SUMMARY
Macrophage infiltration into tumors has been correlated with poor clinical outcome in multiple cancer types. Therefore, new tools to image tumor-associated macrophages could be valuable for diagnosis and prognosis of cancer. Herein we describe the synthesis and characterization of a cathepsin S-directed, quenched activity-based probe (qABP), BMV083. This probe makes use of an optimized non-peptidic scaffold leading to enhanced in vivo properties relative to previously reported peptide-based probes. In a syngeneic breast cancer model, BMV083 provides high tumor specific fluorescence that can be visualized using noninvasive optical imaging methods. Furthermore, analysis of probe labeled cells demonstrates that the probe primarily targets macrophages with an M2 phenotype. Thus, BMV083 is a potential valuable new in vivo reporter for tumor-associated macrophages that could greatly facilitate the future studies of macrophage function in the process of tumorigenesis.
INTRODUCTION
The cysteine cathepsin family contains 11 members that play important roles in protein breakdown in the lysosome (Turk, et al., 2001). Several family members have also been shown to play functional roles in the onset and progression of diverse pathologies ranging from Alzheimer’s disease to cancer (Conus and Simon, 2010; Mohamed and Sloane, 2006; Mueller-Steiner, et al., 2006; Reiser, et al., 2010). Because the activity of these proteases is highly regulated and dependent on posttranslational maturation of the proenzyme, tools that can report on their activity levels have been essential to understanding their biological function in disease pathology. In particular, a number of activity-based probes (ABPs) have been developed that allow the direct profiling of cysteine cathepsin activity levels in vivo. Recently, we developed a near-infrared fluorophore labeled quenched ABP (NIRF qABP), GB137, containing a peptide recognition scaffold linked to an acyloxymethyl ketone (AOMK) electrophile (Blum, et al., 2007). This intrinsically non-fluorescent probe produces a fluorescent signal upon nucleophilic displacement of the quencher group by the active site cysteine of the proteolytically active cysteine cathepsin (Figure 1A). Because high levels of cysteine cathepsin activity are associated with many tumor types, the probe can be used for direct noninvasive optical imaging applications. Furthermore, because the probes are activity-dependent, they can also be used to noninvasively measure the efficacy of small-molecule therapeutics. In contrast to fluorogenic cathepsin substrates that have also been successfully used for live animal imaging experiments (Gounaris, et al., 2008; Mahmood and Weissleder, 2003), with the most recent addition being a cathepsin S selective substrate (Caglic, et al., 2011), the irreversible nature of the ABPs allows subsequent down-stream identification of the targeted protease and correlation with histological and cell biological assessments of the in vivo tumor microenvironment.
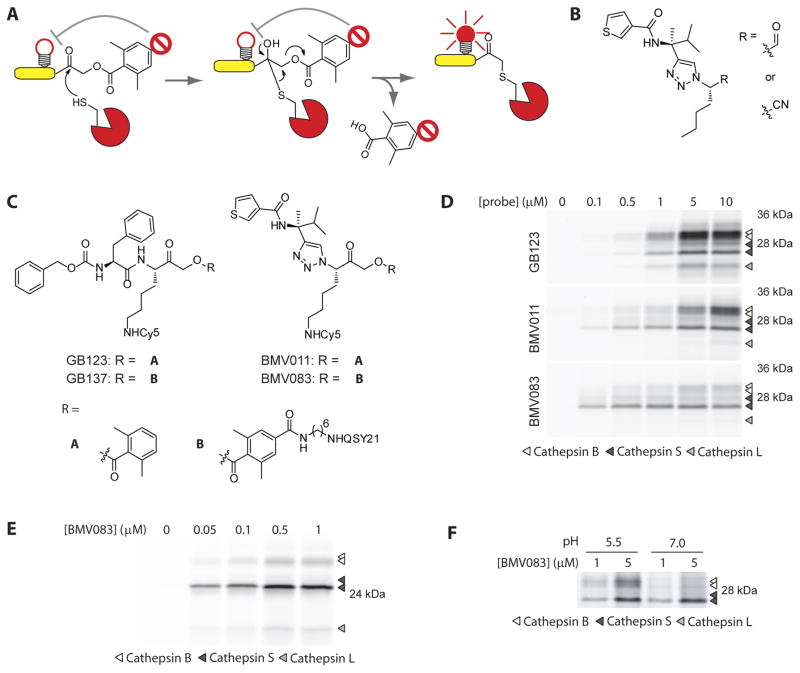
Figure 1.
Non-peptidic cysteine cathepsin activity-based probes. A) Schematic presentation of the mechanism of action of a quenched ABP. B) Structure of the cathepsin S selective aldehyde and nitrile inhibitors reported by the Ellman lab. C) Structures of the peptidic activity-based probes GB123 and the quenched GB137 and the non-peptidic probes BMV011 and the quenched BMV083. D) Labeling profile of GB123, BMV011 and BMV083 in living RAW cells. Cells were exposed to the indicated concentrations of probe for 3 hr, before being harvested, washed and lysed. 40 μg total protein was resolved on 15% SDS-PAGE and fluorescently labeled proteins were visualized by in-gel fluorescence scanning. E) Labeling profile of BMV083 in living human primary macrophages. Cells were exposed to the indicated concentrations of BMV083 for 3 hr, before being harvested, washed and lysed. 40 μg total protein was analyzed as described above. F) BMV083 labeling of RAW cell lysate (35 μg total protein) at pH 5.5 and 7.0 with indicated concentrations of probe for 1 hr. Labeled proteins were analyzed as described above. See also supplemental figures S1–S4.
The primary in vivo targets of our first generation qABP were cathepsin B, S and L (Blum, et al., 2005; Blum, et al., 2007). Although important roles in tumor development have been described for all three of these cysteine cathepsins (Gocheva and Joyce, 2007), cathepsin B and L, like most members of the cysteine cathepsin family are ubiquitously expressed (Conus and Simon, 2010). Cathepsin S however, is most abundantly expressed in antigen presenting cells (APCs) where it plays a major role in MHC II antigen presentation (Zavasnik-Bergant and Turk, 2006). Macrophages are professional APCs and are therefore key players in immunity. They have a variety of functions depending on their activation state - classically activated (M1) or alternatively activated (M2). Macrophages can also be classified into three groups based on their homeostatic functions; host defense (classically activated macrophages), wound healing (wound healing macrophages) and immune regulation (regulatory macrophages) (Mosser and Edwards, 2008). However, macrophages display a high degree of plasticity and activation states can change in response to stimuli from their environment. Furthermore, macrophages can have a blend of characteristics of multiple groups. One such type of macrophage is the tumor-associated macrophage (TAM), which displays characteristics of both wound-healing and regulatory macrophages and plays important roles in tumorigenesis by promoting angiogenesis, tumor growth and invasiveness. These macrophages are recruited to the tumor site and are stimulated by factors in the tumor microenvironment, including the cytokine interleukin-4 (IL-4) which induces cysteine cathepsin activity (Gocheva, et al., 2010). In human studies TAM infiltration in tumors has been associated with poor prognosis, for example in high-risk breast cancers (Mukhtar, et al., 2011).
Development of imaging tools to identify TAM infiltration in tumors could lead to clinical applications for treatment and prognosis of cancer. Because of its confined expression, probes that are designed to target cathepsin S are likely to provide improved contrast for areas with stimulated macrophages compared to more broad-spectrum probes that also target other cysteine cathepsins that have a broader expression profile. Herein we describe the synthesis and characterization of a cathepsin S-directed, non-peptidic NIRF qABP with improved in vivo properties relative to previously reported peptide-based probes. We use this optimized cathepsin S probe for noninvasive optical imaging of a syngeneic mouse model of breast cancer. Fluorescence-activated cell sorting (FACS) experiments identified specific subsets of myeloid derived cells with an M2 macrophage phenotype as the in vivo cellular source of cysteine cathepsin activity responsible for probe fluorescence. These data demonstrate the potential value of our new ABP for the classification of tumor-associated myeloid derived cells based on in vivo protease activity.
RESULTS AND DISCUSSION
Synthesis and in vitro characterization of BMV083
We set out to develop ABPs that would have a high degree of selectivity for cathepsin S that could be used for noninvasive imaging applications. Furthermore, we aimed to give the new generation probe a more drug-like, non-peptidic character to improve stability, bioavailability and half-life in circulation. The Ellman lab recently used the substrate activity screening (SAS) approach to discover 1,4-disubstituted-1,2,3-triazole-based non-natural aldehyde and nitrile inhibitors with high selectivity for cathepsin S over cathepsin B and L (Figure 1B) (Patterson, et al., 2006; Wood, et al., 2005). Triazoles, easily accessible through the Huisgen cycloaddition, have extensively been used as stable peptide bond mimics because of their similar electronic and structural characteristics. We therefore used these non-peptidic triazole-based inhibitors as a starting point for design of our cathepsin S-directed ABPs (the synthesis of ABPs can be found in the supplemental information, supplementary figures S1–S3). To determine the effect of the change of scaffold, we synthesized the fluorescent ABP BMV011 by replacing the norleucine P1 substituent by a lysine side chain to introduce the Cy5 fluorophore and the reversible electrophile by the irreversible 2,6-dimethyl benzoic AOMK (Figure 1C). We compared the labeling profile of BMV011 with that of the peptide-based probe GB123 (Blum, et al., 2007) by exposing RAW cells (a mouse leukaemic macrophage cell line) in culture to increasing concentrations of the probes. BMV011 and GB123 showed comparable labeling potency for cathepsin S (EC50= 1.0μM and = 1.3μM respectively). However, the change in probe scaffold reduced labeling of cathepsin B (EC50(BMV011) = 6.0μM, EC50(GB123) = 1.6μM) and cathepsin L (Figure 1D, top and middle panel), making BMV011 more selective for cathepsin S. Encouraged by these results we synthesized the qABP BMV083, the non-peptidic analogue of GB137 (Figure 1C). The introduction of the quencher moiety further reduced the potency for cathepsin B, similar to the reduction of potency for cathepsin B seen for GB137 compared to GB123. This is probably due to steric hindrance with the occluding loop in the prime site pocket of cathepsin B. The primary target of BMV083 in RAW macrophages was cathepsin S, with a similar labeling potency as GB137 (supplementary Figure S4), and only minimal labeling of cathepsin L at high probe concentrations. Primary human monocyte derived macrophages exposed to increasing concentrations of BMV083 showed a labeling profile similar to that observed in mouse macrophages with cathepsin S being the primary target over cathepsins B and L (Figure 1E). Cathepsin S, in contrast to other cysteine cathepsin family members, has a wide pH optimum and is proteolytically active at both acidic and neutral pH (Kirschke, et al., 1989). Consistent with this finding, we observed equal labeling of cathepsin S by BMV083 in RAW cell lysate at acidic or neutral pH, and substantially diminished labeling of cathepsin B at neutral pH (Figure 1F).
Having determined the cell permeability and labeling specificity of BMV083, we wanted to visualize the dynamics of cysteine cathepsin labeling (unquenching) in living cells using confocal microscopy and correlate this signal with target labeling as assessed by SDS-PAGE analysis of the cell extracts. We exposed 4T1 mouse mammary adenocarcinoma cells to BMV083 and imaged at various time points (Figure 2A). Cy5 fluorescence signal increased over time and was primarily localized to a vesicular staining pattern. Fluorescent probe signals could be eliminated by pretreatment of the cells with a pan-cysteine cathepsin inhibitor (GB111-NH2) (Blum, et al., 2007) suggesting that probe activation was mediated by active cysteine cathepsin targets. Gel-based analysis of BMV083 labeling over the same time course showed that the time dependant increase in Cy5 fluorescence activation correlated with labeling of cathepsin B and L. Furthermore, inhibition of target labeling by pretreatment with GB111-NH2 also correlated with loss of imaging signals in the cells (Figure 2B). In agreement with its expression profile, no active cathepsin S was labeled in these cancer cells, which could explain the slow imaging and labeling kinetics.
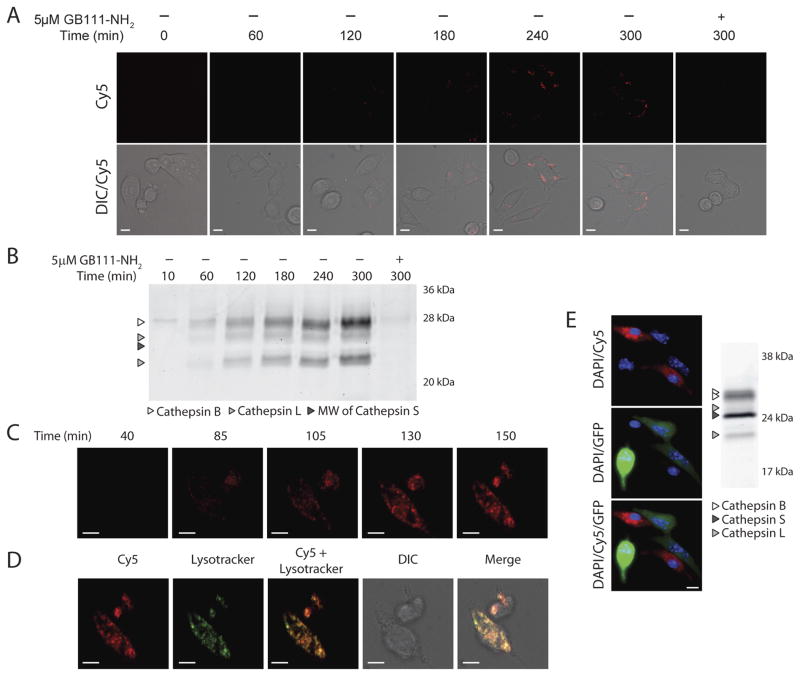
Figure 2.
Live cell imaging of probe labeling. A–D) Live cell confocal microscopy time course of BMV083 cysteine cathepsin labeling. A) 4T1-luc-GFP cells were exposed to 1 μM BMV083 and imaged over time. Where indicated cells were preincubated with the cysteine cathepsin inhibitor GB111-NH2 (5 μM) for 1 hr before addition of probe (Scale bar 10μm). B) 4T1-luc-GFP cells were exposed to 1 μM BMV083 and harvested at indicated time points. 40 μg total protein was resolved on 15% SDS-PAGE and fluorescently labeled proteins were visualized by in-gel fluorescence scanning (MW = molecular weight). C) Mouse primary bone-marrow derived macrophages were exposed to 1 μM BMV083 and imaged over time (Scale bar 10μm). D) After final time point lysotracker was added (Scale bar 10μm). E) 4T1-luc-GFP cells and mouse primary bone-marrow derived macrophages (2 to 1 ratio) were co-cultured. After 90 hr the cells were exposed to 1 μM BMV083 for 2 hr. Cells were imaged (Red, Cy5 fluorescence; Green, GFP; Blue, DAPI staining. Scale bar 10μm) and harvested. 40 μg total protein was resolved on 15% SDS-PAGE and fluorescently labeled proteins were visualized by in-gel fluorescence scanning.
To determine if the probe could more effectively target cells with high cathepsin S levels we next performed cell based imaging studies in primary mouse bone-marrow derived macrophages (BMMs; Figure 2C and D). We found that in BMMs BMV083 is more rapidly unquenched compared to the 4T1 cells. Virtually all of the probe signal in the BMMs could be co-localized with a marker of lysosomal compartments (Figure 2D). Based on these findings, we wanted to see if our cathepsin S-directed probe would be able to selectively label the cathepsin S expressing cells in the presence of cells that predominantly express cathepsins B and L. Therefore we co-cultured BMMs with 4T1 cancer cells expressing both GFP and luciferase (4T1-luc-GFP) and imaged after probe treatment. Indeed, probe labeling was almost exclusively confined to the non-GFP expressing BMMs (Figure 2E). Consistent with the imaging experiment, SDS-PAGE analysis of the labeled cysteine cathepsins in the co-culture showed an intense fluorescent cathepsin S signal that was not present in the culture of the 4T1 cells alone (Figure 2B) and a labeling pattern very similar to that observed in the RAW mouse macrophage cell line (Figure 1D). Thus, the majority of labeling is derived from the macrophage population with only minimal labeling of cat B and L from the tumor cells.
In vivo properties of BMV083 in tumor bearing mice
Having determined the target preference and cell permeability of BMV083 in vitro, we next set out to establish the in vivo performance of the non-peptidic qABP compared to the original peptidic GB137. We subcutaneously allografted athymic Balb/c nude mice with 4T1-luc-GFP cells. When tumors were established, we injected two cohorts of tumor bearing mice via tail vein with equimolar amounts of either GB137 or BMV083 and noninvasively imaged the mice for Cy5 fluorescence over time (Figure 3A and B. Images of the whole mice are depicted in supplemental figure S5A). As reported previously in xenograft tumor models, GB137 showed specific accumulation in tumors starting at early time points (Figure 3A) (Blum, et al., 2007). The non-peptidic counterpart BMV083 however, showed increased tumor specific activation of fluorescence resulting in substantially better contrast at all time points compared to GB137 (Figure 3A and B). The tumor specific fluorescence intensity for BMV083, like GB137, peaked around the 4 hour time point (Figure 3C). Interestingly, unlike the GB137 tumor specific fluorescence, which decreased over time, the BMV083 signal in tumors remained constant until the end of the time course. This could be due to enhanced stability of the free probe in vivo resulting in extended labeling of newly synthesized pools of cysteine cathepsin. After the completion of the noninvasive optical imaging time course, we excised tumors and performed ex vivo imaging followed by homogenization and analysis of the fluorescently labeled protein content by SDS-PAGE (Figure 3D). Here too, BMV083 had significantly increased overall cysteine cathepsin labeling compared to GB137. The in vivo targets for GB137 were primarily cathepsin B (38.0 ± 5.7% (n=5)) and L (33.6 ± 2.3% (n=5)) and to a lesser extent cathepsin S (28.4 ± 1.8% (n=5)). Whereas in cultured RAW cells BMV083 almost exclusively labels cathepsin S in the presence of the more abundant cathepsin B (Figure 1D), in vivo the primary target for BMV083 was cathepsin S (61.4 ± 5.8% (n=5)) with cathepsin B being the secondary target (38.6 ± 5.2% (n=5)). To determine the overall activity levels of the cysteine cathepsins in the 4T1 allografted tumors, we labeled tumor homogenates with the BODIPY TMR labeled general cysteine cathepsin probe BMV099 (Figure 3D; For structure and characterization of BMV099 see Figure S6). Interestingly, this analysis indicated that the most active cysteine cathepsin in this tumor model is cathepsin B. Therefore the relatively higher level of cathepsin B labeling by BMV083in this model is likely due to the fact that this is the most active cathepsin found in the tumor tissues. This result also stresses the fact that the selectivity of any inhibitor or substrate-based reporter highly depends on the experimental setup and the relative activity levels of other related (off-)targets in the tissue, cell or sample being analyzed. The advantage of ABPs over substrate-based reporters is that the covalent irreversible nature of the labeling allows for biochemical validation and characterization of the in vivo observed probe fluorescence. Therefor, we could correlate the enhanced labeling signals by BMV083 in the live animals with whole tissue fluorescence and relative levels of cysteine cathepsin labeling observed for each probe by SDS-PAGE (Figure 3E).
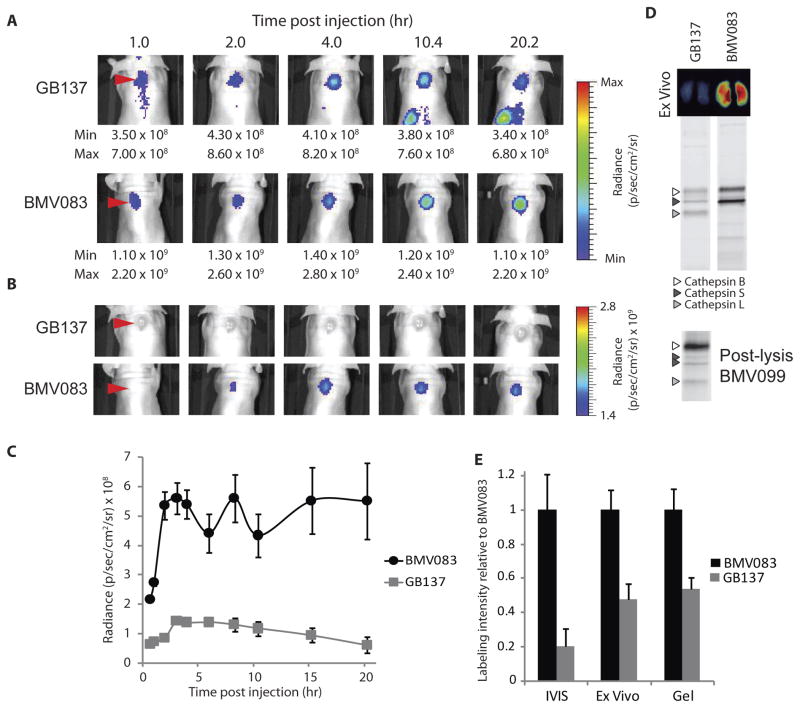
Figure 3.
Noninvasive optical imaging of cysteine cathepsin activity with quenched activity-based probes in tumor bearing mice. A–C) 4T1-luc-GFP cells were injected subcutaneously on the back of athymic BALB/c nude mice, 8 days before imaging. GB137 or BMV083 (20 nmol) was injected (intravenous via tail vein) and fluorescent images of the living mice were taken over time. Red arrowheads indicate the tumor. A) Optimal fluorescence contrast for individual time points. Maximum and minimum of scale bar depicted below image. B) Comparison of GB137 and BMV083 fluorescence at fixed intensity. C) Time dependent tumor specific fluorescence (tumor - background) for BMV083 and GB137 treated mice (n = 5, data represent mean values ± standard errors). D) Ex vivo fluorescence imaging of cross-sections of excised tumors (upper panel). Tumor tissue was homogenized and 40 μg total protein was resolved on 15% SDS-PAGE. In vivo fluorescently labeled proteins were visualized by in-gel fluorescence scanning (middle panel). 4T1 tumors were homogenized and the active cysteine cathepsins were labeled with BMV099 (1 μM for 1 hr, 80 μg total protein) before the proteins were resolved on 15% SDS-PAGE. Fluorescently labeled proteins were visualized by in-gel fluorescence scanning (lower panel). E) Fluorescence intensity of end point of non-invasive optical imaging (shown in A), ex vivo tumor imaging and in-gel fluorescence labeling (shown in D). Intensity relative to BMV083 is depicted (n = 5, data represent mean values ± standard errors). See also supplemental figures S5 and S6.
The fact that GB137 and BMV083 have similar labeling potencies in vitro yet the non-peptidic probe was a more effective label in vivo indicates that the non-peptidic BMV083 has better in vivo properties compared to the peptidic GB137. Furthermore, liver and kidney fluorescence and cysteine cathepsin labeling were very similar for both probes (supplemental figure S5B), indicating that BMV083 might circulate longer before being scavenged by these organs for clearance.
We next tested BMV083 in a more physiologically relevant orthotopic mouse breast cancer model. For this model, we implanted the 4T1 cell line isolated from a spontaneously arising BALB/c mammary tumor into the mammary fat pad of BALB/c mice with an intact immune system (Tao, et al., 2008). This is especially important since host-tumor interactions and immune cell infiltration in the tumor microenvironment are important factors in tumor development. We implanted 4T1-luc-GFP cells in the number 2 and 7 mammary fat pad and tumor growth was monitored. We then intraperitoneally injected tumor-bearing mice with luciferin 10 hours after probe injection followed by imaging for bioluminescence and probe derived Cy5 fluorescence (Figure 4A). The cancer cell derived luciferase bioluminescence demarcated the tumor boundaries and BMV083 activation was localized to the same area, indicating elevated cysteine cathepsin activity in the 4T1 tumor microenvironment. Interestingly, compared to the allograft model, fluorescence analysis of the SDS-PAGE resolved protein content revealed cathepsin S as the major in vivo target for BMV083 with only minor labeling of cathepsin B (Figure 4B, cathepsin S represents 82.0 ± 0.7% (n=6) of total labeling intensity). Ex vivo labeling of the active cysteine cathepsin content in whole tumor homogenates with BMV099 indicated higher cathepsin S levels relative to cathepsin B compared to the xenograft model (Figure 3D) which likely explains why BMV083 labeled primarily cathepsin S in these tumors(Figure 4B). We performed a biodistribution study showing that similar to what has been reported for GB137, the primary target organs of BMV083 are the liver and spleen (Figure 4C, upper panel). Ex vivo labeling of the active cysteine cathepsin content in whole organ homogenates with BMV099 indicated a high degree of cysteine cathepsin activity in the organs that are strongly labeled in vivo (Figure 4C). Interestingly, although there are high levels of cysteine cathepsin activity in the kidney, it is not effectively targeted by BMV083 indicating specific partitioning of the probe to certain organs. On a similar note, the total cysteine cathepsin activity in the tumor was relatively low, however the BMV083 signal in tumors is relatively high, again suggesting partitioning of the probe to specific tissues.
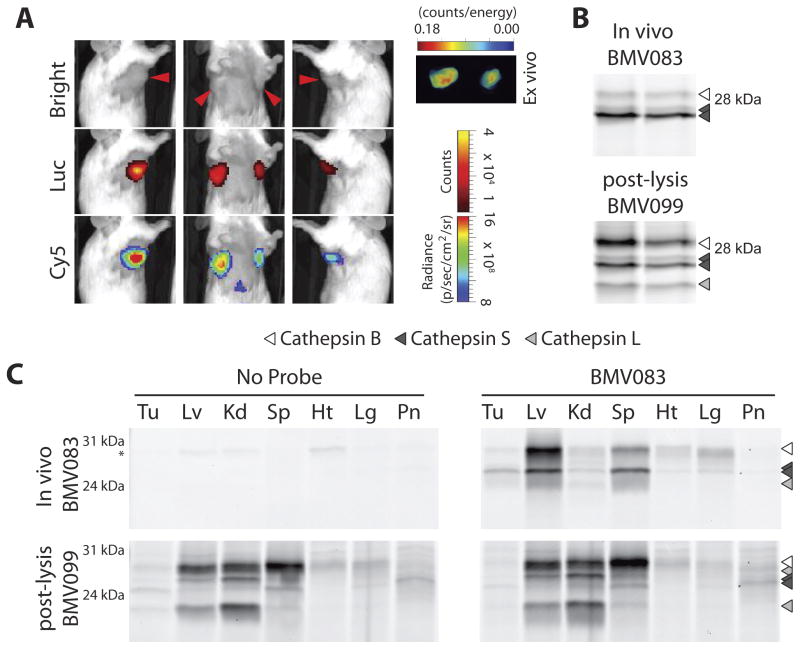
Figure 4.
Noninvasive optical imaging of cysteine cathepsin activity in a syngeneic orthotopic model for breast cancer. A) 4T1-luc-GFP cells were implanted into mammary fat pad number 2 and 7 of female BALB/c mice. On day 15, BMV083 (20 nmol) was injected (intravenous via tail vein). 10 hr post injection the mice were injected with luciferin (intraperitoneal) and fluorescence and luminescence images of the living mice were taken (Top panel, bright field image. Red arrowheads indicate the tumors; middle panel, luciferase bioluminescence (Luc); lower panel, BMV083 signal (Cy5). Ex vivo fluorescence image of excised tumors is shown. B) BMV083 labeling in tumors. The tumors were homogenized and the active cysteine cathepsins were labeled with BMV099 (1 μM for 1 hr, 40 μg total protein) before the proteins were resolved on 15% SDS-PAGE. Fluorescently labeled proteins were visualized by in-gel fluorescence scanning (top panel, invivo BMV083 labeling; bottom panel, ex vivo BMV099 labeling). C) In vivo BMV083 distribution. Organs were homogynized and the active cysteine cathepsins were labeled with BMV099 (1 μM for 1 hr, 40 μg total protein) before the proteins were resolved on 15% SDS-PAGE. Fluorescently labeled proteins were visualized by in-gel fluorescence scanning (top panel, invivo BMV083 labeling; bottom panel, ex vivo BMV099 labeling. *auto-fluorescent band. Tu = tumor, Lv = liver, Kd = kidney, Sp = spleen, Ht = hart, Lg = lungs, Pn = pancreas). See also supplemental figure S6.
Cellular source of BMV083 labeling in tumor bearing mice
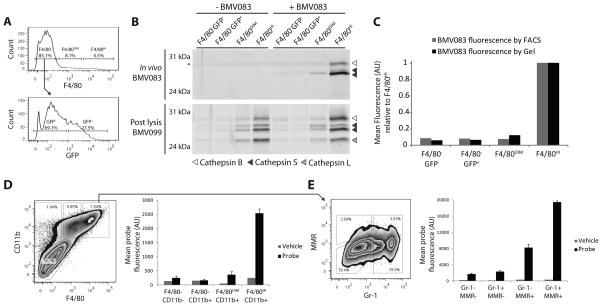
Fluorescent ABPs have previously been used to identify the cell populations within the tumor microenvironment that posses high levels of cysteine cathepsin activity. For example, fluorescence microscopy and immunofluorescence identified macrophages (F4/80+ cells) as the source of cysteine cathepsin activity in the RIP1-Tag2 mouse model of pancreatic islet cell cancer and the MMTV-PyMT mouse model of mammary tumorigenesis treated with a non-quenched Cy3B labeled DCG04 analogue, ‘Cath-ABP’ (Gocheva, et al., 2010). In the same study, tumor derived single cell suspensions were labeled with the fluorescent Cath-ABP ex vivo and analyzed by FACS. Again, macrophages were identified as the primary source of cysteine cathepsin activity, although a non-macrophage population was also found to take up probe. The authors suggested that this population might include cancer cells. Macrophage activation is a plastic and dynamic process that may change during processing of the tissue to generate single cell suspensions for labeling and FACS analysis. Labeling of the tumor-derived cells in vivo is likely to be more physiologically relevant because the cysteine cathepsin targets are labeled in their native activation state. We therefore set out to determine the cellular source of BMV083 derived fluorescence in vivo. Since macrophages have previously been shown to possess high cysteine cathepsin activity in the tumor and the fact that the amount of F4/80 expression is indicative of the macrophage activation state (M2 type macrophages have high F4/80 expression levels compared to a lower F4/80 expression on M1 type macrophages)(Fujiu, et al., 2011), we analyzed four populations of cells from the 4T1-luc-GFP syngeneic orthotopic tumors for BMV083 fluorescence. These included two populations of F4/80 expressing cells (F4/80DIM and F4/80HI), cells that did not express GFP (F4/80−GFP−) and a population containing the GFP expressing tumor cells (F4/80−GFP+) (Figure 5A). The F4/80HI population was responsible for the majority of BMV083 activation, with a more than twelvefold increase in Cy5 mean fluorescence compared to vehicle treatment (Figure 5C, grey bars). To confirm this observation, we isolated the four cell populations in high purity using flow cytometric cell sorting, lysed equal cell numbers per population and labeled the active cysteine cathepsins ex vivo with BMV099 followed by gel analysis of in vivo and ex vivo fluorescently labeled cysteine cathepsins (Figure 5B and C). This ex vivo labeling confirmed that in vivo probe labeling correlated with the absolute levels of cysteine cathepsins that were expressed and active in each cell population and was not the result of unequal uptake of the probe by the different cell populations. Consistent with the overall mean fluorescence levels, the F4/80HI population showed the most robust labeling of cathepsin S and to a lesser extent cathepsin B, similar to the labeling profile observed in whole tumor homogenates (Figure 4B). We observed weaker labeling intensities in the other three populations, with a slightly more pronounced labeling in the F4/80DIM population. We also observed a similar trend when analyzing the same gel for ex vivo BMV099 labeled cysteine cathepsins, indicating that the highest cysteine cathepsin activity was in the F4/80HI population. Interestingly, the degree of F4/80 expression seems to correlate with cysteine cathepsin activity, which suggests that cysteine cathepsin activity could be indicative of differences in activation phenotypes of myeloid-derived cells.
Figure 5.
FACS analysis to determine cellular source of cysteine cathepsin activity in 4T1 syngeneic breast tumors. 4T1 cells were implanted into mammary fat pad number 2 and 7 of female BALB/c mice. On day 15, BMV083 (20 nmol) was injected (intravenous via tail vein). 10 hr post injection tumors were excised. A–C) Single cell suspensions of excised tumors were analyzed by FACS for F4/80 and GFP expression and probe fluorescence. 4 populations (F4/80−GFP−, F4/80−GFP+, F4/80DIM and F4/80HI) were analyzed and sorted. A) F4/80 histogram (top panel) and GFP histogram of F4/80− population (lower panel). B) Equal cell numbers per population (depicted in A) of vehicle and BMV083 treated mice were lysed and the active cysteine cathepsins were labeled with BMV099 (1 μM for 1 hr) before the proteins were resolved on 15% SDS-PAGE. Fluorescently labeled proteins were visualized by in-gel fluorescence scanning (top panel, in vivo BMV083 labeling; lower panel, ex vivo BMV099 labeling, *auto-fluorescent band). C) Bar graph of quantification of mean Cy5 fluorescence per population (depicted in A) and in-gel fluorescence intensity of fluorescently labeled cysteine cathepsins (depicted in B) of vehicle and BMV083 treated mice. D, E) Single cell suspensions of excised tumors were analyzed by FACS for CD11b, F4/80, MMR and Gr-1 expression and probe fluorescence. D) FACS plot and bar graph of mean Cy5 fluorescence of four populations (CD11b−F4/80−, CD11b+F4/80−, CD11b+F4/80DIM and CD11b+F4/80HI) of vehicle and BMV083 treated mice. E) FACS plot and bar graph of mean Cy5 fluorescence of four populations (MMR−Gr-1−, MMR−Gr-1+, MMR+Gr-1− and MMR+Gr-1+) of CD11b+F4/80HI population depicted in D. See also supplemental figure S6.
To further investigate which sub-populations of macrophages have the highest levels of cysteine cathepsin activity in the tumor microenvironment we included three markers to identify the activation state of the labeled myeloid-derived cells. CD11b+Gr-1+ myeloid-derived suppressor cells (MDSCs) have been shown to possess high cysteine cathepsin activity in the APCΔ468 mouse model of hereditary polyposis (Gounaris, et al., 2008). It is also generally believed that tumor-associated macrophages (TAMs) possess an M2 like macrophage character (Mukhtar, et al., 2011). We therefore used the macrophage mannose receptor (MMR, also known as CD206), which is elevated in M2 compared to M1 macrophages as a marker for the different macrophage populations (Mukhtar, et al., 2011). Because of recent reports concerning the immunogenicity of GFP in Balb/c mice (Bosiljcic, et al., 2011), we also implanted 4T1 cells lacking the GFP reporter in the number 2 and 7 mammary fat pad of Balb/c mice. We systemically administered BMV083 via tail vein injection and excised tumors at 10 hours post injection to generate single cell suspensions. Consistent with the results in the 4T1-luc-GFP tumors, we observed the highest levels of probe activation in the F4/80HI cell population as determined by FACS analysis (CD11b+F4/80HI, Figure 5D). To further characterize the probe positive cells, we analyzed the CD11b+F4/80HI population for Gr-1 and MMR expression (Figure 5E). This analysis determined that BMV083 labeling occurs most predominantly in CD11b+F4/80HI cells that express MMR. Furthermore, of the CD11b+F4/80HIMMR+ cells, the Gr-1 expressing population showed more than 2 fold higher probe accumulation compared to the Gr-1 negative population. This data indicates that the CD11b+F4/80HIMMR+Gr-1+ cells are the major cellular source of cathepsin S activity in the 4T1 tumor microenvironment. CD11b+F4/80+Gr-1+ cells in mouse models of cancer have been defined as myeloid-derived suppressor cells (MDSCs). In response to TLR4 ligands released from damaged tumor cells, MDSCs inhibit antitumor T cell responses by inducing apoptosis in activated T cells (Liu, et al., 2010).
The data presented here shows that the cathepsin S-directed non-peptidic NIRF qABP BMV083 has improved in vivo properties compared to previously developed ABPs. In the syngeneic 4T1 mouse model for breast cancer, this probe primarily reports on the cysteine cathepsin activity in the tumor promoting M2 type CD11b+F4/80HIMMR+ macrophages. Of these, the Gr-1 expressing MDSCs constitute the highest degree of probe activation. Because infiltration of these cell types into the tumor microenvironment plays important roles in tumor development and the fact that their occurrence is correlated with poor prognosis and negative effects on tumor immunotherapy, BMV083 could be of prognostic value. Furthermore, BMV083 may find application in the study of the role of cathepsin S in other pathologies such as atherosclerosis, inflammatory disorders and rheumatoid arthritis. (Conus and Simon, 2010; Quillard, et al., 2011; Reiser, et al., 2010)
SIGNIFICANCE
Cells of myeloid origin are major contributors to the development of many disease states. Novel in vivo imaging tools for macrophages could greatly facilitate the study of their role in disease pathology (Quillard, et al., 2011). The consequence of macrophage infiltration in tumor development has been extensively studied in the last few decades. Some consider cancer-related inflammation the seventh hallmark of cancer (Colotta, et al., 2009), while others consider tumor-promoting inflammation a hallmark enabling characteristic (Hanahan and Weinberg, 2011). Regardless, it is clear that tumor-associated macrophages play important roles in tumor development and have been shown to promote angiogenesis, tumor growth and invasiveness (Mantovani, et al., 2008; Qian and Pollard, 2010). Furthermore, macrophage infiltration into tumors has been correlated with poor clinical outcome in several cancers (Bingle, et al., 2002; Kurahara, et al., 2011; Steidl, et al., 2010). The activation state acquired by the infiltrating macrophage in the tumor microenvironment determines its effect on the tumor. Macrophages with the M2 phenotype exhibit tumor promoting characteristics, whereas M1 macrophages are able to eradicate tumors. The macrophage activation state is plastic and tumor promoting macrophages can be converted into tumor killing M1 macrophages, for example, by local treatment with Granulocyte Macrophage Colony-Stimulating Factor (Eubank, et al., 2009). Non-invasive imaging tools that report on the tumor promoting M2 type tumor-associated macrophage content could therefore be of prognostic value. This manuscript presents the synthesis and characterization of BMV083, a non-peptidic cathepsin S-directed NIRF qABP. This probe shows improved in vivo properties compared to our previous generation of peptidic quenched probes, with higher tumor specific fluorescence in non-invasive optical imaging experiments. Furthermore, the primary cellular source of probe labeling in the tumor was determined to be macrophages with an M2 phenotype, making BMV083 a potential in vivo reporter for tumor-associated macrophages that primarily promote tumor growth.
Experimental Procedures
General
All resins and reagents were purchased from commercial suppliers and used without further purifications. All solvents used were HPLC grade. All water-sensitive reactions were preformed in anhydrous solvents under positive pressure of argon. Reactions were analyzed by LC-MS using an API 150EX single-quadrupole mass spectrometer (Applied Biosystems). Reverse-phase HPLC was conducted with an ÅKTA explorer 100 (Amersham Pharmacia Biotech) using C18 columns. NMR spectra were recorded on a Varian 400 MHz (400/100) or a Varian Inova 600 MHz (600/125 MHz) equipped with a pulsed field gradient accessory. Chemical shifts are given in ppm (δ) relative to tetramethylsilane as an internal standard. Coupling constants are given in Hz.Fluorescent gels were scanned using a Typhoon 9400 flatbed laser scanner (GE Healthcare). Statistical analysis was performed using Microsoft Excel, and s.e.m. was calculated by dividing the s.d. by the square root of n. Fluorescent images were acquired on a Zeiss Axiovert 200 M inverted microscope equipped with a 10×, 40× and 63× objective (Carl Zeiss). Slidebook software was used to control the microscope and camera and for data analysis (Intelligent Imaging Innovations).
ABP synthesis
For the synthesis and characterization of BMV011, BMV083 and BMV099 see the Supplementary Information.
Cell culture and labeling of living cells
RAW cells were cultured in DMEM (GIBCO) supplemented with 10% fetal bovine serum (FBS; GIBCO), 100 units/mL penicillin and 100 μg/mL streptomycin (GIBCO). 4T1 cells (ATCC) and 4T1-luc-GFP cells were cultured in RPMI (GIBCO) supplemented with 10% fetal bovine serum (FBS; GIBCO), 100 units/mL penicillin and 100 μg/mL streptomycin (GIBCO). Primary mouse bone-marrow derived macrophages (BMDMs) were isolated and cultured as previously described (Broz, et al., 2010). Briefly, bone marrow was isolated from femurs of 6- to 8-week old C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME). BMDMs were differentiated in DMEM (GIBCO) with 10% FBS and 10% MCSF-containing cell supernatant for 6 days. The cells were then frozen in FBS with 10% DMSO and thawed prior to use. All cells were cultured in a 5% CO2 humidified incubator at 37 °C. Co-cultures of 4T1-luc-GFP cells and BMDMs were established by seeding cells in a 2 to 1 ratio and culture in DMEM supplemented 10% heat inactivated fetal bovine serum (GIBCO), 100 units/mL penicillin and 100 μg/mL streptomycin (GIBCO) for 90 hr. For the derivation of primary human macrophages leukocyte reduction system chambers were obtained from anonymous blood donations at the Stanford Blood Center. Peripheral blood mononuclear cells were isolated by Ficoll gradient centrifugation, then monocytes were purified on an AutoMACS (Miltenyi) by positive selection using CD14 microbeads (Miltenyi). Monocytes were differentiated to macrophages by culturing for nine days in Iscove’s Modified Dulbecco’s Medium supplemented with 10% AB human serum (Invitrogen), 100U/mL penicillin and 100μg/mL streptomycin (Invitrogen). For intact cell labeling, cells were exposed to probe or inhibitor (1 hr preincubation) where indicated (1000x in DMSO) for the indicated times. The cells were harvested, washed with PBS and resuspended in hypotonic lysis buffer (50 mM PIPES pH 7.4, 10 mM KCl, 5 mM MgCl2, 2 mM EDTA, 4 mM DTT, and 1% NP-40) and put on ice for 15 min, centrifuged at 4°C and supernatants were collected. 40 μg total protein was denatured with SDS-sample buffer, resolved by SDS-PAGE (15%) and labeled proteases were visualized by scanning the gel with a Typhoon imager (GE Healthcare). Labeling intensities were quantified using Image J software. For lysate labeling of active cysteine cathepsins, cells were lysed in citrate buffer (50mM Citrate pH 5.5 or pH 7.0, 5 mM DDT, 0.5% CHAPS, 0.1% Triton X). Total protein (35 or 40 μg) was labeled with the indicated probe at indicated concentration for 1 hr at 37 °C before analysis as described above. For live cell confocal microscopy, cells were seeded in a 8-well Lab-Tek chambered coverglass system.
Animal models
All animal care and experimentation was conducted in accord with current National Institutes of Health and Stanford University Institutional Animal Care and Use Committee guidelines. For the 4T1 allograft model female athymic nude mice (nu/nu) (6–8 weeks, Charles River Laboratories) were injected subcutaneously with 3·106 4T1-luc-GFP cells in PBS under isoflurane anesthesia and tumor growth was monitored. On day 8, GB137 or BMV083 (20 nmol; 0.8 nmol g−1) was administered via tail vein in 100 μL volume (60% DMSO in PBS). After injection, mice were imaged noninvasively at indicated time points using an IVIS 100 system. After the last time point the mice were anesthetized with isofluorane and killed by cervical dislocation. Tumors were removed, imaged ex vivo using an FMT 2500 (Perkin Elmer) and the tissue was homogenized in muscle lysis buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate in PBS, pH 7.4). Proteins (40 μg) were resolved by SDS-PAGE (15%) and labeled proteases were visualized by scanning the gel with a Typhoon imager (GE Healthcare). Labeling intensities were quantified using Image J software. For the orthotopic mouse breast cancer model female BALB/c mice (6–8 weeks, The Jackson Laboratory) were injected in the fat pad number 2 and 7 with 1·105 4T1-luc-GFP or 4T1 cells (ATCC) in PBS and tumor growth was monitored. On day 15, the mice were injected with BMV083 and analyzed as described above. For ex vivo labeling of active cysteine cathepsins, tissues were removed and homogenized in citrate buffer (50mM Citrate pH 5.5, 5 mM DDT, 0.5% CHAPS, 0.1% Triton X). Total protein (40 μg) was labeled with BMV099 (1μM final concentration) for 1 hr at 37 °C before analysis as described above.
Antibody Staining and Flow Cytometry
The tumor tissue was dissected from the mouse, sliced into 1–2 mm3 pieces with a razor blade and digested at 37 °C for approximately 2 hr with frequent pipetting in Advanced DMEM/F12 (Invitrogen) with 2 mM Glutamax (Invitrogen), 120 μg/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin-B (PSA), with 200 units/ml Collagenase type III (Worthington, Lakewood, NJ) and 100 units/ml DNase I (Worthington). After the digestion reached completion, cells were filtered with 40-μm nylon mesh (BD Biosciences) washed and resuspended at a density of 10·106 cells/mL in cold HBBS (Invitrogen) with PSA and 2% heat inactivated fetal calf serum. To reduce nonspecific binding, cells suspended in staining buffer were blocked on ice for 10 minutes with rat IgG (Sigma) 10 mg/mL at 1:1000. Cells were then stained, in the dark, on ice for 30 minutes with optimal antibodies concentrations, which was determined by titration experiments. Antibodies used include PE-Cy7 anti-mouse F4/80 (BM8), PE-Cy5 anti-mouse CD11b (M1/70) (ebioscience) and FITC anti-mouse MMR (C068C2), PE anti-mouse Gr-1 (RB6-8C5) (BioLegend). Flow Cytometry was performed with a BD FacsAria II with FacsDiva software. Data analysis was performed using Flowjo. For all experiments, side scatter and forward scatter profiles (area and width) were used to eliminate debris and cell doublets. Dead cells were eliminated by excluding DAPI+ cells (Molecular Probes).
Supplementary Material
Acknowledgments
We thank J. Lee for help with synthesis, A. W. Puri for assistance with macrophage isolation and M. A. Child and E. Due for critical discussions. We also thank T. Doyle at the Stanford Small Animal Facility at Stanford for assistance with optical imaging studies and S. R. Lynch at the Stanford University Department of Chemistry NMR facility for assistance. This work was supported by NIH grants R01 EB005011 and R01 AI078947 (to MB). MV is supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research (NWO).
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nature chemical biology. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nature chemical biology. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- Bosiljcic M, Hamilton MJ, Banath JP, Lepard NE, McDougal DC, Jia JX, Krystal G, Bennewith KL. Myeloid suppressor cells regulate the lung environment--letter. Cancer Res. 2011;71:5050–5051. doi: 10.1158/0008-5472.CAN-10-3982. author reply 5052–5053. [DOI] [PubMed] [Google Scholar]
- Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglic D, Globisch A, Kindermann M, Lim NH, Jeske V, Juretschke HP, Bartnik E, Weithmann KU, Nagase H, Turk B, et al. Functional in vivo imaging of cysteine cathepsin activity in murine model of inflammation. Bioorg Med Chem. 2011;19:1055–1061. doi: 10.1016/j.bmc.2010.10.028. [DOI] [PubMed] [Google Scholar]
- Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- Conus S, Simon HU. Cathepsins and their involvement in immune responses. Swiss Med Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- Eubank TD, Roberts RD, Khan M, Curry JM, Nuovo GJ, Kuppusamy P, Marsh CB. Granulocyte macrophage colony-stimulating factor inhibits breast cancer growth and metastasis by invoking an anti-angiogenic program in tumor-educated macrophages. Cancer Res. 2009;69:2133–2140. doi: 10.1158/0008-5472.CAN-08-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Manabe I, Nagai R. Renal collecting duct epithelial cells regulate inflammation in tubulointerstitial damage in mice. J Clin Invest. 2011;121:3425–3441. doi: 10.1172/JCI57582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris E, Tung CH, Restaino C, Maehr R, Kohler R, Joyce JA, Ploegh HL, Barrett TA, Weissleder R, Khazaie K. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS One. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Kirschke H, Wiederanders B, Bromme D, Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem J. 1989;264:467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, Sakoda M, Ueno S, Natsugoe S, Takao S. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. J Surg Res. 2011;167:e211–219. doi: 10.1016/j.jss.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Liu YY, Sun LC, Wei JJ, Li D, Yuan Y, Yan B, Liang ZH, Zhu HF, Xu Y, Li B, et al. Tumor cell-released TLR4 ligands stimulate Gr-1+CD11b+F4/80+ cells to induce apoptosis of activated T cells. J Immunol. 2010;185:2773–2782. doi: 10.4049/jimmunol.1000772. [DOI] [PubMed] [Google Scholar]
- Mahmood U, Weissleder R. Near-infrared optical imaging of proteases in cancer. Mol Cancer Ther. 2003;2:489–496. [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Mukhtar RA, Nseyo O, Campbell MJ, Esserman LJ. Tumor-associated macrophages in breast cancer as potential biomarkers for new treatments and diagnostics. Expert Rev Mol Diagn. 2011;11:91–100. doi: 10.1586/erm.10.97. [DOI] [PubMed] [Google Scholar]
- Patterson AW, Wood WJ, Hornsby M, Lesley S, Spraggon G, Ellman JA. Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method. J Med Chem. 2006;49:6298–6307. doi: 10.1021/jm060701s. [DOI] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillard T, Croce K, Jaffer FA, Weissleder R, Libby P. Molecular imaging of macrophage protease activity in cardiovascular inflammation in vivo. Thromb Haemost. 2011;105:828–836. doi: 10.1160/TH10-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl C, Lee T, Shah SP, Farinha P, Han G, Nayar T, Delaney A, Jones SJ, Iqbal J, Weisenburger DD, et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N Engl J Med. 2010;362:875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Fang M, Alroy J, Sahagian GG. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer. 2008;8:228. doi: 10.1186/1471-2407-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WJ, Patterson AW, Tsuruoka H, Jain RK, Ellman JA. Substrate activity screening: a fragment-based method for the rapid identification of nonpeptidic protease inhibitors. J Am Chem Soc. 2005;127:15521–15527. doi: 10.1021/ja0547230. [DOI] [PubMed] [Google Scholar]
- Zavasnik-Bergant T, Turk B. Cysteine cathepsins in the immune response. Tissue Antigens. 2006;67:349–355. doi: 10.1111/j.1399-0039.2006.00585.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.