Abstract
Endothelin-1 (ET-1), a powerful vasoconstrictor peptide, is produced by activated hepatic stellate cells (HSC) and promotes cell proliferation, fibrogenesis, and contraction, the latter of which has been thought to be mechanistically linked to portal hypertension in cirrhosis. Interferon-γ (IFNγ), a Th1 cytokine produced by T cells, inhibits stellate cell proliferation, fibrogenesis, and muscle-specific gene expression. Whether IFNγ-induced inhibitory effects are linked to regulation of ET-1 expression in activated stellate cells remains unknown. Here we examined IFNγ's effects on preproET-1 mRNA expression and the signaling pathways underlying this process. We demonstrated that preproET-1 mRNA expression in HSCs was prominently increased during cell culture-induced activation; IFNγ significantly inhibited both preproET-1 mRNA expression and ET-1 peptide production. Similar results were found in an in vivo model of liver injury and intraperitoneal administration of IFNγ. PreproET-1 promoter analysis revealed that IFNγ-induced inhibition of preproET-1 mRNA expression was closely linked to the AP-1 and Smad3 signaling pathways. Furthermore, IFNγ reduced JNK phosphorylation, which tightly was associated with decreased phosphorylation of downstream factors c-Jun and Smad3 and decreased binding activity of c-Jun and Smad3 in the preprpET-1 promoter. Importantly, IFNγ reduced both c-Jun mRNA and protein levels. Given the important role of ET-1 in wound healing, our results suggest a novel negative signaling network by which IFNγ inhibits preproET-1 expression, highlighting one potential molecular mechanism for IFNγ-induced host immunomodulation of liver fibrogenesis.
Keywords: liver, cytokine, gene expression, signal pathway
endothelin-1 (ET-1), a 21 amino-acid bioactive peptide, exerts important biological effects on multiple physiological and pathological processes in liver (for a review, see Refs. 7, 24). Under normal conditions, the sinusoidal endothelium is the major source of ET-1, which appears to be important in intrahepatic circulatory homeostasis. However, during liver injury, ET-1 synthesis shifts to activated hepatic stellate cells (HSCs) (21, 27), themselves an apparent target of ET-1 by virtue of their expression of ET-1 A and B receptors (14, 21). These transmembrane guanine nucleotide-binding protein-coupled receptors stimulate a variety of downstream signaling pathways including MAPKs, PI3-kinase/Akt, Jun, and others (5), which modulate multiple biological processes including cell proliferation, survival, and contraction (24). Thus ET-1 has been thought to be a potent agonist in liver fibrosis. However, liver fibrogenesis is prominently modulated by the host immune response, for example, interferon-γ (IFNγ) knockout deficient mice developed more liver fibrosis, while Th1 cytokine dominant mice or animals with administration of IFNγ exhibited reduced liver fibrosis (26, 31). Those findings suggest that ET-1 expression may be regulated through IFNγ. Whether IFNγ-mediated inhibitory effects on HSC links to regulation of ET-1 expression remains elusive.
ET-1 biosynthesis has been extensively studied (7). Regulation of ET-1 expression appears to be complicated (32). It has been shown that preproET-1 expression is regulated by both transcriptional and posttranscriptional mechanisms (29, 20). The preproET-1 gene promoter contains several putative cis-acting elements, including AP-1 and Smad. AP-1 is required for constitutive ET-1 promoter activity. In contrast, Smad binding appears to be required for TGF-β-induced ET-1 expression via activation of the Smad signaling pathway (29). The 3′ UTR region of preproET-1 contains three AUUUA motifs, which destabilize ET-1 transcripts through an AUF1-proteasome pathway (20). The preproET-1 mRNA is labile with a half-life of < 1 h (15). Such rapid turnover of ET-1 mRNA allows for stringent control over its expression in response to extracellular environmental stimuli.
We have recently shown that IFNγ inhibits muscle-specific gene expression in HSCs through targeting SRF (30). However, ET-1 appears not to be a SRF target gene since no functional SRF binding site in the preproET-1 promoter has been identified. In the present study, we have demonstrated that the preproET-1 gene is a novel target of IFNγ in activated HSCs, and the mechanism by which IFNγ inhibits preproET-1 expression appears to be via suppression of JNK phosphorylation, which leads to decreased phospho-c-Jun and phospho-Smad3, resulting in decreased preproET-1 transcription. Furthermore, IFNγ may inhibit preproET-1 transcription via targeting c-Jun expression. Our results highlight a novel mechanism whereby IFNγ modulates myofibroblast-mediated liver fibrosis through targeting preproET-1 expression.
MATERIALS AND METHODS
Animals and cell culture.
HSCs were isolated from retired breeder Sprague-Dawley rats (Charles River Laboratory) as described previously (30). Isolated HSCs were subjected to activation by incubation in 199OR medium containing 20% serum (10% horse serum and 10% calf serum; Invitrogen, Carlsbad, CA) for 4–5 days. Activated HSCs were then subjected to starvation in 0.2% serum 199OR medium overnight before exposure to IFNγ (500 IU/ml for luciferase assays and 1,000 IU/ml for other experiments) (PBL Biomedical) as described before (10, 12), or pharmacological inhibitors against MEK/ERK (U0126), p38-MAPK (SB203580), JNK (SP600125), and TGF-β type I receptor (SB431542) (Calbiochem). For in vivo experiment, Alzet osmotic pumps (model 1002) with murine IFNγ (140,000 IU per pump, which releases 10,000 IU/day for 14 days, PeproTech) or carrier control (1 × PBS containing 1% BSA) were implanted into the peritoneal cavity of C57/black 6 male mice (22–25 g). The animals were then gavaged with carbon tetrachloride (CCL4) once a week for 2 wk (26, 31, 16). Animals were cared for and experiments were performed in accordance with National Institutes of Health (NIH) guidelines. All experimental procedures were approved by the University of Texas Southwestern Institutional Animal Care and Use Committee.
Plasmids and luciferase assay.
The preproET-1 gene promoter was cloned from a mouse HSC genomic DNA. The specific PCR primers were designed according to the sequence from GenBank database (NT_039580. Mus strain C56BL/6J Chromosome 13). Sense primer: 5′-CAGAGGTCCCTCAGCTGAAGG-3′ and antisence primer: 5′-AGATCTCAGCG CGGTCCTCAG-3′, which span 2,252 bp of the preproET-1 5′ flank region between +131 and −2121 bp. The cloned ppET-1 promotor fragment was ligated into pGL3 basic luciferase reporter vector (Promega) at HindIII and XhoI sites. The luciferase constructs harboring truncated ppET-1 gene promoter fragments were generated by PCR method and mutant AP-1 (from TGACTA to TATCTA), and/or Smad3 (from CAGAC to TACAT) sites were created by site-directed mutagenesis (Stratagene). All constructs were confirmed by sequencing (University of Texas Southwestern Sequencing Core). JNK1/2-KM (dominant active form) and JNK1/2-DN (dominant negative form) expression plasmids were obtained from Dr. Lin (The University of Chicago, Chicago, IL).
Activated HSCs were transiently transfected by using Lipofectamine 2000 (Invitrogen). The cells were exposed to IFNγ (500 IU/ml) for 2 days, and luciferase activity was assayed by using dual luciferase assay system (Promega). Transfections were performed in triplicates, and the experiments were repeated three times. The promoter activity was presented as a fold change relative to the activity of promoter-less luciferase basic reporter (mean ± SD, n = 3, P < 0.05 for comparison of IFNγ to control). For cotransfection experiments, 1 μg of each plasmid was used (total 2 μg of plasmid DNA per transfection).
RNase protection assay and RT-PCR.
RNase protection assay was performed as described (31). Briefly, a cRNA probe used to detect preproET-1 mRNA was generated by TA cloning (Invitrogen) of a 213 bp fragment, encoding the first 71 amino acids of ppET-1. The primer sequences were as follows: sense 5′-ATGGATTATTTTCCCGTGATC-3′; antisense 5′-GATGTCCAGGTGGCAGAAGTA-3′. 32P-labeled probes were generated with an in vitro transcription system (Promega) and purified by NucTrap column (Stratagene). A control probe (GAPDH) was obtained from Ambion; and tRNA was used as negative control. The specific signals were quantitated by image analysis (Syngene, Frederick, MD). The average raw volume from the first control samples (IFNγ −) in each experiment was arbitrarily set to 1 or 100% and experimental data were presented as relative abundance (30).
RT-PCR (real-time PCR) was performed as in Shi and Rockey (30). The primer sequences for c-jun were as follows: sense 5′-ACGACCTTCTACGACGATGCC-3′; antisense 5′-TGCCCATTGCTGGACT GGATG-3′. For ppET-1, sense 5′-GACATCATCTGGGTCAACACTC-3′; antisense 5′-CATCTAACTGCCTGGTCTGTG-3′ (8). The data were presented graphically as relative quantification (2−ΔΔCt) and resultant PCR products were subjected to 1.5% agarose gel electrophoresis.
Immunoblot.
HSCs were lysed in RIPA buffer containing protease inhibitors (Roche) as described (30). Samples were subjected to SDS-PAGE and immunoblotting with specific antibody against ERK, phospho-ERK, p38, phospho-p38, JNK, phospho-JNK, c-Jun, phospho-c-Jun, Smad3, phosphor-Smad3 (Cell Signaling), smooth muscle α-actin (Sigma), and collagen type 1 (Rockland, PA). Following incubation with secondary antibody, specific signals were visualized by using an enhanced chemiluminescence detection kit (Pierce). Specific bands were quantitated by image analysis (Bio Image System, Syngene). The raw value for controls (IFNγ −) in the first lane of each experiment was arbitrarily set to 100, and the data were presented as percent change (30).
ET-1 peptide measurement.
Conditioned cell culture medium was harvested and cleared with centrifugation at 500 g for 7 min. The supernatants were subjected to sandwich ELISA kits according to manufacturer's instruction (Assay Designs) and as previously described (36).
EMSA.
EMSA was performed as described (30). The sequence of the sense strand oligonucleotide for preproET-1 promoter harboring Smad3 site was 5′-CTGGATTGTCAGACGGCGGGCGTC-3′. The sequence of the sense strand oligonucleotide for preproET-1 promoter harboring AP-1 site was 5′-GTTGCCTGTGGGTGACTAATCACACAATAAC-3′ and the mutant preproET-1 AP-1 was 5′-GTTGCCTGTGGGCAACTAATCACACAATAAC-3′. To verify specific DNA-protein interaction, a cold probe or a specific antibody (Cell Signaling) were added in some of the reactions and preincubated for 30 min before incubation with radioactive-labeled probe.
Statistical analysis.
All data are presented as the average of three individual experiments plus standard deviation (mean ± SD). Student's t-tests were used for statistical analysis; values of P < 0.05 were considered significant.
RESULTS
IFNγ inhibits activation-mediated ET-1 synthesis through downregulation of preproET-1 transcription.
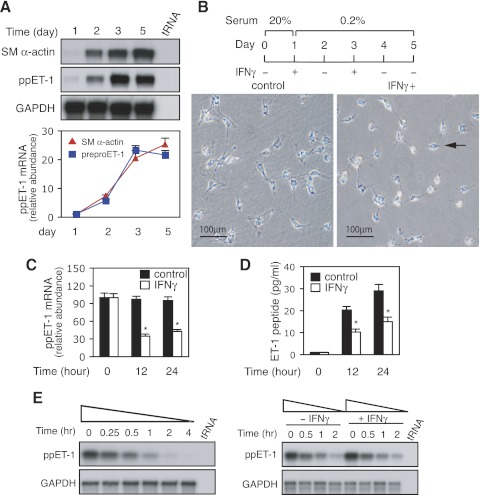
In a (primary) cell culture model system of stellate cell activation that closely recapitulates in vivo stellate cell activation, we examined the correlation between smooth muscle α-actin (SM α-actin) and preproET-1 mRNA expression over time. As shown in Fig. 1A, both SM α-actin and preproET-1 mRNA were expressed in increasing proportions over time. This expression pattern strongly suggested that preproET-1 expression was closely associated with HSC activation (Fig. 1A). We next examined the effect of IFNγ on hepatic stellate cell activation. As shown in Fig. 1B, activated HSCs were spread and had an activated appearance on the 5th day of culture. This activation process was prominently blocked by exposure to IFNγ (Fig. 1B), and HSCs were rounded and appeared refractile (but were still viable as evidenced by exclusion of propidium iodide). Also, we examined the effect of IFNγ on preproET-1 expression in activated HSCs. As shown in Fig. 1C, IFNγ exposure for 12 h led to a reduction in preproET-1 mRNA expression. The inhibitory effect remained prominent at later time points also (Fig. 1C). As expected, ET-1 peptide synthesis was significantly reduced in HSCs after IFNγ exposure (Fig. 1D). Of note, ET-2 mRNA, and ET-3 mRNA (measured by RT-PCR) were detected at levels of < 10% of the abundance of ET-1 mRNA. Further expression of ECE-1 mRNA and protein was not affected by IFNγ exposure. These results suggest that ET-1 is a novel target of IFNγ in activated HSCs and the mechanism of IFNγ's effect is likely to be at the level of preproET-1 regulation.
Fig. 1.
Interferon-γ (IFNγ) inhibits endothelin-1 (ET-1) expression in hepatic myofibrolasts. A: hepatic stellate cells (HSCs) were cultured as in materials and methods and total RNA was isolated at the indicated times. Smooth muscle (SM)-α-actin and preproET-1 (ppET-1) mRNA levels were detected by RNase protection assay (RPA) and signals from specific bands were normalized to control GAPDH mRNA. Top: representative assay; bottom: quantitative data (n = 3 for each time point). B: freshly isolated HSCs were cultured in 20% serum medium for 1 day and then incubated in 0.2% serum medium with or without IFNγ (1,000 IU/ml; top) and phase images (bottom) were taken. The arrow indicates an inactivated HSC following IFNγ exposure. C and D: activated HSCs were incubated in 0.2% serum medium overnight and then exposed to IFNγ (1,000 IU/ml) for 0, 12, or 24 h. Total RNA was isolated and ppET-1 mRNA was detected as in materials and methods (C; n = 3; *P < 0.05 for IFNγ vs. controls); the conditioned supernatants were collected at the indicated times; ET-1 peptide was measured by ELISA and normalized to the total protein content from the monolayer cells (D; n = 3; *P < 0.05 for IFNγ vs. controls). E: stellate cells were incubated in 0.2% serum medium with actinomycin D (10 μg/ml) for the indicated time points. Total RNA was isolated, ppET-1 mRNA was detected by RPA (left). In (right), stellate cells were cultured (left), with the exception that cells were exposed to IFNγ (1,000 IU/ml) for the indicated times. A representative example of 2 different experiments is shown.
Since mRNA stability plays a critical role in determining gene expression (11), we further evaluated preproET-1 mRNA stability in our system. Following actinomycin D exposure for 15 min, preproET-1 mRNA was almost completely degraded after 2 h (Fig. 1E). Next, we examined whether IFNγ had an effect on preproET-1 mRNA decay. As shown in Fig. 1E, IFNγ did not enhance preproET-1 mRNA degradation. The results indicated that IFNγ-mediated downregulation of preproET-1 mRNA expression occurs mainly through transcriptional regulation.
IFNγ exerts a prominent negative effect on preproET-1 promoter activity.
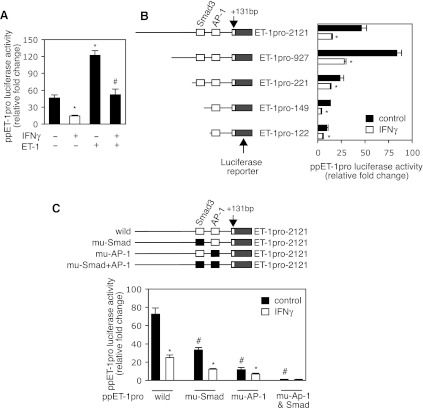
To explore the mechanisms by which IFNγ inhibits preproET-1 expression, we first examined IFNγ's effect on preproET-1 transcription. As shown in Fig. 2A, IFNγ reduced preproET-1 promoter activity up to two to threefold; in contrast, ET-1 robustly increased its own promoter activity. However, the ET-1-mediated increase in preproET-1 promoter activity was almost completely abolished by IFNγ exposure. Next, we generated a series of deletion mutants in the preproET-1 promoter to explore preproET-1 transcriptional responses to IFNγ (Fig. 2B). IFNγ inhibited luciferase activity in all truncated preproET-1 promoter constructs, suggesting that IFNγ-induced inhibitory effects on preproET-1 promoter activity may be mediated through multiple pathways. Since previous studies were shown that AP-1 and Smad3 are critical for preproET-1 promoter activity (18, 29), we hypothesized that the IFNγ-induced inhibitory effect on preproET-1 promoter activity might be associated with one or both of them. Mutation of the AP-1 site led to a remarkable reduction in promoter activity (Fig. 2C), suggesting that AP-1 is critical in regulating preproET-1 promoter activity in stellate cells. Of note, compared with the twofold reduction of the promoter activity caused by IFNγ with the wild-type ppET-1promoter construct, IFNγ induced only a 0.7-fold decrease in the promoter activity with the mutated AP-1 binding site promoter construct. A similar phenomenon was found with the Smad3 mutant, although it was not as prominent as that in the AP-1 mutation construct. Furthermore, mutation of both AP-1 and Smad3 binding sites in the preproET-1 promoter essentially abolished all promoter activity. These findings suggested that both AP-1 and Smad3 signaling pathways are required to induce preproET-1 transcription and that IFNγ-induced inhibitory effects on preproET-1 promoter activity appear to closely associate with AP-1 and Smad3 pathways.
Fig. 2.
Transcriptional analysis of IFNγ-induced effects on the ppET-1 promoter. A: HSCs were transduced with a ppET-1 promoter (ppET-1pro) luciferase reporter plasmid and then exposed to IFNγ (500 IU/ml) or ET-1 (20 nM) for 48 h. Cell lysates were assayed for luciferase activity (n = 3; *P < 0.05 vs. control and #P < 0.05 vs. control with ET-1). B: a series of the truncated ppET-1 promoter luciferase reporter plasmids were generated (left) and stellate cells were transfected and exposed to IFNγ as in A (n = 3; *P < 0.05 vs. control). C: a ppET-1 promoter luciferase reporter plasmid harboring a mutant (mu) AP-1 binding site or Smad3 binding site or both was prepared (top) and stellate cells were transfected and exposed to IFNγ as in A (n = 3; *P < 0.05 for IFNγ vs. control and #P < 0.05 for wild-type ppET-1 luciferase reporter plasmid vs. site mutants).
JNK-c-jun pathway plays a key role in IFNγ-induced inhibition of preproET-1 transcription.
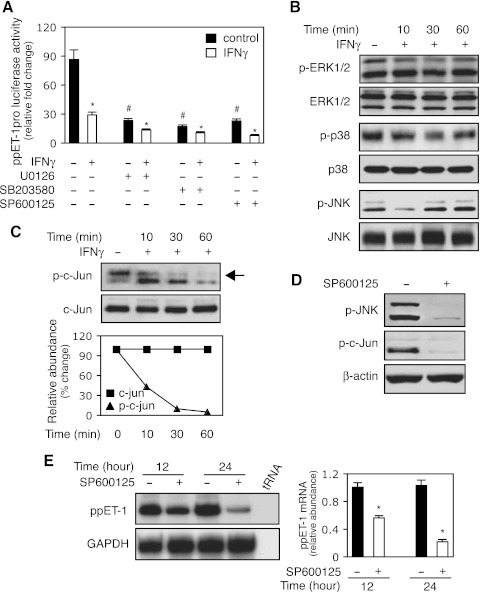
Since the preproET-1 promoter has an AP-1 binding site (29) and MAP kinase mediates a variety of cell activities (5), we hypothesized that IFNγ-induced inhibition of ET-1 expression might proceed through the MAK kinase pathway. Inhibition of ERK, P38, or JNK phosphorylation with specific inhibitors led to prominent reduction in preproET-1 promoter activity, which was further reduced by IFNγ (Fig. 3A). This finding implied that all three MAP kinases could be important in the regulation of ET-1 expression and mediation of IFNγ's inhibitory effect. Next, we examined MAP kinase family member phosphorylation following IFNγ exposure. Importantly, p-JNK was prominently reduced early after IFNγ exposure, while the phosphorylation of ERK and P38 did not appear to be significantly affected (Fig. 3B). Therefore, we explored JNK further. Since c-Jun is downstream of JNK, we hypothesized that given interferon's effect on p-JNK, there would likely also be an effect on phospho-c-Jun. Thus, we examined IFNγ's effect on c-Jun phosphorylation over several time points (Fig. 3C); phospho-c-Jun was reduced shortly after exposure to IFNγ and prominently decreased through 60 min of exposure (Fig. 3C). To confirm that the decrease in c-Jun phosphorylation was a result of reduced JNK phosphorylation, we used a specific JNK inhibitor to block JNK phosphorylation (Fig. 3D); as expected, it also blocked c-Jun phosphorylation (Fig. 3D). Finally, we examined the correlation between JNK phosphorylation and preproET-1 expression by using the JNK inhibitor. As shown in (Fig. 3E), preproET-1 mRNA expression was significantly reduced following JNK inhibition, which was similar to the effect with IFNγ treatment (Fig. 1C). Taken together, these data suggested that JNK-c-Jun pathway might play an important role in IFNγ-induced inhibition of preproET-1 mRNA expression.
Fig. 3.
The JNK/c-Jun pathway mediates IFNγ-induced inhibition of ppET-1 transcription. A: HSCs were transduced with a ppET-1 promoter/luciferase reporter construct (ppET-1–2121+131) and preexposed to U0126 (10 μM, MEK/ERK inhibitor), SB203580 (10 μM, p38-MAPK inhibitor) or SP600125 (10μM, JNK inhibitor) for 30 min followed by the addition of IFNγ (500 IU/ml). Luciferase activity was measured (n = 3; *P < 0.05 vs. corresponding control with no IFNγ; #P < 0.05 vs. no IFNγ). B: HSCs were incubated in 0.2% serum medium with or without IFNγ (1,000 IU/ml) for the indicated times, and whole cell extracts were subjected to immunoblotting with antibodies to detect phospho-ERK1/2 (p-ERK1/2), ERK1/2, phospho-p38 (p-p38), p38, phospho-JNK (p-JNK) and JNK. Representative images of 3 others are shown. C: stellate cells were incubated in 0.2% serum medium with or without IFNγ (1,000 IU/ml) as in (B), and whole cell lysates were subjected to immunoblot with anti-phospho-c-Jun (p-c-Jun) or c-Jun antibodies. Quantitative data are shown (bottom, n = 3). D: stellate cells were exposed to SP600125 (SP, 10 μM) for 30 min, and cell lysates were immunoblotted with p-JNK (top) or p-c-Jun (middle). E: HSCs were incubated in 0.2% serum medium with or without SP600125 (10μM) for the indicated times. PreproET-1 mRNA was detected as in materials and methods, and signals corresponding to specific bands were normalized to control GAPDH mRNA signals. The quantitative data are presented graphically (deviation = 3; *P < 0.05 vs. control).
JNK plays an important role in IFNγ's inhibitory effect on preproET-1 promoter activity.
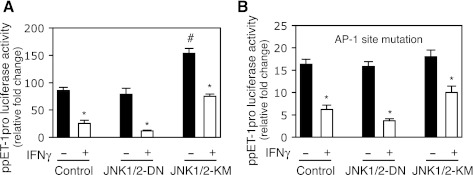
To further explore the functional importance of JNK in IFNγ-induced inhibition of preproET-1 promoter activity, we overexpressed a dominant active or negative form of JNK in HSCs. As expected, preproET-1 promoter activity was significantly increased by overexpression of the dominant active JNK1/2 (JNK1/2-KM) (Fig. 4A). However, overexpression of the dominant negative JNK1/2 (JNK1/2-DN) did not obviously affect preproET-1 promoter activity. Notably, overexpression of the dominant active JNK1/2 significantly blocked IFNγ-induced inhibition of preproET-1 promoter activity (0.9-fold reduction) compared with the control (twofold reduction), but overexpression of the dominant negative JNK1/2 prominently potentiated IFNγ-induced inhibition of preproET-1 promoter activity (3.2-fold reduction) (Fig. 4A). Since AP-1 was essential to preproET-1 promoter activity (Fig. 2C), we explored whether JNK-mediated effect on preproET-1 promoter activity was AP-1 dependent. As shown in Fig. 4B, mutation of the AP-1 site in the preproET-1 promoter led to dramatically decreased promoter activity (similar to that shown in Fig. 2C) and overexpression of the dominant active JNK1/2 failed to significantly increase preproET-1 promoter activity. Moreover, IFNγ-induced inhibitory effect on the preproET-1 promoter activity was obviously compromised compared with that in Fig. 4A. Taken together, these data suggested that JNK was a key factor to mediate IFNγ-induced inhibition of preproET-1 promoter activity via AP-1, but in addition to AP-1, other factors might also mediate IFNγ's effect via JNK pathway.
Fig. 4.
JNK mediates IFNγ-induced inhibition of ppET-1 promoter activity. A: HSCs were cotransfected with ppET-1 promoter luciferase reporter plasmid (ppET1–2121+131) with expression plasmid containing dominant-negative JNK1/2 (JNK1/2-DN) or dominant-active JNK1/2 (JNK1/2-KM) or control expression vector for overnight, exposed to IFNγ (500 IU/ml) for 2 days. The luciferase activity was assayed (n = 3; *P < 0.05 vs. no IFNγ; #P < 0.05 vs. control). B: cotransfection was performed as the same as in A except ppET1–2121+131 construct was replaced with the ppET1–2121+131 containing AP-1 site mutation (n = 3; *P < 0.05 vs. no IFNγ).
IFNγ-induced inhibition of ET-1 expression through a TGF-β-Smad pathway.
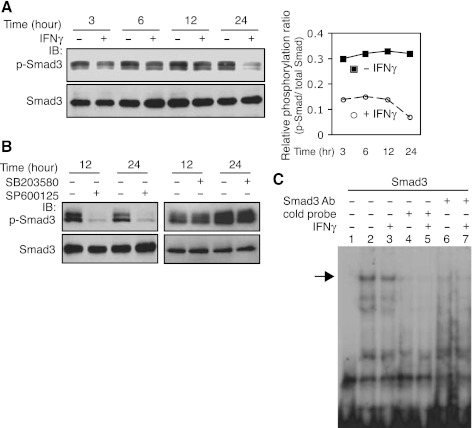
Previous data suggest that IFNγ antagonizes TGF-β's effect on collagen type 1 gene expression via a TGF-β-Smad pathway (13). Additionally, the TGF-β-Smad pathway appears to play a role in regulation of preproET-1 expression in endothelial cells (29). These previous data led us to postulate that IFNγ might inhibit TGF-β-Smad-mediated preproET-1 transcriptional activation in the current system. To explore this possibility, we first examined the effect of IFNγ on Smad3 signaling. Total Smad3 levels were not affected following IFNγ exposure, but Smad3 phosphorylation was reduced at all time points compared with controls (Fig. 5A), which led to a relative decrease in phosphorylation [i.e., the ratio between p-Smad and total Smad in IFNγ-treated stellate cells was reduced (Fig. 5A)]. Since phosphorylation appears to be an important prerequisite for Smad3 binding activity (19), we further examined Smad3 binding activity in the preproET-1 promoter. As shown in Fig. 5C, a weaker shifted band (lane 3) was detected in the sample with IFNγ exposure for 16 h compared with the control (lane 2). To confirm the specificity of binding, an antibody against Smad3 (lanes 6 and 7) and the cold probe (lanes 4 and 5) were also examined. Notably, the DNA-protein complexes in lanes 2 and 3 were essentially depleted by Smad3 antibody or cold probe. We further postulated that reduced Smad3 phosphorylation was likely a result of the effect of IFNγ on JNK phosphorylation (Fig. 3). We found that inhibition of JNK phosphorylation with a JNK specific inhibitor (SP600125) substantially abrogated Smad3 phosphorylation (Fig. 5B). However, blocking Erk phosphorylation did not reduce Smad3 phosphorylation (Fig. 5B), suggesting that JNK, but not Erk, activates Smad3 signaling in our system. Taken together, these data indicate that targeting endogenous TGF-β signaling through a JNK-Smad pathway is an important route for IFNγ-mediated inhibition of preproET-1 mRNA expression in stellate cells.
Fig. 5.
IFNγ inhibits Smad3 phosphorylation and binding in the ppET-1 promoter. A: HSCs were cultured in 0.2% serum medium and exposed to IFNγ (1,000 IU/ml) for the indicated time periods. Phospho-Smad3 (p-Smad3) and total Smad3 were detected by immunoblotting and the signals were quantitated as in materials and methods. The relative phosphorylation ratio is determined by p-Smad vs. total Smad and was presented graphically (right). B: p-Smad3 or Smad3 were detected after exposure to SP600125 (SP, 10μM) (left) or SB203580 (SB, 10 μM) (right) for the indicated time periods by immunoblotting. C: HSCs were cultured in 0.2% serum medium and exposed to IFNγ (1,000 IU/ml) for 16 h. Nuclear extracts were used for EMSA (specific shifted band is indicated by an arrow). Lane 1 is a control (no nuclear extract); cold probe: lanes 4 and 5; anti-Smad3 antibody: lanes 6 and 7.
IFNγ directly inhibits c-Jun expression.
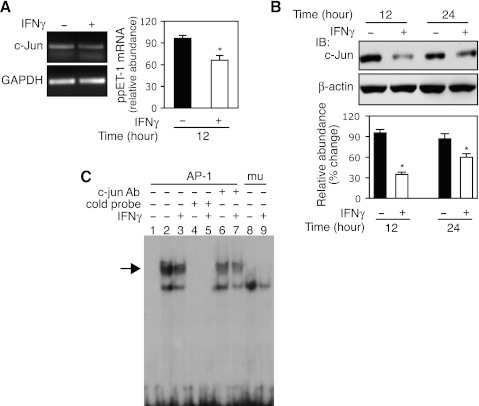
c-Jun is an important component of the AP-1 transcription complex, which is integrally related to cell proliferation and differentiation (34). We hypothesized that IFNγ might directly target c-Jun, which in turn might mediate IFNγ's inhibitory effect on preproET-1 mRNA expression. As shown in Fig. 6A, c-Jun mRNA was decreased ∼ 30% compared with the control following IFNγ exposure for 12 h. We also examined c-Jun protein expression following IFNγ (Fig. 6B). Interestingly, c-Jun protein levels were prominently reduced at 12 h, and IFNγ maintained the inhibitory effect through 24 h. Furthermore, we examined c-Jun binding activity in the preproET-1 promoter (Fig. 6C). As expected, IFNγ exposure led to reduced binding of c-Jun (lane 3) compared with no IFNγ exposure (lane 2). The specific binding was confirmed by using cold probe (lanes 4 and 5), specific antibody against c-Jun (lanes 6 and 7), and AP-1 site mutant probe (lanes 8 and 9). Notably, the cold probe or AP-1 site mutant probe completely abrogated the DNA-protein complex formation as that occurred in lanes 2 and 3, which was prominently reduced by anti-c-Jun antibody (indicated by arrow). Taken together, these data demonstrated that IFNγ signals to c-Jun to negatively regulate preproET-1 transcription.
Fig. 6.
IFNγ directly targets c-Jun. A: following incubation in 0.2% serum medium overnight, HSCs were exposed to IFNγ (1,000 IU/ml) for 12 h, and total RNA was subjected to RT-PCR for c-Jun expression analysis [a representative image (left) and quantitative data (right); n = 3; *P < 0.05 vs. control]. B: HSCs were exposed to IFNγ (1,000 IU/ml) as indicated time points and whole cell lysates were subjected to immunoblotting (n = 3; *P < 0.05 vs. control). C: HSCs were exposed to IFNγ (1,000 IU/ml) for 16 h and nuclear extracts were subjected to EMSA (the specific shifted band is highlighted with an arrow). Lane 1 is a control (no nuclear extract); cold probe: lanes 4 and 5; anti-c-Jun antibody: lanes 6 and 7; mutant probe lanes 8 and 9. The experiments in B and C were repeated twice.
IFNγ downregulates preproET-1 mRNA expression in vivo.
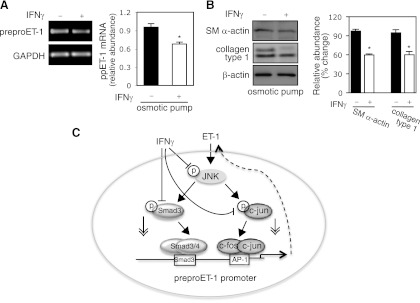
Previous studies demonstrated that IFNγ reduced liver fibrogenesis in animal (rat or mouse) models (26, 16). To explore the in vivo effect of IFNγ, we examined whether IFNγ inhibits preproET-1 mRNA expression in stellate cells following CCl4-induced liver injury. Using osmotic pumps (26), IFNγ was consistently released for 2 wk during CCl4 exposure. As shown in Fig. 7A, preproET-1 mRNA levels in isolated stellate cells from mice receiving IFNγ was significantly decreased compared with control. Smooth muscle α-actin, a molecular marker for stellate cell activation, as well as col1α1 (collagen type 1) was also reduced in stellate cells from mice exposed to IFNγ compared with control (Fig. 7B). In aggregate, our data link IFNγ's negative regulatory effect to preproET-1 transcription during stellate cell activation (Fig. 7C).
Fig. 7.
IFNγ reduces ppET-1 expression during liver injury. A: osmotic pumps containing IFNγ (140,000 IU/100 μl) or carrier (BSA-PBS/100 μl) were implanted into the peritoneal cavity of the mice and then gavaged with CCl4 once a week for 2 wk, beginning 2 days after surgery. Stellate cells were isolated and harvested as in materials and methods and a representative image including specific bands is shown. On the right, ppET-1 mRNA was measured by RT-PCR (n = 5, *P < 0.05 vs. control). B, left: cell lysates were immunoblotted to detect smooth muscle a actin, collagen type 1 (col1α1), and β-actin. On the right, specific bands were scanned, normalized to β-actin, and presented graphically (n = 5, *P < 0.05 vs. control). C: a diagram highlights putative IFNγ-endothelin-1 signaling pathways.
DISCUSSION
The importance of ET-1 in wound healing has been well established (2, 35). However, in liver wound healing the molecular mechanisms underlying preproET-1 regulation remain unclear (24, 32). In this study, we have demonstrated a novel signaling pathway in which ET-1 synthesis is downregulated by IFNγ in HSCs. We have shown that IFNγ inhibits JNK phosphorylation and consequently leads to decreased c-Jun and Smad3 phosphorylation as well as reduced total c-Jun expression and diminished Smad3 and c-Jun binding activity in the preproET-1 promoter, which are required for ET-1 transcriptional activation. Our results link IFNγ signaling to a preproET-1 transcriptional regulation through JNK, Smad3 and c-Jun (Fig. 7C).
ET-1 together with ET-2 and ET-3 comprise the endothelin family of 21 amino acid peptides produced in various cells and tissues, especially endothelial and epithelial lineages. These endothelins are derived from three individual genes (7). Compared with ET-1, ET-2, and ET-3 mRNA levels were almost undetectable in our system, suggesting that ET-1 is the major species of endothelins produced by stellate cells, which further serve as an autocrine target of ET-1 (Fig. 1A). ET-1 not only induces stellate cell proliferation, but also increases extracellular matrix synthesis (such as collagen type I expression) (24). Simultaneously, activated HSCs also respond to IFNγ - including reduced fibrogenesis, reduced proliferation, and inhibition of the smooth muscle gene program typical of myofibroblasts (3, 28). Our data extend these effects to include an inhibitory effect on preproET-1 mRNA expression (Fig. 1, C and D), which likely contributes to IFNγ's antiproliferative and anti-fibrogenic effects (Fig. 1B).
We emphasize that our data suggest one potential mechanism for IFNγ's anti-fibrogenic effect in stellate cells. Indeed, it is widely accepted that IFNγ inhibits ECM/collagen expression in stellate cells by virtue of a number of effects on stellate cells (3, 15, 28). In fact, IFNγ appears to have diverse signaling effects in stellate cells, including beyond those on the ET-1 system.
Transcriptional regulation of preproET-1 expression involves multiple transcription factors (32). However, AP-1 and Smad3 appear to be critical in our system since simultaneous mutation of their binding sites in the preproET-1 promoter essentially abolished all promoter activity, which is very similar to a previous study in bovine aortic endothelial cells (18). Compared with Smad3, AP-1 appears to be more important in mediating the IFNγ-induced inhibitory effect on preproET-1 transcriptional activation since mutation of AP-1 binding site in the promoter alone prominently compromised IFNγ's effect (Fig. 2, B and C). Although all members of the MAPK signaling family (i.e., ERK and P38) appeared to associate with preproET-1 transcriptional activation (Fig. 3A), we found that p-JNK was prominently reduced by IFNγ Fig. 3B. Not unexpectedly, we also found IFNγ potently inhibited phospho-c-Jun (Fig. 3), consistent with previous data, and emphasizing that c-Jun is downstream of JNK (22). Interestingly, overexpression of a dominant-negative JNK greatly facilitated IFNγ-induced inhibition of preproET-1 promoter activity (Fig. 4). Furthermore, inhibition of JNK phosphorylation by a pharmaceutical inhibitor led to a reduction of preproET-1 mRNA expression (Fig. 3, D and E) similar to that induced by IFNγ. Our results likely provide a novel insight into the mechanism of decreased liver fibrosis in JNK1-deficient mice or pan-JNK inhibition in vivo models (17). Notably, ET-1 can robustly increase its own promoter activity (Fig. 2A), which coincides with cotransfection of a dominant-active JNK (Fig. 4A). It has been demonstrated that ET-1 signals to JNK (5). Such signal transduction feedback loop from ET-1 to JNK and JNK to preproET-1 transcription appears to play a critical role in HSC activation. Our highly reproducible data link IFNγ's inhibitory effect on ET-1 expression with JNK-c-Jun pathway.
TGF-β-Smad3 pathway played an important role in regulation of preproET-1 expression (Fig. 2C) (29). Inhibition of endogenous TGF-β signaling by IFNγ appears to be an important mechanism in IFNγ-induced inhibition of preproET-1 transcriptional activation. It was demonstrated that phosphorylation of Smad3 is an essential step for Smad3's DNA binding activity and TGF-β signaling (19). Indeed, IFNγ inhibited Smad3 phosphorylation, which further led to a reduced Smad3 binding in the preproET-1 promoter (Fig. 5C). Previous studies indicated that JNK could physically interact with Smad3 and led to Smad3 phosphorylation (33). Thus, IFNγ-induced inhibition of Smad3 phosphorylation likely came from the effect of IFNγ on JNK since we blocked JNK activity and resulted in essential abrogation of Smad3 phosphorylation (Fig. 5D).
Both c-fos and c-Jun are key components of the AP-1 transcription factor complex, which is critical for regulation of many genes, including collagen type 1 and ET-1 (29, 9). IFNγ has been shown to negatively regulate c-fos expression (23). We here demonstrated that c-Jun was also negatively regulated by IFNγ in our system (Fig. 6). Thus, IFNγ-induced total c-Jun reduction and decreased c-Jun binding activity further support the position that AP-1 is a major molecular target of IFNγ-JNK-c-Jun pathway in preproET-1 transcriptional regulation.
Our data have therapeutic implications. For example, it has been previously demonstrated that abrogation of each the IFNγ and ET-1 signaling systems are capable of reducing fibrosis in animal models. For example, it was shown that inhibition of ET-A receptors (6) or both ET-A and -B receptors (25) inhibited fibrosis. It has also been well established that IFNγ inhibits fibrosis in animal models (26, 3, 31, 16); thus whether a blocking IFNγ and ET-1 in a combined fashion is more effective than either alone is an intriguing and attractive possibility.
It has been well demonstrated that ET-1 is important in regulation of cell proliferation, muscle-specific gene expression, and cellular contraction, which are all closely associated with the pathological wound healing responses, vasculopathies, and even cancer progression (1, 4, 24). Our data in a model of liver wound healing (both in vitro and in vivo), which demonstrate not only a potent effect of IFNγ on ET-1, but also on wound healing, suggest a novel mechanism for the effect of IFNγ (i.e., through ET-1). The data also highlight an important signaling network linking IFNγ to preproET-1 transcription through JNK-Smad3 and c-Jun pathways (Fig. 7C), which may provide a potentially important molecular mechanism for IFNγ's effect in wound healing.
GRANTS
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK-50574 (to D. C. Rockey).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
T.L., Z.S., and D.C.R. conception and design of research; T.L. and Z.S. performed experiments; T.L., Z.S., and D.C.R. analyzed data; T.L., Z.S., and D.C.R. interpreted results of experiments; T.L., Z.S., and D.C.R. prepared figures; T.L., Z.S., and D.C.R. drafted manuscript; T.L., Z.S., and D.C.R. edited and revised manuscript; T.L., Z.S., and D.C.R. approved final version of manuscript.
ACKNOWLEDGMENTS
The mouse preproET-1 promoter sequence has been submitted to GenBank, accession no. EU095329.
REFERENCES
- 1. Abraham D, Dashwood M. Endothelin–role in vascular disease. Rheumatology (Oxford) 47, Suppl 5: v23– v24, 2008. [DOI] [PubMed] [Google Scholar]
- 2. Alam I, Bass NM, Bacchetti P, Gee L, Rockey DC. Hepatic tissue endothelin-1 levels in chronic liver disease correlate with disease severity and ascites. Am J Gastroenterol 95: 199– 203, 2000. [DOI] [PubMed] [Google Scholar]
- 3. Baroni GS, D'Ambrosio L, Curto P, Casini A, Mancini R, Jezequel AM, Benedetti A. Interferon γ decreases hepatic stellate cell activation and extracellular matrix deposition in rat liver fibrosis. Hepatology 23: 1189– 1199, 1996. [DOI] [PubMed] [Google Scholar]
- 4. Bhalla A, Haque S, Taylor I, Winslet M, Loizidou M. Endothelin receptor antagonism and cancer. Eur J Clin Invest 39, Suppl 2: 74– 77, 2009. [DOI] [PubMed] [Google Scholar]
- 5. Bouallegue A, Daou GB, Srivastava AK. Endothelin-1-induced signaling pathways in vascular smooth muscle cells. Curr Vasc Pharmacol 5: 45– 52, 2007. [DOI] [PubMed] [Google Scholar]
- 6. Cho JJ, Hocher B, Herbst H, Jia JD, Ruehl M, Hahn EG, Riecken EO, Schuppan D. An oral endothelin-A receptor antagonist blocks collagen synthesis and deposition in advanced rat liver fibrosis. Gastroenterology 118: 1169– 1178, 2000. [DOI] [PubMed] [Google Scholar]
- 7. Davenport AP, Maguire JJ. Endothelin. Handb Exp Pharmacol 176: 295– 329, 2006. [DOI] [PubMed] [Google Scholar]
- 8. de Frutos S, Duling L, Alo D, Berry T, Jackson-Weaver O, Walker M, Kanagy N, Gonzalez Bosc L. NFATc3 is required for intermittent hypoxia-induced hypertension. Am J Physiol Heart Circ Physiol 294: H2382– H2390, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedman SL. Transcriptional regulation of stellate cell activation. J Gastroenterol Hepatol 21, Suppl 3: S79– S83, 2006. [DOI] [PubMed] [Google Scholar]
- 10. Fujita T, Maesawa C, Oikawa K, Nitta H, Wakabayashi G, Masuda T. Interferon-γ down-regulates expression of tumor necrosis factor-α converting enzyme/a disintegrin and metalloproteinase 17 in activated hepatic stellate cells of rats. Int J Mol Med 17: 605– 616, 2006. [PubMed] [Google Scholar]
- 11. Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113– 126, 2007. [DOI] [PubMed] [Google Scholar]
- 12. Ghosh AK, Yuan W, Mori Y, Chen S, Varga J. Antagonistic regulation of type I collagen gene expression by interferon-γ and transforming growth factor-β. Integration at the level of p300/CBP transcriptional coactivators. J Biol Chem 276: 11041– 11048, 2001. [DOI] [PubMed] [Google Scholar]
- 13. Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-γ interferes with transforming growth factor-β signaling through direct interaction of YB-1 with Smad3. J Biol Chem 278: 43470– 43479, 2003. [DOI] [PubMed] [Google Scholar]
- 14. Housset C, Rockey DC, Bissell DM. Endothelin receptors in rat liver: lipocytes as a contractile target for endothelin 1. Proc Natl Acad Sci USA 90: 9266– 9270, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inoue A, Yanagisawa M, Takuwa Y, Mitsui Y, Kobayashi M, Masaki T. The human preproendothelin-1 gene. Complete nucleotide sequence and regulation of expression. J Biol Chem 264: 14954– 14959, 1989. [PubMed] [Google Scholar]
- 16. Jeong WI, Park O, Radaeva S, Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology 44: 1441– 1451, 2006. [DOI] [PubMed] [Google Scholar]
- 17. Kluwe J, Pradere JP, Gwak GY, Mencin A, De Minicis S, Osterreicher CH, Colmenero J, Bataller R, Schwabe RF. Modulation of hepatic fibrosis by c-Jun-N-terminal kinase inhibition. Gastroenterology 138: 347– 359, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee ME, Dhadly MS, Temizer DH, Clifford JA, Yoshizumi M, Quertermous T. Regulation of endothelin-1 gene expression by Fos and Jun. J Biol Chem 266: 19034– 19039, 1991. [PubMed] [Google Scholar]
- 19. Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci USA 94: 10669– 10674, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mawji IA, Robb GB, Tai SC, Marsden PA. Role of the 3′-untranslated region of human endothelin-1 in vascular endothelial cells. Contribution to transcript lability and the cellular heat shock response. J Biol Chem 279: 8655– 8667, 2004. [DOI] [PubMed] [Google Scholar]
- 21. Pinzani M, Milani S, De Franco R, Grappone C, Caligiuri A, Gentilini A, Tosti-Guerra C, Maggi M, Failli P, Ruocco C, Gentilini P. Endothelin 1 is overexpressed in human cirrhotic liver and exerts multiple effects on activated hepatic stellate cells. Gastroenterology 110: 534– 548, 1996. [DOI] [PubMed] [Google Scholar]
- 22. Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-jun mediated by MAP kinases. Nature 353: 670– 674, 1991. [DOI] [PubMed] [Google Scholar]
- 23. Radzioch D, Varesio L. c-fos mRNA expression in macrophages is downregulated by interferon-γ at the posttranscriptional level. Mol Cell Biol 11: 2718– 2722, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rockey DC. Hepatic fibrosis, stellate cells, and portal hypertension. Clin Liver Dis 10: 459– 479, vii–viii, 2006. [DOI] [PubMed] [Google Scholar]
- 25. Rockey DC, Chung JJ. Endothelin antagonism in experimental hepatic fibrosis. Implications for endothelin in the pathogenesis of wound healing. J Clin Invest 98: 1381– 1388, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rockey DC, Chung JJ. Interferon γ inhibits lipocyte activation and extracellular matrix mRNA expression during experimental liver injury: implications for treatment of hepatic fibrosis. J Investig Med 42: 660– 670, 1994. [PubMed] [Google Scholar]
- 27. Rockey DC, Fouassier L, Chung JJ, Carayon A, Vallee P, Rey C, Housset C. Cellular localization of endothelin-1 and increased production in liver injury in the rat: potential for autocrine and paracrine effects on stellate cells. Hepatology 27: 472– 480, 1998. [DOI] [PubMed] [Google Scholar]
- 28. Rockey DC, Maher JJ, Jarnagin WR, Gabbiani G, Friedman SL. Inhibition of rat hepatic lipocyte activation in culture by interferon-γ. Hepatology 16: 776– 784, 1992. [DOI] [PubMed] [Google Scholar]
- 29. Rodriguez-Pascual F, Redondo-Horcajo M, Lamas S. Functional cooperation between Smad proteins and activator protein-1 regulates transforming growth factor-β-mediated induction of endothelin-1 expression. Circ Res 92: 1288– 1295, 2003. [DOI] [PubMed] [Google Scholar]
- 30. Shi Z, Rockey DC. Interferon-γ-mediated inhibition of serum response factor-dependent smooth muscle-specific gene expression. J Biol Chem 285: 32415– 32424, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi Z, Wakil AE, Rockey DC. Strain-specific differences in mouse hepatic wound healing are mediated by divergent T helper cytokine responses. Proc Natl Acad Sci USA 94: 10663– 10668, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stow LR, Jacobs MEL, Wingo CS, Cain BD. Endothelin-1 gene regulation. FASEB J 25: 16– 28, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velden JL, Alcorn JF, Guala AS, Badura EC, Janssen-Heininger YM. c-Jun N-terminal kinase 1 promotes transforming growth factor-β1-induced epithelial-to-mesenchymal transition via control of linker phosphorylation and transcriptional activity of Smad3. Am J Respir Cell Mol Biol 44: 571– 581, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vesely PW, Staber PB, Hoefler G, Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat Res 682: 7– 12, 2009. [DOI] [PubMed] [Google Scholar]
- 35. Xu SW, Howat SL, Renzoni EA, Holmes A, Pearson JD, Dashwood MR, Bou-Gharios G, Denton CP, du Bois RM, Black CM, Leask A, Abraham DJ. Endothelin-1 induces expression of matrix-associated genes in lung fibroblasts through MEK/ERK. J Biol Chem 279: 23098– 23103, 2004. [DOI] [PubMed] [Google Scholar]
- 36. Zhan S, Chan CC, Serdar B, Rockey DC. Fibronectin stimulates endothelin-1 synthesis in rat hepatic myofibroblasts via a Src/ERK-regulated signaling pathway. Gastroenterology 136: 2345– 2355 e2341–e2344, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]