Abstract
Although several major histocompatibility complex (MHC)-wide single-nucleotide polymorphism (SNP) studies have been performed in populations of European descent, none have been performed in Asian populations. The objective of this study was to identify human leukocyte antigen (HLA) loci associated with multiple sclerosis (MS) in a Japanese population genotyped for 3534 MHC region SNPs. Using a logistic regression model, two SNPs (MHC Class III SNP rs422951 in the NOTCH4 gene and MHC Class II SNP rs3997849, susceptible alleles A and G, respectively) were independently associated with MS susceptibility (204 patients; 280 controls), two (MHC Class II SNP rs660895 and MHC Class I SNP rs2269704 in the NRM gene, susceptible alleles G and G, respectively) with aquaporin-4− (AQP4−) MS susceptibility (149 patients; 280 controls) and a single SNP (MHC Class II SNP rs1694112, susceptible allele G) was significant when contrasting AQP4 + against AQP4− patients. Haplotype analysis revealed a large susceptible association, likely DRB1*04 or a locus included in the DRB1*04 haplotype, with AQP4− MS, which excluded DRB1*15:01. This study is the largest study of the HLA’s contribution to MS in Japanese individuals.
Keywords: multiple sclerosis, HLA, Japanese, MHC, AQP4
Introduction
Multiple sclerosis (MS) is the prototypic disease of central nervous system (CNS) myelin and is considered to be autoimmune in origin. Although the exact cause of MS is unknown, there is an unequivocal, if partial, genetic contribution to its pathogenesis.1,2 Although several loci with modest replicable effects on MS susceptibility have been identified (mostly in studies of populations of European descent), the only genetic region identified with a large, consistently replicated effect in all populations is the human leukocyte antigen (HLA) region on chromosome 6p21.3. The primary signal arises from the HLA-DRB1 gene in the Class II segment of the locus, more specifically the *15:01 allele of DRB1, but complex hierarchical allelic effects, copy number and cis–trans regulatory interactions across the entire region, including a protective signal in the Class I region, have been reported as well.3–9
MS is relatively rare in Asians, but clinical heterogeneity is worth noting. Some have a disease, termed conventional MS (CMS), which is indistinguishable from MS in western countries (disseminated lesions in the CNS), whereas others have a variant, termed opticospinal MS (OSMS), which involves predominantly the optic nerve and spinal cord.10 The exact relationship between CMS and OSMS is uncertain; OSMS might represent a true variant of CMS or a phenocopy that is biologically unrelated to CMS. HLA data suggest that the two forms are immunogenetically distinct. In studies of the HLA in Japanese MS populations, CMS was associated with HLA-DRB1*15:01,10 whereas OSMS was associated with the centromeric HLA-DPB1 locus,10–12 both Class II major histocompatibility complex (MHC) genes. Recently, autoantibodies against the cell membrane water channel aquaporin-4 (AQP4), a specific biomarker for neuromyelitis optica (NMO), were identified in a proportion of patients with OSMS, leading to a reclassification of this entity based on seropositivity to AQP4. Because the distinction between MS and NMO (an inflammatory disease affecting only the optic nerves and spinal cord) and their etiologies is not clear, especially in the Japanese population where there is a higher relative prevalence of OSMS,13 a well-defined biomarker such as AQP4 seropositivity may be more useful for stratification for genetic analyses. Indeed, in Japanese populations, HLA-DRB1*12 was found to be a risk factor for anti-AQP4 antibody-positive patients, but not antibody-negative MS.14 However, a comprehensive investigation of the entire HLA region in Japanese MS has yet to be performed.
The advent of large-scale single-nucleotide polymorphism (SNP)-based genotyping allowed broad analyses of the HLA region in MS. For example, several whole-genome association studies have been completed in individuals of European descent,15–23 which included thousands of SNPs in the HLA region. Most of these studies validated the strong Class II HLA-DRB1*15:01 association with disease susceptibility, as well as HLA Class I20,24 and a DRB1*03:01-DQB1*02:01 (ref. 23) associations. In addition, a few studies have focused exclusively on SNPs in the HLA region using customized arrays, confirming the presence of a disease locus in the Class I region.6,9,24 To date, significant Class I associations have not been identified in Japanese MS. The unique patterns of linkage disequilibrium (LD) between different ethnic groups have represented a powerful means to delineate causative disease-associated variants in the HLA region.25–27 The objective of this study is to identify HLA loci associated with MS in a Japanese data set using a high-density SNP array. In addition, because of the complexities of defining disease subclasses and the resulting sizes of those subclasses in this population, a biomarker (presence or absence of autoantibodies against AQP-4) was used instead to stratify the population for further refinement of genetic associations.
Results
Population parameters
A total of 280 healthy controls (HC) and 204 individuals with MS with data for 3534 HLA region SNPs remained after all quality control (QC) steps (Table 1). In all, 46% of the HC and 75% of the individuals with MS were women (gender was included as a covariate for all analyses). A total of 55 of all patients were positive for anti-AQP4 antibody (AQP4 +). Among 193 cases that fulfill the revised McDonald criteria,28 51 cases (26.4%) are positive for the anti-AQP4 antibody.
Table 1.
Clinical phenotypes
| Anti-AQP4Ab+ (n = 55)
|
Anti-AQP4Ab− (n = 149)
|
|||
|---|---|---|---|---|
| NMOa+ (n = 38) | NMO− (n = 17) | NMO+ (n = 8) | NMO− (n = 141) | |
| McDonald criteriab (+)(n = 193) | 35 | 16 | 8 | 134 |
| CMS (n = 110) | 9 | 9 | 1 | 91 |
| OSMS (n = 52) | 25 | 3 | 7 | 17 |
| Othersc (n = 31) | 1 | 4 | 0 | 26 |
| McDonald criteria (−) and | 3 | 1 | 0 | 7 |
| criteria for CISd (+) (n = 11) | ||||
Abbreviations: Ab, antibody; AQP4, aquaporin-4; CIS, clinically isolated syndrdome; CMS, conventional multiple sclerosis; NMO, neuromyelitis optica; OSMS, opticospinal multiple sclerosis.
The revised criteria for NMO by Wingerchuk et al.50
The revised McDonald criteria by Polman et al.28
Those cases who fulfill the revised McDonald criteria, but did not meet the criteria for CMS or OSMS.
CIS criteria by Dalton et al.41
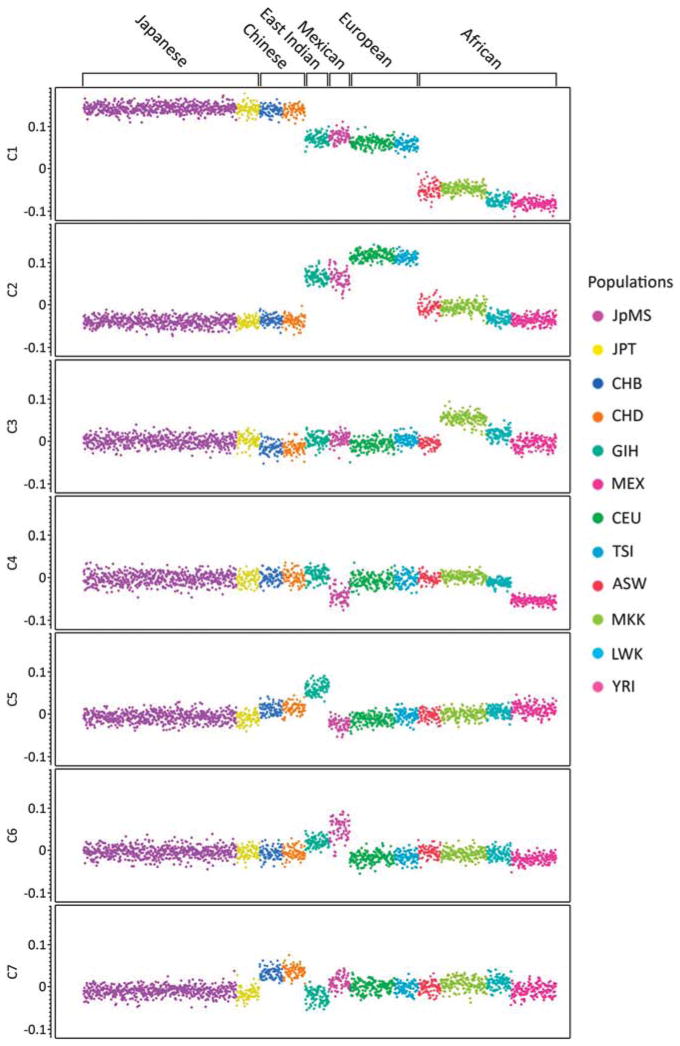
Multidimensional scaling was used to determine the ethnic relationship between the Japanese MS cohort and 12 HapMap populations. By plotting the first by the second dimension, all patients and controls cluster with the HapMap Japanese and Chinese populations (Supplementary Figure 1), verifying that they are indeed East Asian. Plotting all informative dimensions (n =7) separately, patients and controls always cluster with the HapMap Japanese population, including dimension 7, which separates the Chinese from the Japanese populations (Figure 1), verifying that the experimental sample is non-Chinese East Asian (genotyping data for other East Asian populations was not available).
Figure 1.
Plot of each of the first seven multidimensional scaling dimensions individually. The Japanese MS (JpMS) sample always clusters with the HapMap Japanese population, including dimension 7, which separates the Chinese from the Japanese populations. The y axis is the value for each of the first seven dimensions. ASW, African ancestry in Southwest USA; CEU, CEPH (NW European); CHB, Han Chinese in Beijing, China; CHD, Chinese in Denver, Colorado; GIH, Gujarati Indians in Houston, Texas; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, California; MKK, Maasai in Kinyawa, Kenya; TSI, Tuscan in Italy; YRI, Yoruban in Ibadan, Nigeria.
Association analyses
The two SNPs typically used to infer DRB1*15 status (rs3135388 and rs9271366 (ref. 29)) were not useful for determining *15:01 carrier status and copy number in this data set. The SNP rs3135388 is fixed for the G allele in the current population, as well as the HapMap Japanese population, and rs9271366 captured in our data set both the *15:01 and *15:02 alleles. The association between rs9271366 and all MS and AQP4− MS was not significant (comparison-wise P = 0.33 and 0.15, respectively). Across the (classical) HLA-typed individuals, 48% of the *15 alleles in the data set were *15:02, indicating that the *15:02 alleles are likely diluting the risk effect of the *15:01 alleles for this particular SNP. Therefore, only DRB1*15:01 presence or absence data, from DRB1*15:01-specific primers,25 was used for the experimental sample and was moderately associated with MS (P = 0.014) and AQP4− MS (P = 0.01); however, this association was modest compared with the SNP associations (see below: trend test, P = 10−6–10−7).
Two SNPs were associated with MS using the iterative model (Table 2). Rs422951 in the Class III region had the most significant association with MS (P = 2.9 × 10−6; odds ratio (OR) = 0.4; 32 296 360 bp) (Supplementary Figure 2A). This SNP results in a missense mutation in NOTCH4. After fitting rs422951 into the model, rs3997849 was the most significant SNP (P = 8.1 × 10−5; OR = 0.5; 32 790 379 bp). The SNP rs3997849 is in the HLA Class II region, closest to the HLA-DQA2 gene (26 761 bp away). When fitting both SNPs as covariates in the model, no other SNPs were significant for MS at false discovery rate (FDR) P<0.1.
Table 2.
SNPs significantly (FDR P = 0.1) associated with MS in the Japanese sample
| Comparison | N SNPs in model | SNP | Raw P-value | FDR P | Odds ratio | Susceptible allele | Position | Class | Closest gene | Distance to closest gene (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| MS vs cont. | 0 | rs422951 | 2.94E–06 | 6.08E–03 | 0.4 | A | 32 296 360 | III | NOTCH4 | 0 |
| MS vs cont. | 1 | rs3997849 | 8.07E–05 | 5.59E–02 | 0.5 | G | 32 790 379 | II | HLA-DQA2 | 26 761 |
| AQP4− vs cont. | 0 | rs660895 | 6.23E–07 | 6.61E–04 | 2.23 | G | 32 685 357 | II | HLA-DRB1, HLA-DRB5 | 19 817 |
| AQP4− vs cont. | 1 | rs2269704 | 1.64E–04 | 8.49E–02 | 0.27 | G | 30 764 931 | I | NRM29, NRM | 0 |
| AQP4+ vs AQP4− | 0 | rs1694112 | 1.34E–05 | 4.74E–02 | 3.3 | G | 32 757 641 | II | HLA-DQB1 | 15 197 |
Abbreviations: AQP, aquaporin-4; FDR, false discovery rate; LD, linkage disequilibrium; MS, multiple sclerosis; SNP, single-nucleotide polymorphism.
The most significant SNP from each iteration was fit as a covariate for the remaining iterations (N SNPs in Model) to remove associations resulting from LD with that SNP. Gender was an additional covariate in all iterations. Minor allele is reference for odds ratios. MS vs Cont = all patients (n = 204) vs controls (n = 280); AQP4− vs controls = AQP4− patients (n = 149) vs controls (n = 280); AQP4+ vs AQP4− = AQP4+ patients (n = 55) vs AQP4− patients (n = 149).
Two SNPs were significant for AQP4− patients vs controls using the iterative model (Table 2). SNP rs660895 was significant (P = 6.2 × 10−7; OR = 2.23; 32 685 357 bp) with no SNPs in the model (Supplementary Figure 2B). This SNP is in the Class II region and is closest to the HLA-DRB1 and HLA-DRB5 genes (19 817 bp away). After fitting rs660895 into the model, rs2269704 was the most significant SNP (P = 1.64 × 10−4; OR = 0.27; 30 764 931 bp). This SNP is in the Class I region and is in an intron of the NRM29/NRM gene. After fitting these two SNPs, no other SNPs were significant for AQP4− MS susceptibility at FDR P<0.1.
Only a single SNP, rs1694112, was significant (P = 1.34 × 10−5; OR = 3.30; 32 757 641 bp) when contrasting AQP4− vs AQP4 + patients (Supplementary Figure 2C). This SNP is in the Class II region and is closest to the HLA-DQB1 gene (15 197 bp away). No SNPs were significant (FDR P>0.1) when contrasting AQP4 + patients vs controls.
Haplotype analyses
Haplotype analyses were performed to further clarify the roles of the significant SNPs in tagging causative variation (Table 3). For MS, using a haplotype dosage model, three Class II–class III haplotypes (rs422951–rs3997849: G–G, G–A, A–A) were at least suggestively (comparison-wise P<0.1) associated in the resistant direction (OR <1), and the remaining haplo-type (rs422951–rs3997849: A–G) was significantly associated in the susceptible direction. All three ‘resistant’ haplotypes are significantly associated with resistance when fit together in the multivariate model. For AQP4− demyelinating patients vs controls, two haplotypes (rs660895–rs2269704: A–G, A–A) were significantly associated in the resistant direction, whereas a single haplotype (rs660895–rs2269704: G–G) was associated with susceptibility. Both resistant haplotypes were highly significant when fit in the multivariate model.
Table 3.
Haplotype associations with MS susceptibility
| Comparison | SNPs | Haplotype | OR | P-value | Frequency |
|---|---|---|---|---|---|
| (a) Individual haplotype dosage | |||||
| MS vs controls | rs422951–rs3997849 | GG | 0.41 | 3.46E–06 | 0.18 |
| MS vs controls | rs422951–rs3997849 | AG | 2.45 | 5.17E–10 | 0.6 |
| MS vs controls | rs422951–rs3997849 | GA | 0.33 | 8.46E–02 | 0.01 |
| MS vs controls | rs422951–rs3997849 | AA | 0.62 | 5.18E–03 | 0.21 |
| AQP4− vs controls | rs660895–rs2269704 | GG | 2.32 | 9.04E–08 | 0.3 |
| AQP4− vs controls | rs660895–rs2269704 | AG | 0.71 | 2.12E–02 | 0.6 |
| AQP4− vs controls | rs660895–rs2269704 | GA | 0.43 | 2.76E–01 | 0.01 |
| AQP4− vs controls | rs660895–rs2269704 | AA | 0.22 | 1.93E–06 | 0.09 |
| Comparison | SNPs | Haplotype | OR | P-value |
|---|---|---|---|---|
| (b) Multivariate model | ||||
| MS vs controls | rs422951–rs3997849 | GG | 0.33 | 4.01E–08 |
| MS vs controls | rs422951–rs3997849 | GA | 0.25 | 3.06E–02 |
| MS vs controls | rs422951–rs3997849 | AA | 0.49 | 4.49E–05 |
| AQP4− vs controls | rs660895–rs2269704 | AG | 0.5 | 1.22E–05 |
| AQP4− vs controls | rs660895–rs2269704 | AA | 0.13 | 1.54E–09 |
Abbreviations: AQP, aquaporin-4; MS, multiple sclerosis; OR, odds ratio; SNP, single-nucleotide polymorphism.
Haplotypes formed from the two SNPs significant for each for MS and AQP4− MS. (a) Association of dosage of each haplotype with MS susceptibility (MS vs controls) or AQP4− MS susceptibility (AQP4− vs controls). (b) Multivariate model with all three susceptible haplotypes for all MS and both significant susceptible haplotypes for AQP4− MS. Gender was fit as a covariate in all models.
Haplotype tagging of DRB1 alleles
As the HLA-DRB1 gene, and specifically the *15:01 allele, has been shown in populations of European descent and in Japanese to have an association with MS susceptibility, the two-locus haplotypes associated with MS and the single SNP associated with AQP4− vs AQP4 + patients were scrutinized for their ability to tag DRB1 alleles in the subset of individuals (n = 218) with DRB1 data (Supplementary Table 1; Supplementary Figure 3). For the MS group, the most significantly associated and only susceptible haplotype was AG (rs422951–rs3997849). This haplotype captures 100% of the DRB1*15:01 alleles, which may explain part of the association with MS, but also captures most of the *08:03, *08:02 and *04 alleles. The association of this haplotype with MS could be due to any single allele or combinations of these DRB1 alleles. Interestingly, the 53% of the A–A resistant haplotypes contain DRB1*15:02 alleles, and all of the *15:02 alleles are captured by this haplotype. The GG haplotype, which is the most associated resistant haplotype, is fairly evenly split between three DRB1 alleles: *01:01, *13:02 and *09:01. The G–A haplotype is at such a low frequency in the population (1.4%) that interpretations of DRB1 allele tagging for this haplotype are likely not meaningful.
For the AQP4− vs HC comparisons, the most significantly associated and only susceptible haplotype was GG (rs660895–rs2269704). Interestingly, this haplo-type excludes DRB1*15:01, but includes all of the *08:02 and *12:02 alleles and most of the *04 alleles. The most associated resistant haplotype (A–A) mostly contains *13:02 alleles (sensitivity = 75%; positive predictive value = 75%). The A–G haplotype, which is mildly associated with resistance, captures all of the *15:01 and *15:02 alleles, and the G–A haplotype is too rare to make sound conclusions. Finally, the SNP associated with AQP4− vs AQP4 + does not capture any single DRB1 allele well enough to draw DRB1-specific conclusions.
Discussion
The HLA region has repeatedly shown a strong association with MS in studies of individuals of European descent.2 The HLA-DRB1*15:01 allele, as observed through classical HLA typing and *15:01 tagging SNP studies, is the likely source of the major HLA effect in individuals of European descent. In the Japanese population, which has a higher frequency of OSMS than populations of European descent, the *15:01 allele was found to be associated with CMS only.10 This study investigated the association between 6040 HLA region SNPs with MS in 204 Japanese patients and 280 Japanese controls.
A total of 2506 SNPs were removed from the analysis owing to QC. The majority (65%) of these SNPs were removed because of minor allele frequency <0.05. This is not surprising considering that most known SNPs were discovered in non-Asian populations, and that there is less genetic variation in the Japanese population as compared with many of populations in which the SNPs were identified. The total number of SNPs passing QC and remaining in the analysis was 3534. The reduction in the total number of SNPs from QC decreased the coverage of the MHC region to an average of 707 SNPs per Mb.
Although the number of individuals were modest for an association study (n = 280, 204, 149 and 55 for controls, all patients, AQP4− and AQP4 + patients, respectively), this study is the largest of its kind in Japanese MS. For the main analysis with 204 patients, the power, calculated using the Power for Genetic Association Analyses program30 (co-dominant model, disease prevalence = 0.001, disease allele frequency = 0.2, marker allele frequency = 0.2, effective degrees of freedom = 3429 (calculated using the EDF program included with Power for Genetic Association Analyses), α = 0.1), was good to detect large effects (power = 0.97 for relative risk = 3), but not moderate effects (power = 0.4 for relative risk = 2). Power was considerably less for the other analyses with fewer patients, as expected (data not shown). Consequently, as in any study of the genetics of a complex disease, this study likely does not capture all biologically associated loci or loci with small effects if they are present. The results and discussion herein therefore pertain to those effects that the study had the power to identify.
Fitting the most significant SNP from an MHC-wide analysis as a covariate in subsequent MHC-wide analyses identified two SNPs that were significantly (FDR P = 0.1) associated with MS. It was decided a priori to use an FDR P = 0.1 to strike a balance between power and false negatives. At this significance level, 90% of all significant associations are expected to be true positives. The sequential method allows the determination of multiple SNPs that are associated independent of the LD they share with a more significant SNP. The most significantly associated SNP (rs422951) was in the Class III region and results in a missense mutation (Thr→Ala) in the NOTCH4 gene. NOTCH4 is involved in cell differentiation, proliferation and apoptosis, and has been implicated as a schizophrenia-associated locus.31 Although possible associations have been identified for other autoimmune diseases,9 a NOTCH4 genetic association independent of DRB1 has not been previously shown for MS. However, the NOTCH signaling pathway may be important in T-cell activation, oligodendrocyte differentiation, remyelination and has been suggested as a target for treatment of MS.32 Therefore, the association of the NOTCH4 missense mutation with MS may be of great importance to identifying treatments for Japanese MS patients. The secondarily MS-associated SNP (rs3997849) is closest to the HLA-DQA2 gene in the Class II region. This gene encodes a protein involved in antigen presentation, and although previously studied in MS in a population of European descent,33 the gene has not been previously shown to be associated with MS independent of DRB1.
Classification of patients into MS or NMO disease is complex in the Japanese population. Many patients fall into both OSMS and NMO groups, and several non-NMO patients are AQP4 seropositive (Table 1). Therefore, AQP4 seropositivity, rather than disease class, was used to stratify the data for subsequent analysis. Two SNPs were significantly associated with AQP4− MS. The SNP rs660895 was the primary associated SNP, and is closest to the DRB1 gene. DRB1 is the strongest and most replicated associated gene with MS in populations of European descent.2 When fitting rs660895 in the model, rs2269704, which is located in an intron of the Class I NRM29/NRM gene, was the most significantly associated SNP. This gene is a nuclear envelope membrane protein, and no evidence of previous associations of this gene with MS could be identified. Finally, contrasting AQP4 = and AQP4−, MS patients identified a single significantly associated SNP (rs1694112), which is closest to the HLA-DQB1 gene.
It should be noted that both SNPs significant for all MS, rs422951 and rs3997849, were both significant for AQP4− patients vs controls (FDR P = 0.003 and 0.06, respectively) in the same direction, and rs660895 and rs2269704 in all MS (FDR P = 0.006 and 0.01, respectively). Thus, it can be concluded that the differences in top associations between the two analyses, all demyelinating and AQP4− demyelinating, is likely due to random statistical fluctuations that occur when making a small modification to a data set (removing 55 AQP4 + individuals), rather than true differences being identified by subsetting the data.
Although it is interesting, and possibly informative, to discuss the genes closest to the identified significant SNPs, the extensive and intricate LD patterns in the MHC leads to the possibility that these SNPs may be tagging a causative mutation further away than the closest genes. As DRB1 is the classical MS-associated gene in individuals of European descent and evidence of association has been reported in Japanese MS, haplo-types formed by the SNPs were scrutinized for the ability to tag DRB1 alleles in 218 Japanese individuals for whom four-digit DRB1 data were available. For the analysis of all MS, the single susceptible haplotype contained all of the DRB1*15:01 alleles. However, because this haplotype also tagged several other DRB1 alleles well, it is not possible to determine which of the DRB1 alleles are responsible for the association. The most highly associated resistant haplotype (G–G) mostly contained *01:01, *13:02 and *09:01 alleles. DRB1*01 and *09 alleles have previously been shown to confer resistance to MS and AQP4− MS,14 and it is likely the same association identified in this data set.
For the AQP4− haplotype analyses, G–G and A–G haplotypes made up 90% of the total haplotypes. The most strongly associated haplotype was G–G (P = 9.04 × 10−8; OR = 2.32). Seventy-five percent of this haplotype contains *04 alleles. The association of this haplotype is particularly striking when considering that it excludes *15:01, and considering that the *15:01 effect is included in the non-GG haplotypes in the analysis (*15:01 is found exclusively with the A–G haplotype). This haplotype is tagging a non-*15:01 allele, which has a large effect, which could be larger than the *15:01 effect, on AQP4− MS susceptibility in the Japanese population. The very modest associations between *15:01 presence/absence and general MS and AQP4− MS also support the observation that the *15:01 haplotype may have a reduced role in Japanese MS as compared with MS in western populations. Isobe et al.14 also identified a *04 association with AQP4− MS, and it is therefore this allele group that is the likely source of the susceptible association or part of a susceptible haplotype.
Although an association of DRB1*15:01 with conventional Japanese MS has been reported previously,10,34,35 a lack of a DRB1*15 association, except when stratifying by *09 or *12, has also been reported in Japanese MS.14 The results of the present study clarify the *15 association. In this study, with a larger sample, presence/absence of *15:01 was found to be modestly associated with MS and AQP4− MS. However, the SNP that tags both *15:01 and *15:02 (rs9271366) was not associated with either disease classification. This result indicates that the non-associated *15:02 alleles are diluting the modest association of *15:01 when only two-digit genotypes are available. If *15:01 is indeed the causative allele in western populations, this finding is surprising considering that *15:02 proteins are expected to present the same antigens as proteins derived from *15:01.36 However, *15:02 was also identified as MS non-associated in two very small previously published studies,37,38 and it has been shown that the two alleles may have differing effects with regard to aplastic anemia.36 The difference in effect of the two alleles may be due to the single amino-acid difference between proteins from the two alleles, LD with another associated mutation or differences in expression between the two alleles. Because of the large association of *15:01 with MS in individuals of European descent and moderate association in other ethnicities, determining the mechanism for the difference in association between *15:01 and *15:02 may greatly increase our understanding of the molecular causes of MS.
Through conditional analysis, Lincoln et al.39 found that the Class II haplotype block and DRB1 accounted for most of the MHC-associated MS susceptibility in two populations of European descent, and that Class III associations could be explained by LD with Class II genes. Other studies have suggested, through haplotype analysis, that DRB1*15:01 interacts with other genes in the Class II region to cause susceptibility, or that DRB1*15:01 is part of a susceptible haplotype, but itself is not the causative genetic factor for the strongest genetic association with MS in individuals of European descent. This study also finds that in the Japanese MS population, the DRB1*15:01 allele is not part of the major MS susceptible haplotype in AQP4− patients.
In conclusion, the objective of this study was to elucidate the effects of the HLA in Japanese MS. This study is the largest study of the HLA’s contribution to MS in Japanese individuals. Haplotype analysis revealed a large susceptible association, likely DRB1*04 or a locus in LD with DRB1*04 alleles, with AQP4− MS, which excluded DRB1*15:01 and other loci sharing a haplotype with DRB1*15:01. Several resistant haplotypes were identified, but it is difficult to say whether these haplotypes truly harbor resistant alleles or whether they only appear resistant when opposed to the susceptible haplotypes. Finally, although a very modest association of DRB1*15:01 with MS was observed, DRB1*15:02 was not associated. Because of the similarities of the proteins from these two alleles, differing only at a single amino acid, further studies to understand the nature of this difference, whether it be functional or haplotypic, could greatly increase our understanding of the molecular mechanisms leading to MS.
Materials and methods
Subjects
All samples from Japanese cases were collected in the Neurology Departments of the University Hospitals of the South Japan MS Genetics Consortium, which comprises the following six universities, all located in southwestern Japan: Kyushu University, Yamaguchi University, Ehime University, Hiroshima University, Kinki University and Osaka University. The final data set consisted of DNAs from 280 control (HC) and 204 patients with MS (193 cases who fulfilled the revised McDonald criteria28 and 11 cases who at least met the criteria of clinically isolated syndrome41 and were suggestive of MS), genotyped on a custom Infinium iSelect HD Custom Genotyping BeadChip (Illumina Inc., San Diego, CA, USA) for 6040 MHC region SNPs. SNPs were selected by previously described methods,27,42,43 and included an additional 4431 non-chromosome six SNPs genotyped for assessment of population stratification. Anti-AQP4 antibody was measured in all patients using green uorescent proteinAQP4 fusion protein-transfected human embryonic kidney cells as described previously.14 All participants gave written informed consent. This study was approved by the UCSF institutional review board, and the institutional ethical committees at each university of the South Japan MS Genetics Consortium.
DRB1 genotypes were available for 218 Japanese individuals. The HLA-DRB1 alleles of the subjects were determined by hybridization of sequence-specific oligonucleotide probes in specific amplicons, as described elsewhere.44 In addition, 264 controls and 203 patients were typed for DRB1*15:01 presence/absence using validated gene-specific TaqMan assays as described by Caillier et al.25
Quality control
SNPs
SNPs were removed from the data set for missing genotypes greater than 5%, violation of HardyWeinberg equilibrium (P<0.001), or having a minor allele frequency less than 5%. SNPs were also removed if they were significantly (P<0.001) differently missing between all patients and controls (PLINK—test-missing) or if they were non-randomly missing (P<0.001) with respect to their expected genotypes derived from nearby SNPs in LD (PLINK—test-mishap). After all QC, 3534 HLA region SNPs remained. All genomic positions reported correspond to NCBI SNP build 129.45
Samples
All individuals with 10% or more missing genotypes were removed from analysis. To verify ethnicity, multidimensional scaling was used to cluster the experimental sample with data from 12 HapMap populations. Data for 705 non-chromosome six SNPs were common between all data sets and used for the analysis. Plots were generated for the first component onto the second component, and for each informative component (components 1–7) separately.
To identify population stratification, principal components were calculated for each individual using the 3668 non-chromosome six SNPs remaining after QC. Based on the scree plot, the first three components were considered informative (Supplementary Figure 4). Visual examination of a three-dimensional plot of the first three components identified three obvious outliers (Supplementary Figure 5). T2 statistic analysis (Supplementary Figure 6) identified six outliers (including the three identified by visual examination), and data for all six samples were removed from the study.
Statistical analyses
Association analyses
Logistic regression (PLINK—logistic) was used to determine the association between HLA SNPs and MS, AQP4 + MS and AQP4− MS. An additive genetic model was assumed and gender included as a covariate for all analyses. Following the methods of McElroy et al.,27 for each trait multiple rounds of analyses were performed, with each successive round including the most significant SNPs from the previous rounds, until no SNPs were significant at an FDR46 of 0.1. This method facilitates the identification of multiple associated SNPs that are not redundantly associated with the trait through LD.
Haplotype analyses
Haplotypes were estimated for the significant SNPs (PLINK—hap). Haplotype effects were determined by weighted logistic regression of disease status onto haplotype dosage, with gender as a covariate to identify specific haplotypes that may be tagging trait-associated loci. Four-digit HLA-DRB1 genotypes were available for a subset of individuals (n = 218). To investigate how haplotypes of significant SNPs relate to HLA-DRB1 allelic polymorphism, haplotype frequencies were estimated by maximum-likelihood implemented in an Expectation Maximization algorithm.47 Positive predictive values and sensitivities of the SNPs to predict HLA-DRB1 alleles at the haplotype level were computed. All analyses were completed using PLINK (version 1.07),48 JMP Genomics 4.1 (SAS Institute Inc., Cary, NC, USA) and R (version 2.9),49 unless otherwise noted.
Supplementary Material
Acknowledgments
We thank the MS patients and controls who participated in this study. We thank RR Lincoln, R Guerrero, H Mousavi and A Santaniello for sample and database management. This work was supported by grants from the National Institute of Health U19AI067152, RO1NS46297 and National Multiple Sclerosis Society RG3060C8. JPM is a post-doctoral fellow supported by the National MS Society. NI is a post-doctoral fellow supported by The Association for Preventive Medicine of Japan. The members of the South Japan Multiple Sclerosis Genetic Consortium are J Kira, N Isobe, T Matsushita and S Yoshimura (Kyushu University), S Kusunoki and K Miyamoto (Kinki University), S Sakoda and Y Nakatsuji (Osaka University), T Kohriyama, K Ochi and M Matsumoto (Hiroshima University), T Kanda (Yamaguchi University) and T Miki and Y Kawano (Ehime University).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- 1.Ebers GC, Yee IM, Sadovnick AD, Duquette P. Conjugal multiple sclerosis: population-based prevalence and recurrence risks in offspring. Canadian Collaborative Study Group. Ann Neurol. 2000;48:927–931. [PubMed] [Google Scholar]
- 2.McElroy JP, Oksenberg JR. Multiple sclerosis genetics. Curr Top Microbiol Immunol. 2008;318:45–72. doi: 10.1007/978-3-540-73677-6_3. [DOI] [PubMed] [Google Scholar]
- 3.Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet. 2003;72:710–716. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 5.Brassat D, Salemi G, Barcellos LF, McNeill G, Proia P, Hauser SL, et al. The HLA locus and multiple sclerosis in Sicily. Neurology. 2005;64:361–363. doi: 10.1212/01.WNL.0000149765.71212.0A. [DOI] [PubMed] [Google Scholar]
- 6.Cree BA, Rioux JD, McCauley JL, Gourraud PA, Goyette P, McElroy J, et al. A major histocompatibility Class I locus contributes to multiple sclerosis susceptibility independently from HLA-DRB1*15:01. PLoS One. 2010;5:e11296. doi: 10.1371/journal.pone.0011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, et al. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 8.Link J, Lorentzen AR, Kockum I, Duvefelt K, Lie BA, Celius EG, et al. Two HLA class I genes independently associated with multiple sclerosis. J Neuroimmunol. 2010;226:172–176. doi: 10.1016/j.jneuroim.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kira J. Multiple sclerosis in the Japanese population. Lancet Neurol. 2003;2:117–127. doi: 10.1016/s1474-4422(03)00308-9. [DOI] [PubMed] [Google Scholar]
- 11.Fukazawa T, Kikuchi S, Miyagishi R, Miyazaki Y, Yabe I, Hamada T, et al. HLA-dPB1*0501 is not uniquely associated with opticospinal multiple sclerosis in Japanese patients. Important role of DPB1*0301. Mult Scler. 2006;12:19–23. doi: 10.1191/135248506ms1252oa. [DOI] [PubMed] [Google Scholar]
- 12.Matsushita T, Matsuoka T, Isobe N, Kawano Y, Minohara M, Shi N, et al. Association of the HLA-DPB1*0501 allele with anti-aquaporin-4 antibody positivity in Japanese patients with idiopathic central nervous system demyelinating disorders. Tissue Antigens. 2009;73:171–176. doi: 10.1111/j.1399-0039.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 13.Kira J. Neuromyelitis optica and opticospinal multiple sclerosis: mechanisms and pathogenesis. Pathophysiology. 2011;18:69–79. doi: 10.1016/j.pathophys.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Isobe N, Matsushita T, Yamasaki R, Ramagopalan SV, Kawano Y, Nishimura Y, et al. Influence of HLA-DRB1 alleles on the susceptibility and resistance to multiple sclerosis in Japanese patients with respect to anti-aquaporin 4 antibody status. Mult Scler. 2010;16:147–155. doi: 10.1177/1352458509355067. [DOI] [PubMed] [Google Scholar]
- 15.ANZgene. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 16.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 17.Baranzini SE. The genetics of autoimmune diseases: a networked perspective. Curr Opin Immunol. 2009;21:596–605. doi: 10.1016/j.coi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Association scan of 14 500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comabella M, Craig DW, Camina-Tato M, Morcillo C, Lopez C, Navarro A, et al. Identification of a novel risk locus for multiple sclerosis at 13q31. 3 by a pooled genome-wide scan of 500 000 single nucleotide polymorphisms. PLoS One. 2008;3:e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.IMSGC. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 22.Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42:495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caillier SJ, Briggs F, Cree BA, Baranzini SE, Fernandez-Vina M, Ramsay PP, et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol. 2008;181:5473–5480. doi: 10.4049/jimmunol.181.8.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McElroy JP, Cree BA, Caillier SJ, Gregersen PK, Herbert J, Khan OA, et al. Refining the association of MHC with multiple sclerosis in African Americans. Hum Mol Genet. 2010;19:3080–3088. doi: 10.1093/hmg/ddq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ‘McDonald Criteria’. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 29.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case–control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glessner JT, Hakonarson H. Common variants in polygenic schizophrenia. Genome Biol. 2009;10:236. doi: 10.1186/gb-2009-10-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurynczyk M, Selmaj K. Notch: a new player in MS mechanisms. J Neuroimmunol. 2010;218:3–11. doi: 10.1016/j.jneuroim.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Duvefelt K, Anderson M, Fogdell-Hahn A, Hillert J. A NOTCH4 association with multiple sclerosis is secondary to HLA-DR*1501. Tissue Antigens. 2004;63:13–20. doi: 10.1111/j.1399-0039.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 34.Fukazawa T, Kikuchi S, Miyagishi R, Niino M, Yabe I, Hamada T, et al. CTLA-4 gene polymorphism is not associated with conventional multiple sclerosis in Japanese. J Neuroimmunol. 2005;159:225–229. doi: 10.1016/j.jneuroim.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Miyagishi R, Niino M, Fukazawa T, Yabe I, Kikuchi S, Tashiro K. C–C chemokine receptor 2 gene polymorphism in Japanese patients with multiple sclerosis. J Neuroimmunol. 2003;145:135–138. doi: 10.1016/j.jneuroim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Sugimori C, Yamazaki H, Feng X, Mochizuki K, Kondo Y, Takami A, et al. Roles of DRB1 *1501 and DRB1 *1502 in the pathogenesis of aplastic anemia. Exp Hematol. 2007;35:13–20. doi: 10.1016/j.exphem.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Kira J, Kanai T, Nishimura Y, Yamasaki K, Matsushita S, Kawano Y, et al. Western versus Asian types of multiple sclerosis: immunogenetically and clinically distinct disorders. Ann Neurol. 1996;40:569–574. doi: 10.1002/ana.410400405. [DOI] [PubMed] [Google Scholar]
- 38.Ma JJ, Nishimura M, Mine H, Saji H, Ohta M, Saida K, et al. HLA-DRB1 and tumor necrosis factor gene polymorphisms in Japanese patients with multiple sclerosis. J Neuroimmunol. 1998;92:109–112. doi: 10.1016/s0165-5728(98)00189-1. [DOI] [PubMed] [Google Scholar]
- 39.Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, Tiislar M, et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet. 2005;37:1108–1112. doi: 10.1038/ng1647. [DOI] [PubMed] [Google Scholar]
- 40.Lincoln MR, Ramagopalan SV, Chao MJ, Herrera BM, Deluca GC, Orton SM, et al. Epistasis among HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci determines multiple sclerosis susceptibility. Proc Natl Acad Sci USA. 2009;106:7542–7547. doi: 10.1073/pnas.0812664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalton CM, Brex PA, Miszkiel KA, Hickman SJ, MacManus DG, Plant GT, et al. Application of the new McDonald criteria to patients with clinically isolated syndromes suggestive of multiple sclerosis. Ann Neurol. 2002;52:47–53. doi: 10.1002/ana.10240. [DOI] [PubMed] [Google Scholar]
- 42.de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 43.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuji K, Aizawa M, Sasazuki T, editors. 11th International Histocompatibility Workshop Reference Protocol for the HLA DNA Typing Technique. HLA 1991: Proceedings of the 11th International Histocompatibility Workshop and Conference; Oxford University Press; 1992. [Google Scholar]
- 45.NCBI. Database of Single Nucleotide Polymorphisms (dbSNP): dbSNP Build ID. National Center for Biotechnology Information, National Library of Medicine; Bethesda, MD: p. 129. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 47.Gourraud PA, Genin E, Cambon-Thomsen A. Handling missing values in population data: consequences for maximum likelihood estimation of haplotype frequencies. Eur J Hum Genet. 2004;12:805–812. doi: 10.1038/sj.ejhg.5201233. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
- 50.Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology. 2006;66:1485–1489. doi: 10.1212/01.wnl.0000216139.44259.74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.