Q457 (TM10). Residues H83 and N78 are coordinated with the sugar O2, E102 with O3, W291 with O4, and Q457 with O5. The positively charged K321 (TM7) interacts with N78 (TM1), and there is a hydrogen bond between N78 and the OH of the Y290 (TM6). The Na2 site in hSGLT1, some 10 Å distant from the glucose binding site, is composed of 5 residues: S392, S393, S289, A76, and I79. S393 is highlighted in green in Fig. 3A.
Functional Expression of Mutants
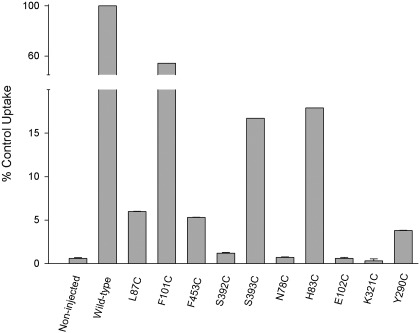
To determine the roles of these predicted gating and ligand coordinating residues in hSGLT1, we mutated each residue in turn to cysteine or alanine and recorded the change in transport kinetics. To demonstrate that the mutants were functional and transported sugar, standard transport assays were conducted where the uptake of radiolabeled αMDG was measured in oocytes expressing WT or mutant hSGLT1. Relative to WT (control), the sugar transport rate 4–6 days after cRNA injection was significantly reduced for most mutants (Fig. 4). For S392C, N78C, E102C, and K321C, transport was not significantly different from that in noninjected oocytes or in WT oocytes in the presence of 500 μM phlorizin, 0.15 ± 0.01% of control (P < 0.05; 1-way ANOVA and Kruskal-Wallis test).
Fig. 4.
α-methyl-d-glucopyranoside (αMDG) uptake into Xenopus oocytes expressing wild-type (WT) or mutant hSGLT1. Results were normalized to the uptake measured in oocytes expressing WT transporters, 243 ± 29 pmol/h per oocyte. Data are means ± SE from individual experiments with 6–8 oocytes per condition; each mutant was tested at least twice in oocytes from different donor frogs. For WT and noninjected oocytes, data are compiled as means ± SE from 7 experiments carried out 4–6 days after cRNA injection. Error bars are SEs when the SE exceeds the size of the symbol.
Electrophysiological methods were used to determine whether the differences in transport rate were due to differences in ligand affinity, turnover number, or transport protein expression levels; results are summarized in Table 2. To estimate kinetic parameters, we used individual oocytes with a high level of expression of each mutant, as gauged by the maximal αMDG-induced current (Imax) and the number of hSGLT1 proteins expressed in the plasma membrane (hSGLT capacitive charge, Qmax). For most mutants the level of protein expression in the membrane was comparable to that for WT, whereas for three mutants, N78A/C, S393A/C, and L87C, the expression was only 10–25% of WT. Two mutants, E102C and K321C, were poorly expressed, i.e., Qmax was smaller than 1 nC, and their kinetics could not be determined (Table 2). On the other hand, with the exception of K321C, all mutants exhibited sugar-induced inward Na+ currents >40 nA when voltage-clamped at −50 mV. In noninjected oocytes, αMDG-induced currents were <1 nA.
Although we generally replaced residues with cysteines, for the SCAM studies described below, we also replaced several residues with alanine (N78, S392, S393) to compare directly with previous vSGLT mutations (6, 45). The kinetics of the cysteine and alanine mutants were found to be quite similar.
Uniporter (leak) currents in the absence of glucose were 5–10% of the maximal sugar-induced currents in all but three mutants. For N78A/C, F453C, and K321A (34), the uniporter currents were 40–50% of the currents induced by 100 mM glucose (not shown).
Kinetics of Na+-αMDG Cotransport
Sugar binding residues.
As shown in Table 2, K0.5αMDG was 10 to >100 mM for these mutants, i.e., >200-fold higher than for WT. The expression levels of E102C and K321C were too low for reliable kinetics analysis, but we have previously reported that the K0.5αMDG for rabbit K321C was 43 mM (34). These results confirm that the vSGLT residues involved in sugar coordination also are important for sugar transport by hSGLT1.
Outer and inner gates.
K0.5αMDG was 1–6 mM for L87C, F101C, and F453C, i.e., 3- to 10-fold higher than for WT (0.3–0.5 mM) (Table 2). K0.5Na was 9 mM for F101C, 21 mM for F453C, and 23 mM for L87C; for WT hSGLT1, K0.5Na was 30 mM. Thus, for these mutants, the apparent affinities for Na+ and sugar were not too dissimilar from those for the WT. The Hill coefficients for activation of the mutant transporters F101C, F453C, and L87C by Na+ were 1.1 ± 0.1 (3), 1.3 ± 0.1 (3), and 1.7 ± 0.2 (3), respectively. Mutation of the inner gate Y290C showed very poor apparent affinity for αMDG, because the αMDG-induced current vs. [αMDG] relation did not saturate at the highest concentration tested (100 mM), i.e., K0.5αMDG >100 mM. An explanation for the low apparent sugar affinity of Y290C is the importance of Y290 in “stacking” with the sugar ring. K0.5Na was ∼2-fold higher than in WT, 66 vs. 30 mM (Table 2).
Na+ binding.
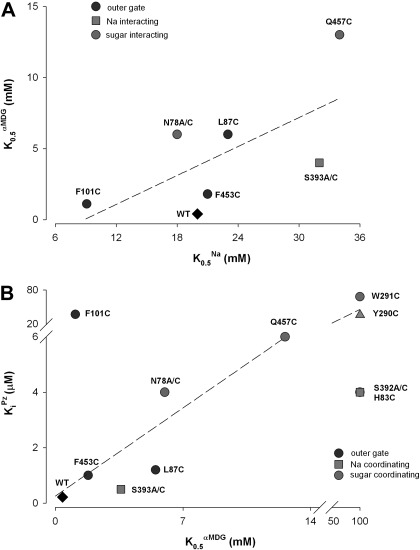
One putative Na+ binding site, the Na2 site, has been identified in vSGLT1 and other members of the LeuT structural fold family (2, 6, 33, 46). Na+ at this site is predicted to be coordinated with two serines on TM8 and carbonyl oxygens at the unwound region of TM1. We mutated both serine residues to cysteine or alanine. As shown in Table 2, for S392A/C there was an increase in K0.5Na from ∼30 to 65 mM and a dramatic increase in K0.5αMDG from 0.5 to >100 mM. This loss of sugar binding may account for the loss of sugar transport by the equivalent vSGLT mutant S365A (6). On the other hand S393C/A led to only modest changes in K0.5Na and K0.5αMDG, suggesting that it is not essential for Na+ coordination. Where it was possible to obtain reliable estimates of K0.5αMDG, we found a linear relationship between K0.5αMDG and K0.5Na (Fig. 5A).
Fig. 5.
A: relation between the half-saturation concentration (apparent affinity constant) for αMDG (K0.5αMDG) and that for Na+ (K0.5Na). Both parameters were determined at a membrane potential (Vm) = −50 mV (Table 1). Note that for F101C, K0.5Na is lower than expected from the K0.5αMDG value. B: relation between KiPz and K0.5αMDG at Vm = −50 mV (data from Table 2). Note that the KiPz value for F101C is 200-fold higher than expected.
Phlorizin Binding Kinetics
Phlorizin is a high-affinity, nontransported, competitive inhibitor for hSGLT1, so we measured the phlorizin Ki for each mutant (Table 2). KiPz ranged from 0.22 μM for WT to as high as 850 μM for rabbit K321C. As shown in Fig. 5B, there is a linear relationship between KiPz and K0.5αMDG for most of the gate and sugar binding residue mutants. Again, the only exception is F101C, where KiPz was 200-fold higher than expected from the K0.5αMDG estimate, suggesting that gate residue F101 is important for phlorizin binding.
Turnover (Imax/Qmax) and Stoichiometry
Under conditions of tight stoichiometric coupling between Na+ and glucose, the catalytic turnover of the transporter may be estimated in single cells from Imax/Qmax, the ratio of the maximum rate of cotransport (Imax) and the maximal cotransporter charge (Qmax), which is an index of the number of transporters in the plasma membrane (51). Imax was obtained from the αMDG-induced currents measured at a saturating external sugar concentration at hyperpolarizing membrane potentials. In Fig. 6 we show that for WT hSGLT1, the Imax-V curves saturate at membrane potentials more hyperpolarizing than −100 mV, and thus the ratio of Imax at −150 mV to Qmax (∼600 nA/∼10 nC), which is the turnover rate, is ∼60 s−1. The Imax-V curves of Q457C and S393C also saturate at voltages more negative than −100 mV (Fig. 6; not shown for S393C), and for these mutants the ratio Imax/Qmax gives a similar value, 50 s−1 (Table 2). We estimated that the Imax/Qmax ratios for H83C, W291C, S392C, F101C, F453C, and Y290C were much greater than 100 s−1. Only minimum ratios were obtained when 1) the Imax-V curves did not saturate within the practical voltage range that is possible in oocyte studies, +50 to −150 mV, e.g., F453C and S292C (Fig. 6); and/or 2) it was not possible to measure Imax because K0.5αMDG was much greater than 100 mM (Table 2).
In an effort to determine if the increased Imax/Qmax was indeed an increase in translocation rate, we next directly determined the coupling of Na+ to sugar transport for those mutants with a high apparent affinity for sugar and high Imax/Qmax ratios by simultaneously measuring Na+ and sugar uptakes in single oocytes under voltage clamp (Table 3). For WT hSGLT1, the coupling ratio was 2:1, whereas it was greater than 2:1 for the gate mutants L87C (2.3), F101C (4.3), and F453C (5.9). As another control, we determined the coupling for, G507C, a mutant with transport kinetics comparable to those of WT (25). We conclude that the high Imax/Qmax ratios observed for the external gate mutants (Table 2) are due to uncoupling of Na+ and sugar transport.
Table 3.
Stoichiometry of Na+-to-glucose transport
| Mutant | Stoichiometry |
|---|---|
| Wild type | 2.0 ± 0.05 (3) |
| L87C | 2.3 ± 0.1 (3) |
| F101C | 4.3 ± 0.2 (3) |
| F453C | 5.9 ± 0.6 (6) |
| G507C | 1.9 ± 0.06 (3) |
Stoichiometry values (means ± SE) were obtained from the ratio of the αMDG-induced inward Na+ current (converted to moles) and the [14C]αMDG uptake in single oocytes expressing SGLT1. The external [14C]αMDG concentration was 0.2 to 1 mM in 100 mM NaCl buffer, and the membrane potential of the oocytes was clamped at −90 mV.
Alkylation of Cysteine Mutants
The effect of alkylating reagents on Na+-glucose cotransport was determined for each mutant by measuring the sugar-induced current before and after exposure of the oocyte to the reagents in 100 mM NaCl buffer. The glucose-induced currents of all gating residues and sugar coordinating mutants (N78, H83, Y290, Q457) were inhibited by >90% (not shown) with rate constants varying between 800 and 10,000 M−1·s−1 (Table 4). This indicates that these cysteines are readily accessible to the extracellular hydrophilic reagents in the [C2Na2] conformation and that this pathway is critical for Na+-sugar cotransport. Transport by S392 and S393 was only partially inhibited by MeMTS, suggesting low accessibility. No functional effects of these MTS reagents were observed on the WT protein even at much higher concentrations, and no results were obtained for the E102C, F452C, or W291C mutants due to low expression.
Table 4.
Rates of MTS inhibition
| Residue | Reagent | κ, M−1·s−1 (MTSEA Equivalent) |
|---|---|---|
| N78C | MTSEA (3 μM) | 1,000 |
| H83C | MTSEA (1 μM) | 4,000 |
| L87C | MTSEA (2 μM) | 10,000 |
| F101C | MTSET (40 μM) | 500 |
| Y290C | MTSET (25 μM) | 800 |
| S392C | MeMTS* | na |
| S393C | MeMTS* | na |
| F453C | MTSET (2 μM) | 2,000 |
| Q457C | MTSEA (0.3 μM) | 14,000 |
Labeling conditions and estimated methanethiosulfonate (MTS) reaction rates: for each reagent we determined the concentration that inhibited hSGLT1 Na+/glucose currents by 50% in 2 min in 100 mM NaCl buffer. Because 2-aminoethyl methanethiosulfonate (MTSEA) is inherently more reactive than 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET), we normalized the MTSET reaction rate to that of MTSEA, e.g., the concentration for ∼50% inhibition of H83C was determined for MTSEA (1 μM) and MTSET (5 μM) in Na buffer for 2 min: κ = 0.5/([MTS]·t). Since the rate was 5-fold higher for MTSEA than MTSET, the rates for MTSET were multiplied by 5 to normalize the results.
Neither MTSEA nor MTSET inhibited S392C or S393C, but 100 μM methyl methanethiosulfonate (MeMTS) inhibited 50%. Low functional expression of E102C, and L452C precluded estimation of rates, and neither MTSEA nor MTSES inhibited W291C. Note that tetramethylrhodamine-6-maleimide(TMR6M) partially inhibited E102C and L452C and TMR5M partially inhibited W291C and L452C.
Conformational Changes
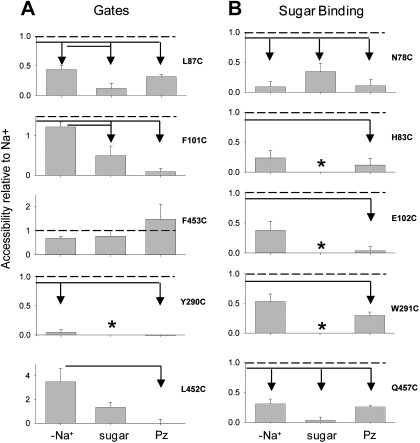
The homology models predict that the exposure of residues in the sugar binding pocket to extracellular MTS reagents changes during the course of the transport cycle (see Fig. 1). Because the probability of a given transport intermediate can be manipulated with substrates, we determined the accessibility of each cysteine to the MTS reagents in four different conformations: outward open [C2Na2], phlorizin-bound [C3Na22Pz], inward open [C5Na2], and ligand-free [C1] and [C6]. This was accomplished by comparing the effects of the MTS reagent on transport after the oocytes were incubated with the reagent in 1) 100 mM NaCl [CNa2], 2) 100 mM NaCl plus 500 μM phlorizin buffer [C3Na2Pz], 3) 100 mM NaCl plus saturating sugar [C5Na2], and 4) ligand-free buffer, 100 mM choline-Cl [C1] or [C6]. MTS concentration and exposure time were adjusted to inhibit ∼50% of the current when incubated in Na+ alone. Accessibility was expressed relative to the inhibition of the Na+-glucose current after incubation in Na+ alone (Fig. 7). For example, incubating the gate mutant L87C with 2 μM MTSEA in 100 mM NaCl [C2Na2] inhibited 50%, whereas incubation in 100 mM NaCl buffer containing 100 mM αMDG [C5Na2] only inhibited 5%; i.e., the accessibility was only 10% of that in NaCl buffer. In the case of three mutants, E102C, W291C, and L452C, accessibility was determined by labeling with TMR6M or TMR5M (12). Figure 7 summarizes the results expressed as a fraction of the inhibition observed in Na+ buffer.
Fig. 7.
Exposure of residues is conformation dependent. Exposure of residues to the external solution was measured by labeling with methanethiosulfonate (MTS) reagents or fluorescent dyes in the presence or absence of substrates. Accessibility is expressed relative to labeling in Na+ under the same conditions. All residues in the sugar and Na binding sites were accessible in Na+, indicating the sugar and Na+ pathways are open. A: residues modeled to form the external gate, L87, F101, and F453; internal gate and stacking with pyranose, Y290; and as a control, L452, modeled to face away from the sugar access pathway. B: sugar binding residues are N78, H83, E102, and W291. Arrows indicate conditions in which accessibility was significantly different from the line origin (Tukey post hoc, 0.05 level). *Mutants with K0.5αMDG > 100 mM; data are not interpretable because the proportion of transporters in the sugar-bound conformation is low. [Graph for Q457C is replotted from data in Hirayama et al. (12).]
In general, the accessibility of the cysteine residues was greater in the [C2Na2] conformation than in the three other conformations. When phlorizin was bound to the transporter [C3Na2Pz], the labeling was either eliminated (F101C, Y290C, L452C, N78C, H83C, and E102C) or dramatically reduced (L87C, W291C, and Q457C). For one mutant, F453C, there were no significant changes in accessibility among the four conformations. Likewise, external sugar, which shifts the conformational state to [C5Na2] (27), buried the cysteine residues into much less accessible positions [the results for mutants with low sugar affinity (>100 mM) were not interpretable because the percentage of protein in [C5Na2] was unknown]. Accessibility was also dramatically reduced in the absence of Na+, with the exception of two outer gate mutants, F101C and F453C. These experiments show that the accessibility of these cysteine mutants to MTS reagents was very sensitive to the conformational state of the protein. The clearest exception is the outer gate residue F453, which is readily accessible in all conformations.
DISCUSSION
One of the fundamental biological processes is energy transduction. Symporters are molecular motors that pump substrates across a membrane by harnessing the potential energy stored in the transmembrane electrochemical gradient. How is this energy transduced into mechanical work? We do know that this mechanism involves conformational changes in the protein, and in this study we have characterized the functional consequences of mutating important residues in the ligand binding sites and how their exposure to the external environment changes with conformation.
Solution of the atomic structure of symporters has been a great advance in our understanding of the molecular details. The “LeuT fold” forms the core of several genetically unrelated Na+ symporter families, suggesting that they share a common mechanism, which is certainly the case for hSGLT1 and hGAT1 (22). To date, this structural family includes four Na+ cotransporters, LeuT, vSGLT, Mhp1, and BetP, and three Na+-independent members, AdiC (7), CaiT (41), and ApcT (42). The four Na+ cotransporters have been crystallized in four different conformations, clearly supporting an alternating access mechanism where the substrate goes through an occluded state as it passes through the protein. The implications are that substrate binding is ordered, and a coordinated opening and closing of the inner and outer gates is responsible for stoichiometric ion and cosubstrate transport. Each structure, however, is only a snapshot representing a single moment in a series of steps that comprise the mechanism, and our goal is to understand the atomic mechanism by which cotransport occurs by integrating structure with kinetics.
Our previous biochemical and biophysical studies have resulted in a quantitative kinetic model of hSGLT1 with a minimum of six conformational states. The probability of each conformation is highly dependent on the membrane potential and on Na+ and glucose concentrations on each side of the membrane (Fig. 1) (25, 37). In the absence of Na+ and sugar, at a resting membrane potential of −50 mV, most of the proteins are in the inward-facing empty (ligand free) conformations ([C1] and [C6]). Under normal physiological conditions (high [Na+] outside, low [Na+] inside, and a resting membrane potential of −50 mV), the transporter is approximately equally distributed between the inward-facing empty ([C6]) and outward-facing Na+-bound conformations ([C2Na2]). When sugar is added to the external solution, SGLT1 is able to undergo the complete transport cycle, with the predominant conformation governed by the slowest transition rate in the cycle, predicted to be the internal release of Na+ from [C5Na2]. If the nontransported competitive inhibitor phlorizin is added to the external medium, the predominant conformation shifts from [C2Na2] to a dead-end state, [C3Na2Pz], a version of the substrate-bound conformation. Thus, by changing external ligands, we can alter the probability of the transporter residing in each conformation (25).
In this study, we examined the role of the hSGLT1 residues identified as the gates and ligand binding residues based on homology to vSGLT (6, 45). Of the 17 residues modeled to be involved in ligand coordination and gating, only one has been previously identified, Q457 (12, 24). Here, we report on 11 of the new mutants that were identified: 5 were not studied because either they were not conserved (A105 and T287) or bonding involved backbone carbonyls (A62, I65, and S389). Each of the 12 conserved residues was individually mutated to cysteine, and the kinetics of the mutated hSGLT1 were measured to determine the functional effect of modification of the side chain. In addition, we determined how exposure of these cysteine residues changed with protein conformation during the transport cycle. The conformational changes are those predicted by homology modeling. Functional effects are consistent with the predicted functional roles, and the accessibility data provide some limits for how exposure of some residues can change in different states. Our data suggests that “gate” residues contribute directly to the coupling between substrate and Na+ transport.
An Interpretation of Changes in Kinetics
What are the implications for a change in binding site structure? Under equilibrium conditions, the energetics of glucose binding to SGLT1 is described by the Gibbs equation: ΔG = −RT·ln(K), relating the Gibbs free energy (G) and the binding or association constant (K). The equation leads to K1/K2 = exp[−Δ(ΔG)/RT], relating the ratio of the binding constants with the change in free energy, where K1 and K2 refer to the binding constants for wild-type and mutant proteins. Thus a loss of one hydrogen bond (e.g., ≈5 kcal/mol) between glucose and the binding site would decrease the sugar binding constant by over 300-fold. The homology model of hSGLT1 indicates that residues H83 and E102 coordinate sugar binding at O2 and O3 (Fig. 3B), so we would expect a mutation that eliminated one of these hydrogen bonds to result in a large increase in sugar K0.5: for H83C, K0.5αMDG >> 100 mM, and for E102C, there is very low affinity since 100 mM αMDG did not generate a current. Two other mutants, Y290C and S392C, also exhibited sugar K0.5 values >>100 mM. The large increase in K0.5αMDG with mutations of sugar coordinating residues (Table 2) is in agreement with the loss in sugar affinity when the equatorial hydroxyl group at C2 or C3 in the pyranose ring is removed (47). When the hydrogen bonds are absent, as in the case of 2-deoxy-d-glucose and 3-deoxy-d-glucose, large (>>200-fold) reductions in apparent affinity for sugar have been reported for hSGLT1 (44).
The high K0.5 value for S392C is surprising because it is not directly involved in sugar binding (Fig. 2B). One explanation is that this is due to a loss in Na+ affinity, and another is that S392 could be involved in aligning the residues that form the sugar binding site. No functional effects were observed with the S393 mutations, and this means that this residue is not likely to be important in coordination of Na+ to SGLT1. Y290 is the “stacking” residue, in which its π electrons interact with the hydrogens comprising the hydrophobic face of the sugar (13, 32). Loss of the multiple weak hydrogen bonds (CH/π interactions) may produce a 200-fold increase in the sugar K0.5. Overall, the loss of hydrogen bonding between the protein and sugar, whether it be the loss of a protein coordinating residue or removal of a hydroxyl group from the sugar, produces a similar large loss in apparent affinity.
A likely explanation for the 10-fold increase in K0.5 for N78C comes from our molecular dynamics simulations and mutational analysis on vSGLT, which predict the hydroxyl at tyrosine 290 is important in controlling release of the sugar from its binding site into the inner vestibule of the protein (45). The aromatic ring of Y290 is placed for interaction with the sugar by a hydrogen bond between N78 and the hydroxyl from Y290, and dissociation of this bond is the action that permits the Y290 side chain to swing away and expose the binding site to the inside of the cell. Loss of this hydrogen bond could account for the 10-fold increase in sugar K0.5.
Several other mutants had a more modest effect on the sugar K0.5. According to the homology model, Q457 forms a hydrogen bond with the sugar at O5, but Q457C shows only a 25-fold reduction in the sugar K0.5. One explanation is that the missing hydrogen bond might be partially compensated by interactions with water. Modification of the predicted outer gate residues L87C and F453C increased K0.5αMDG by only 4- to 10-fold. These relatively small changes in K0.5αMDG are consistent with the prediction that the external gate residues are not directly involved in sugar binding.
It is also important to note that the large changes in sugar K0.5 reported here for ligand binding residues (Table 2) are quite specific, because such changes were not observed for many other SGLT1 mutants we and others have characterized (Fig. 8C). Relatively modest changes in the sugar K0.5 (<10-fold) did occur with A166 and T460 (12, 31). This is now expected, since inspection of the structure suggests that T460 is in the sugar translocation pathway, and mutation of A166 (in TM3, adjacent to TM1 and TM10) affects sugar specificity (31).
When interpreting changes in sugar and Na+ K0.5 values, it is important to remember that these are only apparent binding affinities. Because Na+ and glucose are transported across the membrane simultaneously via a conformational change of the fully loaded protein (C3 → C4, Fig. 1), both K0.5αMDG and K0.5Na are phenomenological parameters dependent on all the rate constants of the transport cycle. Specifically, for the simple six-state kinetic model for SGLT1, K0.5αMDG and K0.5Na are functions of all of the 14 rate constants and 2 dielectric coefficients (see Eqs. A39 and A45 in Ref. 37). The interdependence of the apparent substrate affinities is demonstrated by the linear relationship between K0.5αMDG and K0.5Na (Fig. 5A).
Turnover and Stoichiometry
In the case of tight coupling between Na+ and sugar, the ratio Imax/Qmax in single cells provides a measure of the catalytic turnover rate for SGLT1. For WT hSGLT1, the Na+-glucose stoichiometry is 2:1, and the turnover number (Imax/Qmax) is ≈50 s−1 (Tables 2 and 3) (47). The I-V curves of the outer gate mutants (L87C, F101C, and F453C) and S392C did not saturate with large hyperpolarizing membrane voltages, and so it is problematic to estimate their maximum cotransport rate (Imax). The data, however, suggest that there are large increases in Imax/Qmax for the outer and inner gate mutants and the Na+ binding mutant S392C/A (Table 2). Direct measurement of stoichiometry shows that for at least the outer gate mutants, this is actually due to uncoupling of Na+ and sugar cotransport (Table 3). We conclude that gate side chains are not simply barriers but are important for coordinating conformational changes, and thus efficient coupling of Na+ and sugar transport by hSGLT1.
Cysteine Mutants and Accessibility
The cysteine residues in the transport pathway were employed for accessibility studies using SCAM (17). We found that derivatization of the cysteine by MTSEA or MTSES completely inhibited transport in 7 of the 12 mutants, 4 gate residues and 3 of the sugar coordinating residues (Table 4). The putative Na+ coordinating residues S392C and S393C were poorly accessible to even the smallest MTS reagent (MeMTS) under all experimental conditions, and the low expression of E102C and K321C made definitive determination of the effects of labeling on these mutants difficult. Nevertheless, our results show that the side chain of residues in the sugar transport pathway formed by TM1, TM2, TM6, and TM7 in the outward open conformation (Fig. 8A) is important for cotransport, in contrast to those in other positions in the protein. As summarized in Fig. 8C, only five other cysteine mutants were sensitive to MTS reagents (see also Ref. 12).
Alternating Access
Na+-sugar cotransport by SGLT1 occurs via a series of conformational changes triggered by ion and substrate binding. In a transport cycle, the substrate binding site is alternatively exposed to the external and internal environments, and the substrate is never exposed to both sides simultaneously, alternating access. Comparison of the outward open and inward occluded hSGLT1 homology models suggests large conformational changes, and this is confirmed by the SCAM results with the residues modeled to be in the sugar binding site. All are highly accessible in the [C2Na2] state but differ in their accessibility in other conformations. Although we cannot translate these differences into structural motion, they will provide constraints on how the transitions occur. For example, the outer gate residue F101C is equally accessible in [C6] and [C2Na2], suggesting that the change in conformation between the two states does not change the exposure of F101. In the phlorizin-bound conformation, [C3Na2Pz], F101C is protected, so it is not exposed to the external solution. Y290C was accessible only in [C2Na2], but F453C was exposed to the same degree in all conformations. It does have to be kept in mind that changes in accessibility produced by the addition of ligands may be due to steric hindrance by sugar or phlorizin, but it is unlikely that this accounts for differences in the presence and absence of Na+.
Although all of the binding site residues are exposed in [C2Na2], access to the sites was not equivalent. Reaction rates vary widely; for example, the estimated reaction rate for Q457C to MTSEA was 14,000 M−1·s−1, and that for F101C was 500 M−1·s−1 (Table 3).
How specific are accessibility results to the sugar transport pathway? This can be judged from the results of previous studies of over 30 SGLT1 cysteine mutants (see Fig. 8C) where there were no reported changes in transporter kinetics, thus emphasizing the specificity of the Na+ and sugar binding residues. Only six mutants were either inhibited by MTS reagents (F163C, A166C, I443C, T460C, A468C, D454C, and R499C) and/or labeled by TMR in a conformationally sensitive manner (I443C, Q445C, T460C, G507C, and L527C) (see Ref. 12). Studies are in progress to more finely map the conformational changes using accessibility measurements.
Structural Interpretation of Protein Motion
The SCAM results provide a framework for evaluating the hSGLT1 homology models. The Na+-bound outward-facing open model [C2Na2] is shown viewed from the extracellular compartment in Fig. 8A. A similar view of the closed conformation from the same perspective is shown in Fig. 8B. We have high confidence in the occluded model (Fig. 8B) because hSGLT1 and vSGLT share high amino acid sequence identity and similarity (32 and 60%) and the gate and ligand coordinating residues are conserved. They are also functionally homologous and have similar kinetics and conserved key residues. Furthermore, treatment of homologous mutants with MTS reagents blocks transport, e.g., Q457C (hSGLT1) and Q423C (vSGLT) (24, 49).
The spatial relationship of the residues changes dramatically between the two conformations. On each structural model we have highlighted the location of the cysteine mutants and their accessibility to MTS reagents: green, accessible; red, inaccessible; and pink, not measureable due to low expression and/or incomplete reactivity (Fig. 8). In the outward open model ([C2Na2], Fig. 8A), all of the external and internal gate residues (L87C, F101C, F453C, and Y290) and the sugar coordinating residues are accessible to external MTS reagents. In agreement with the SCAM results, S392C and S393C are accessible from the extracellular face, but only by the smallest reagent, MeMTS, suggesting that the access pathway is highly restricted. In the inward occluded conformation, all of the positions except the outer gate, F453C, are no longer accessible in the model, as observed experimentally (Fig. 6). Thus conformational dependence of the interactions of MTS reagents with hSGLT1 agrees with the predicted structure of hSGLT1 in the outward-facing open and the inward-facing conformations.
Assuming that the phlorizin-bound conformation is an occluded conformation approximating [C3Na2S], there is sufficient space within the inward facing hSGLT1 model to dock a phlorizin molecule into the glucose binding site (not shown). The glucose moiety of phlorizin probably binds to the same sugar site as glucose given the linear relationships between KiPz and K0.5αMDG (Fig. 5B) over several orders of magnitude; i.e., mutating the sugar coordinating residues reduces both sugar and phlorizin binding. KiPz for the F101C mutant deviates from the KiPz vs. K0.5αMDG relationship, suggesting that the A-phenolic ring of phlorizin is in close proximity to F101.
By comparing the outward-facing open and inward-facing occluded or open models of hSGLT1 (Fig. 8), and from insights gained from the structure of vSGLT from inward occluded to inward open conformations (45), we can make predictions about the structural changes that occur during and after external sugar binding. Basically, the motion is reminiscent of a rocking-bundle model [e.g., Forrest and Rudnick (8)], where two groups of helices move as rigid bodies relative to each other as the transporter alternates between outward- and inward- facing conformations. In the transition from outward-facing open to the inward-facing occluded conformations, the “sugar bundle” (TM2, TM6, and TM7) and sugar-interacting residues move closer to the sugar, and the outer gates move to block entry to, or exit from, the sugar binding site. The “hash bundle” consisting of TM3, TM4, TM8, and TM9, moves toward the sugar site, but TM8 contains three residues of the Na2 site, which is preserved in the inward occluded form, so that region must remain stationary. For vSGLT, the subsequent transition from inward occluded to inward open is accomplished by a rotation of the hash bundle away from the Na2 site, and Na+ exits the Na2 site to the cytoplasm. The sugar bundle then rotates in the opposite direction to the hash bundle, and the sugar interacting residues move away from the sugar. Thus, closing of the outside sugar vestibule results in opening of the inside vestibule.
To break it down further, how does sugar bind, and how does this event induce the conformational changes that result in occlusion? How do the sugar coordinating residues come within the 3-Å bonding distance to each of the sugar polar groups? Is it an induced fit? Does this occur as an all-or-none event, or is there an order to the formation of each bond? Does the sugar enter the external hydrophilic site in a preferred orientation, C4 end first? hSGLT transports large glucosides, e.g., indican (5 × 12 × 20 Å), so it is most probable that the C4 end of the sugars and glucosides enter the binding site first. Is there a preferred sequence to the removal of the sugar and protein shells of hydration? Do these bulky glycosides inhibit transport by preventing formation of the occluded state?
Only one of the two putative Na+ binding sites in hSGLT1, the Na2 site, has been tentatively identified by homology to vSGLT and LeuT (6, 50). The Na+ is thought to be coordinated with the hydroxyl side chains of S392 and S393 and with the carbonyl oxygens of A76 and I79. Mutation of S392 produced the expected reduction in Na+ affinity but in addition dramatically reduced the affinity for sugar (K0.5αMDG > 100 mM). On the other hand, mutation of S393 did not profoundly alter the transport kinetics (Table 2), showing that it is not an important Na+ coordinating residue. A surprising finding was that mutants H83C, Y290C, and W291 all have a low affinity for Na+ in addition to their low affinity for sugar. Our interpretation of these results is that S392 is involved in Na+ coordination at the Na2 site and that H83C, Y290C, and W291 form part of the Na1 site as in LeuT (50). However, to definitively the identify Na+ binding sites, it is necessary to obtain higher resolution crystal structures (see Ref. 47 for further discussion).
Additional crystal structures and functional assays are required to describe the full transport cycle of hSGLT1, including the structures of hSGLT1 in the outward open, inhibitor-bound, and ligand-free conformations. Perhaps the most pressing for the LeuT structural family is to understand how external Na+ promotes the formation of the outward open conformation to permit ligand and inhibitor binding. Hints on the structural changes emerge from the accessibility measurements in the presence and absence of Na+ (Fig. 7 and Ref. 12). Compared with the protein in the outward open Na+-bound conformation [C2Na2], there is poor access to N78C (TM1), H83C (TM1), L87C (TM1), Q445C (TM9), Q457C (TM10), T460C (TM10), A468C (TM10), and R499C (TM11) in the absence of Na+ (the [C1] or [C6] conformation). On the other hand, there was no significant difference in accessibility of F101C (TM2), I443C (TM9), F453 (TM10), G507C (TM11), L527C (TM12), and Y528C (TM12) in the two conformations. Overall, from these results we suggest that the conformation of the ligand-free transporter resembles the inward-facing occluded conformation [C4Na2S] (Fig. 8B).
In summary, there were two aims in this study. Our first aim was to correlate the mechanism of hSGLT1 with its structure by focusing on the conserved ligand binding and gate residues identified in the bacterial vSGLT structure. We carried out a comprehensive kinetic analysis of 12 mutants where we measured K0.5 and Imax for both sugar and Na+, the density of mutant transporters in the plasma membrane, the apparent turnover number (Imax/Qmax), Ki for phlorizin inhibition, and the Na+-to-sugar coupling ratio for the outer gate mutants. Strong conclusions are drawn about the mechanism of Na+-sugar cotransport: 1) mutations of sugar binding residues produce dramatic reductions in apparent sugar affinity, in some cases, greater than 200-fold, supporting previous predictions based on the kinetics of deoxy-sugar transport; 2) mutations of one of the two serines in the putative Na2 binding site dramatically reduce apparent Na+ affinity, but a major surprise is that one of the two serines in the Na2 site is not important for Na+ binding; 3) evidence is provided for phlorizin and glucose to bind at the same site and that residue F101 is involved in binding in the aglycone portion of the molecule; and 4) the residues identified as gates, forming the barrier between sugar and the external milieu, have a role in coupling sugar to Na+ transport.
The second aim was to examine the conformational changes of hSGLT1 with Na+-sugar cotransport. Our accessibility studies indicate that external Na+ binding to the transporter results in “opening” of the sugar binding vestibule to the external membrane compartment, and the low apparent affinity of the transporter for sugar in the absence of Na+ is due to inaccessibility of glucose to the binding site. After sugar binding, the outer gates close, and the sugar binding sites become inaccessible or occluded to the external membrane surface. A rate-limiting step for sugar transport is located on the internal membrane surface, since the sugar binding sites are inaccessible under saturating concentrations of external sugar. Opening of the sugar binding vestibule is specific, since studies on numerous residues in other parts of hSGLT1 do not produce the large changes in K0.5 for Na+ and sugar and the same pattern of dependence of accessibility on Na+ and sugar concentrations as residues in the sugar binding vestibule. Finally, the accessibility results are consistent with homology models and provide insight into the dynamics of conformational changes underlying Na+ and sugar cotransport.
GRANTS
This work was supported by National Institutes of Health Grants DK19567 (to E. M. Wright) and GM78844 (to J. Abramson) and American Heart Association Grant 630258N (to J. Abramson).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.S.-R., B.A.H., D.D.L., V.C., J.A., and E.M.W. conception and design of research; M.S.-R., B.A.H., D.D.L., and V.C. performed experiments; M.S.-R., B.A.H., D.D.L., V.C., J.A., and E.M.W. interpreted results of experiments; M.S.-R., B.A.H., D.D.L., V.C., J.A., and E.M.W. edited and revised manuscript; M.S.-R., B.A.H., D.D.L., V.C., J.A., and E.M.W. approved final version of manuscript; B.A.H., D.D.L., and E.M.W. analyzed data; B.A.H., D.D.L., V.C., J.A., and E.M.W. prepared figures; D.D.L. and E.M.W. drafted manuscript.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Salam Faham's early contributions to homology modeling of hSGLT1 and the docking of inhibitors.
Present address of M. Sala-Rabanal: Department of Cell Biology and Physiology, and Center for the Investigation of Membrane Excitability Diseases, Washington University, St. Louis, MO 63110.
Present address of V. Chaptal: Institut de Biologie et Chimie des Protéines, Equipe LPRAC, 7 passage du Vercors, 69007 Lyon, France.
REFERENCES
- 1. The CCP4 suite: programs for protein crystallography Acta Crystallogr D Biol Crystallogr 50: 760–763, 1994 [DOI] [PubMed] [Google Scholar]
- 2.Abramson J, Wright EM. Structure and function of Na+-symporters with inverted repeats. Curr Opin Struct Biol 19: 425–432, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez-Sampedro A, Loo DD, Wright EM, Zampighi GA, Hirayama BA. Coupled sodium/glucose cotransport by SGLT1 requires a negative charge at position 454. Biochemistry 43: 13175–13184, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006, chapt. 5, unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321: 810–814, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature 460: 1041–1043, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA 105: 10338–10343, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frillingos S, Sahin-Toth M, Persson B, Kaback HR. Cysteine-scanning mutagenesis of putative helix VII in the lactose permease of Escherichia coli. Biochemistry 33: 8074–8081, 1994 [DOI] [PubMed] [Google Scholar]
- 10.Hediger MA, Turk E, Wright EM. Homology of the human intestinal Na+/glucose and Escherichia coli Na+/proline cotransporters. Proc Natl Acad Sci USA 86: 5748–5752, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirayama BA, Diez-Sampedro A, Wright EM. Common mechanisms of inhibition for the Na+/glucose (hSGLT1) and Na+/Cl−/GABA (hGAT1) cotransporters. Br J Pharmacol 134: 484–495, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirayama BA, Loo DD, Diez-Sampedro A, Leung DW, Meinild AK, Lai-Bing M, Turk E, Wright EM. Sodium-dependent reorganization of the sugar-binding site of SGLT1. Biochemistry 46: 13391–13406, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Hodel AE, Quiocho FA. Protein-ligand interactions. In: International Tables for Crystallography, Volume F: Crystallography of Biological Macromolecules, edited by Rossmann MG, Arnold E. New York: Springer, 2006, p. 579–587 [Google Scholar]
- 14.Huntley SA, Krofchick D, Silverman M. A glutamine to glutamate mutation at position 170 (Q170E) in the rabbit Na+/glucose cotransporter, rSGLT1, enhances binding affinity for Na+. Biochemistry 45: 4653–4663, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Huntley SA, Krofchick D, Silverman M. Position 170 of rabbit Na+/glucose cotransporter (rSGLT1) lies in the Na+ pathway; modulation of polarity/charge at this site regulates charge transfer and carrier turnover. Biophys J 87: 295–310, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda TS, Hwang ES, Coady MJ, Hirayama BA, Hediger MA, Wright EM. Characterization of a Na+/glucose cotransporter cloned from rabbit small intestine. J Membr Biol 110: 87–95, 1989 [DOI] [PubMed] [Google Scholar]
- 17.Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol 293: 123–145, 1998 [DOI] [PubMed] [Google Scholar]
- 18.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature 459: 347–355, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Lo B, Speight P, Silverman M. Transmembrane IV of the high-affinity sodium-glucose cotransporter participates in sugar binding. Am J Physiol Cell Physiol 295: C64–C72, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Speight P, Silverman M. Reanalysis of structure/function correlations in the region of transmembrane segments 4 and 5 of the rabbit sodium/glucose cotransporter. Biochem Biophys Res Commun 378: 133–138, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Lo B, Silverman M. Cysteine scanning mutagenesis of the segment between putative transmembrane helices IV and V of the high affinity Na+/Glucose cotransporter SGLT1. Evidence that this region participates in the Na+ and voltage dependence of the transporter. J Biol Chem 273: 29341–29351, 1998 [DOI] [PubMed] [Google Scholar]
- 22.Loo DD, Eskandari S, Boorer KJ, Sarkar HK, Wright EM. Role of Cl− in electrogenic Na+-coupled cotransporters GAT1 and SGLT1. J Biol Chem 275: 37414–37422, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Loo DD, Hazama A, Supplisson S, Turk E, Wright EM. Relaxation kinetics of the Na+/glucose cotransporter. Proc Natl Acad Sci USA 90: 5767–5771, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo DD, Hirayama BA, Gallardo EM, Lam JT, Turk E, Wright EM. Conformational changes couple Na+ and glucose transport. Proc Natl Acad Sci USA 95: 7789–7794, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo DD, Hirayama BA, Karakossian MH, Meinild AK, Wright EM. Conformational dynamics of hSGLT1 during Na+/glucose cotransport. J Gen Physiol 128: 701–720, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo DD, Hirayama BA, Meinild AK, Chandy G, Zeuthen T, Wright EM. Passive water and ion transport by cotransporters. J Physiol 518: 195–202, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loo DD, Hirayama BA, Sala-Rabanal M, Wright EM. How drugs interact with transporters: SGLT1 as a model. J Membr Biol 223: 87–106, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Mackenzie B, Loo DD, Wright EM. Relationships between Na+/glucose cotransporter (SGLT1) currents and fluxes. J Membr Biol 162: 101–106, 1998 [DOI] [PubMed] [Google Scholar]
- 30.Martin MG, Turk E, Lostao MP, Kerner C, Wright EM. Defects in Na+/glucose cotransporter (SGLT1) trafficking and function cause glucose-galactose malabsorption. Nat Genet 12: 216–220, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Meinild AK, Loo DD, Hirayama BA, Gallardo E, Wright EM. Evidence for the involvement of Ala 166 in coupling Na+ to sugar transport through the human Na+/glucose cotransporter. Biochemistry 40: 11897–11904, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Morales JC, Reina JJ, Diaz I, Avino A, Nieto PM, Eritja R. Experimental measurement of carbohydrate-aromatic stacking in water by using a dangling-ended DNA model system. Chemistry 14: 7828–7835, 2008 [DOI] [PubMed] [Google Scholar]
- 33.Noskov SY, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J Mol Biol 377: 804–818, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panayotova-Heiermann M, Loo DD, Lam JT, Wright EM. Neutralization of conservative charged transmembrane residues in the Na+/glucose cotransporter SGLT1. Biochemistry 37: 10522–10528, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Panayotova-Heiermann M, Loo DD, Lostao MP, Wright EM. Sodium/d-glucose cotransporter charge movements involve polar residues. J Biol Chem 269: 21016–21020, 1994 [PubMed] [Google Scholar]
- 36.Parent L, Supplisson S, Loo DD, Wright EM. Electrogenic properties of the cloned Na+/glucose cotransporter: I. Voltage-clamp studies. J Membr Biol 125: 49–62, 1992 [DOI] [PubMed] [Google Scholar]
- 37.Parent L, Supplisson S, Loo DD, Wright EM. Electrogenic properties of the cloned Na+/glucose cotransporter: II. A transport model under nonrapid equilibrium conditions. J Membr Biol 125: 63–79, 1992 [DOI] [PubMed] [Google Scholar]
- 39.Quick M, Loo DD, Wright EM. Neutralization of a conserved amino acid residue in the human Na+/glucose transporter (hSGLT1) generates a glucose-gated H+ channel. J Biol Chem 276: 1728–1734, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458: 47–52, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Schulze S, Koster S, Geldmacher U, Terwisscha van Scheltinga AC, Kuhlbrandt W. Structural basis of Na+-independent and cooperative substrate/product antiport in CaiT. Nature 467: 233–236 [DOI] [PubMed] [Google Scholar]
- 42.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325: 1010–1014, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MS, Iwata S, Henderson PJ, Cameron AD. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voss AA, Diez-Sampedro A, Hirayama BA, Loo DD, Wright EM. Imino sugars are potent agonists of the human glucose sensor SGLT3. Mol Pharmacol 71: 628–634, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Watanabe A, Choe S, Chaptal V, Rosenberg JM, Wright EM, Grabe M, Abramson J. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468: 988–991, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J, Ma P, Saidijam M, Patching SG, Hope RJ, Norbertczak HT, Roach PC, Iwata S, Henderson PJ, Cameron AD. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322: 709–713, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright EM, Loo Donald DF, Hirayama Bruce A. Biology of human sodium glucose transporters. Physiol Rev 91: 733–794, 2011 [DOI] [PubMed] [Google Scholar]
- 48.Wright EM, Turk E. The sodium/glucose cotransport family SLC5. Pflügers Arch 447: 510–518, 2004 [DOI] [PubMed] [Google Scholar]
- 49.Xie Z, Turk E, Wright EM. Characterization of the Vibrio parahaemolyticus Na+/glucose cotransporter. A bacterial member of the sodium/glucose transporter (SGLT) family. J Biol Chem 275: 25959–25964, 2000 [DOI] [PubMed] [Google Scholar]
- 50.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437: 215–223, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Zampighi GA, Kreman M, Boorer KJ, Loo DD, Bezanilla F, Chandy G, Hall JE, Wright EM. A method for determining the unitary functional capacity of cloned channels and transporters expressed in Xenopus laevis oocytes. J Membr Biol 148: 65–78, 1995 [DOI] [PubMed] [Google Scholar]