Abstract
Unfolded RNase A is known to contain an equilibrium mixture of two forms, a slow-folding form (U1) and a fast-folding form (U2). If U1 is produced after unfolding by the slow cis-trans isomerization of proline residues about X-Pro imide bonds, then the formation of U1 should be catalyzed by strong acids. Therefore, the rate of formation of U1 has been measured at different HClO4 concentrations. After rapid unfolding of the native protein in concentrated HClO4 at 0°, the slow formation of U1 was measured by use of refolding assays. Catalysis of its formation was found at HClO4 concentrations above 5 M. The uncatalyzed reaction follows apparent first-order kinetics but, in the acid-catalyzed range, two reactions are found. The faster reaction produces two-thirds of the slow-folding species and shows acid catalysis above 5 M HClO4. Catalysis of the slower reaction begins at 8 M HClO4. The faster reaction shows a 100-fold increase in rate at 10.6 M HClO4 over the rate of the uncatalyzed reaction of 5 M. The activation enthalpy of the uncatalyzed reaction has been measured in two sets of unfolding conditions: ΔH‡ is 21.5 kcal/mol (1 kcal = 4.2 × 103 J) in 3.3 M HClO4 and 21.0 kcal/mol in 5 M guanidine HCl, pH 2.5.
Both acid catalysis of the formation of U1 and its high activation enthalpy are consistent with the rate-limiting step being cis-trans isomerization either of X-Pro imide bonds or of peptide bond. The rate of the uncatalyzed reaction is in the range expected for proline isomerization and is 0.1% of that of peptide bond isomerization; thus, the simplest explanation for the formation of U1 is proline isomerization. Earlier data, showing that the kinetic properties of the U1 ⇄ U2 reaction in refolding conditions differ from those of proline isomerization, can be explained if there is kinetic coupling between early steps in the folding of U1 and its conversion to U2.
The existence of two acid-catalyzed reactions that are distinguished by the HClO4 concentration at which catalysis begins suggests that at least two essential proline residues produce slow-folding species of RNase A by isomerization after unfolding. Because protonation of imide bonds is responsible for acid catalysis of proline isomerization, the slower reaction probably involves an imide bond with a low pK. It may be the bond connecting Lys-41 and Pro-42, because the positive charge on Lys-41 could make this bond more difficult to protonate.
Keywords: protein folding
Full text
PDF
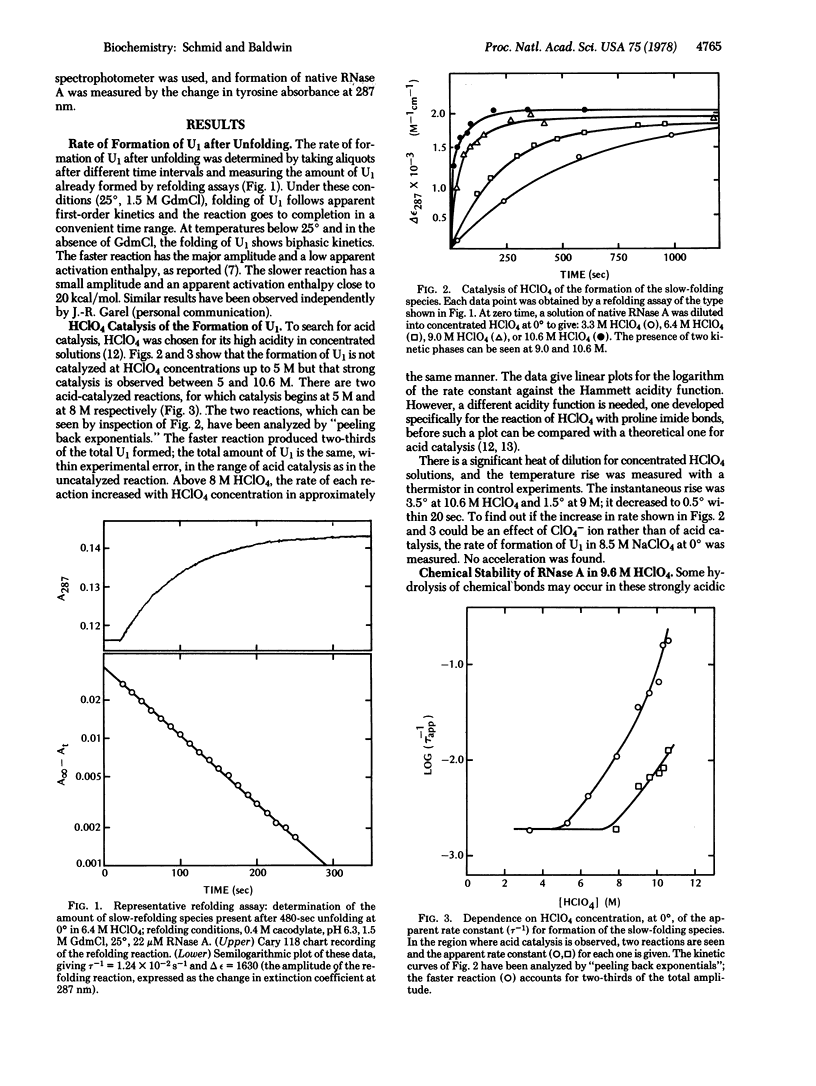
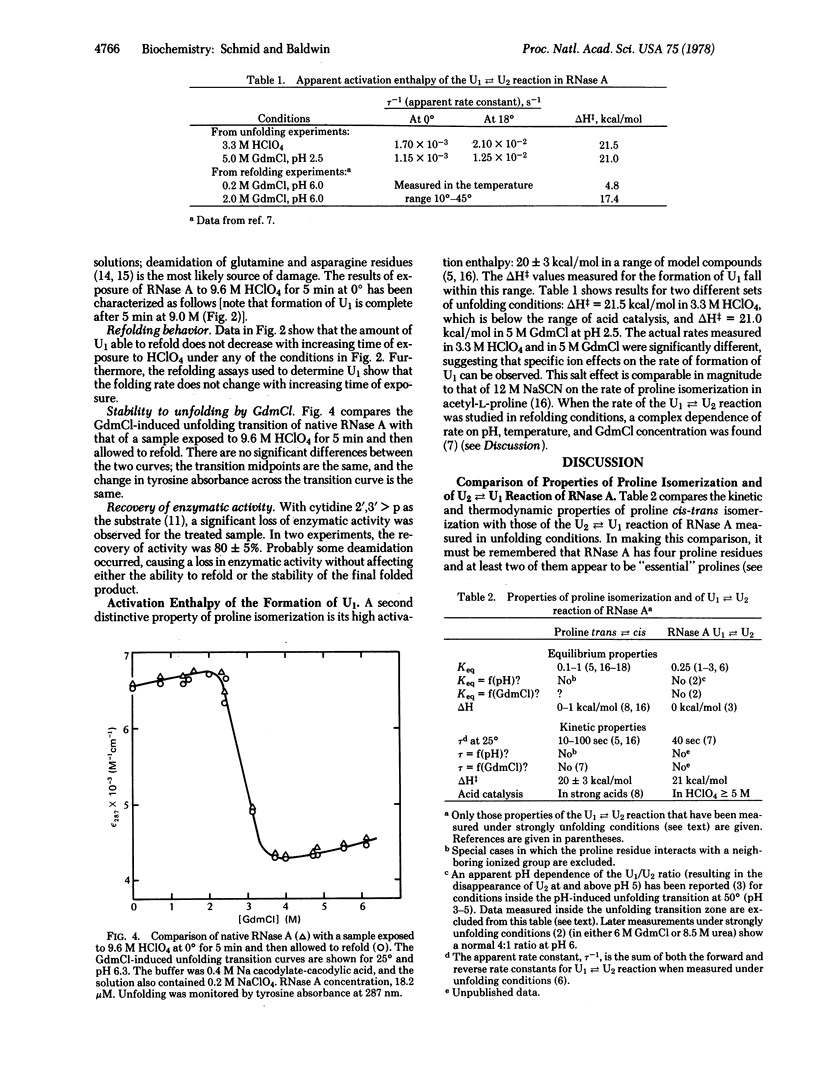


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brandts J. F., Brennan M., Lung-Nan Lin Unfolding and refolding occur much faster for a proline-free proteins than for most proline-containing proteins. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4178–4181. doi: 10.1073/pnas.74.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts J. F., Halvorson H. R., Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975 Nov 4;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- CROOK E. M., MATHIAS A. P., RABIN B. R. Spectrophotometric assay of bovine pancreatic ribonuclease by the use of cytidine 2':3'-phosphate. Biochem J. 1960 Feb;74:234–238. doi: 10.1042/bj0740234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. N., Bovey F. A. Cis-trans equilibrium and kinetic studies of acetyl-L-proline and glycyl-L-proline. Biopolymers. 1977 Jul;16(7):1465–1472. doi: 10.1002/bip.1977.360160707. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Baldwin R. L. A physical difference between the fast- and slow-refolding forms of nitrotyrosyl ribonuclease A: the pK values of the nitrotyrosyl groups. J Mol Biol. 1975 Jun 5;94(4):621–632. doi: 10.1016/0022-2836(75)90326-5. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Baldwin R. L. Both the fast and slow refolding reactions of ribonuclease A yield native enzyme. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3347–3351. doi: 10.1073/pnas.70.12.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel J. R., Baldwin R. L. The heat-unfolded state of ribonuclease A is an equilibrium mixture of fast and slow refolding species. J Mol Biol. 1975 Jun 5;94(4):611–620. doi: 10.1016/0022-2836(75)90325-3. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Nall B. T., Baldwin R. L. Guanidine-unfolded state of ribonuclease A contains both fast- and slow-refolding species. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1853–1857. doi: 10.1073/pnas.73.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grathwohl C., Wüthrich K. Nmr studies of the molecular conformations in the linear oligopeptides H-(L-Ala)n-L-Pro-OH. Biopolymers. 1976 Oct;15(10):2043–2057. doi: 10.1002/bip.1976.360151013. [DOI] [PubMed] [Google Scholar]
- Grathwohl C., Wüthrich K. The X-Pro peptide bond as an nmr probe for conformational studies of flexible linear peptides. Biopolymers. 1976 Oct;15(10):2025–2041. doi: 10.1002/bip.1976.360151012. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J., Baldwin R. L. A quantitative treatment of the kinetics of the folding transition of ribonuclease A. Biochemistry. 1976 Apr 6;15(7):1462–1473. doi: 10.1021/bi00652a017. [DOI] [PubMed] [Google Scholar]
- Manjula B. N., Acharya A. S., Vithayathil P. J. Deamidated active intermediates in the irreversible acid denaturation of ribonuclease-A. Int J Pept Protein Res. 1976;8(3):275–282. doi: 10.1111/j.1399-3011.1976.tb02504.x. [DOI] [PubMed] [Google Scholar]
- Molday R. S., Englander S. W., Kallen R. G. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972 Jan 18;11(2):150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- Nall B. T., Garel J. R., Baldwin R. L. Test of the extended two-state model for the kinetic intermediates observed in the folding transition of ribonuclease A. J Mol Biol. 1978 Jan 25;118(3):317–330. doi: 10.1016/0022-2836(78)90231-0. [DOI] [PubMed] [Google Scholar]
- Robinson A. B., Rudd C. J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr Top Cell Regul. 1974;8(0):247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- SELA M., ANFINSEN C. B. Some spectrophotometric and polarimetric experiments with ribonuclease. Biochim Biophys Acta. 1957 May;24(2):229–235. doi: 10.1016/0006-3002(57)90186-5. [DOI] [PubMed] [Google Scholar]



