Abstract
The farnesoid X receptor (FXR) is a ligand (bile acid)-dependent nuclear receptor that regulates target genes involved in every aspect of bile acid homeostasis. Upon binding of ligand, FXR recruits an array of coactivators and associated proteins, some of which have intrinsic enzymatic activity that modify histones or even components of the transcriptional complex. In this study, we show chromatin occupancy by the Set7/9 methyltransferase at the FXR response element (FXRE) and direct methylation of FXR in vivo and in vitro at lysine 206. siRNA depletion of Set7/9 in the Huh-7 liver cell line decreased endogenous mRNAs of the FXR target genes, the short heterodimer partner (SHP) and bile salt export pump (BSEP). Mutation of the methylation site at K206 of FXR to an arginine prevented methylation by Set7/9. A pan-methyllysine antibody recognized the wild-type FXR but not the K206R mutant form. An electromobility shift assay showed that methylation by Set7/9 enhanced binding of FXR/retinoic X receptor-α to the FXRE. Interaction between hinge domain of FXR (containing K206) and Set7/9 was confirmed by coimmunoprecipitation, GST pull down, and mammalian two-hybrid experiments. Set7/9 overexpression in Huh-7 cells significantly enhanced transactivation of the SHP and BSEP promoters in a ligand-dependent fashion by wild-type FXR but not the K206R mutant FXR. A Set7/9 mutant deficient in methyltransferase activity was also not effective in increasing transactivation of the BSEP promoter. These studies demonstrate that posttranslational methylation of FXR by Set7/9 contributes to the transcriptional activation of FXR-target genes.
Keywords: epigenetics, bile acid metabolism, gene regulation, farnesoid X receptor
farnesoid x receptor (FXR) is a bile acid-dependent nuclear receptor (NR) that binds to a specific DNA-binding motif, an FXR response element (FXRE), or inverted repeat element 1 element, located in promoters of FXR target genes involved in lipid, carbohydrate, and bile acid homeostasis (22). FXR and other NRs serve as biosensors to sample a plethora of lipophilic molecules, which move to the nucleus, bind specifically to the NR, and translate the signal into gene transcription (18, 22). The FXRE consists of two copies of a nucleotide (AGGTCA) sequence, arranged as inverted repeats separated by one nucleotide. FXR, which forms a heterodimer with RXRα, contains several functional domains that are conserved in the nuclear hormone receptor family: amino-terminal transcription activation and DNA-binding domains (DBD). These motifs are connected to the COOH-terminal, ligand-binding domain (LBD) via a polypeptide sequence known as the hinge region. Ligand binding to FXR, interaction with DNA, and presumably release of inhibitory proteins lead to a conformational change in FXR and recruitment of coactivator complexes (2, 9). Moreover, the contributions of two separate domains of the FXR and other NRs are central to activation of transcription: the activation function-1 (AF-1) domain, contained within the NH2-terminal transactivation domain, and the AF-2 domain in the COOH-terminal LBD (22). Alterations in chromatin structure resulting from the ordered recruitment of coactivators and associated modifications convey signals to the basal transcriptional machinery, resulting in activation of gene expression.
Methylation of histones is an important modification that influences gene expression (17). It is well established that lysine methyltransferases, such as Set7/9, and arginine methyltransferases, such as CARM1 methylate-specific residues on H3 and H4 histone tails, often combine with other histone modifying enzymes on the same or different histone tails (2, 4). Previous studies have shown the Set7/9 upregulates transcription by catalyzing histone H3-K4 monomethylation in a SET domain-dependent manner and antagonizing both histone H3-K9 methylation (a repressive mark) and promoter association of the repressive nucleosome remodeling and deacetylase complex (25, 37) . Adding to the level of regulatory complexity, it has been recognized more recently that Set7/9 is also a nonhistone protein methyltransferase catalyzing modification of several transcriptional regulatory proteins, including p53, TAF10, p300/CBP-associated factor (PCAF), estrogen receptor (ER), androgen receptor (AR) and the RelA subunit of NF-κB. (20, 23, 28). In the case of the acetyltransferase PCAF multiple lysine residues are methylated by Set7/9 (24). In a recent peptide array methylation study, nine new nonhistone protein targets were identified, one of which, the MINT protein (Msx2-interacting nuclear target protein), was dimethylated at one lysine residue. The functional significance of these modifications is unknown (7).
Histone tail acetylation and methylation are known to enhance accessibility of genes to transcription factors and the basal transcriptional machinery (33, 39). However, as mentioned above, in addition to modifying histones at the loci of FXR-target genes, FXR itself can be modified by coactivators. For example, Lysine 217 of FXR is the major acetylation site targeted by p300 and SIRT1 (19). Acetylation of FXR increases its stability but inhibits heterodimerization with RXRα, DNA binding, and transactivation activity (18). Since there is a well-conserved consensus recognition site for Set7/9 lysine methylation in the hinge region of FXR, we sought to determine whether FXR was methylated by Set7/9 and how this modification influenced FXR signaling.
MATERIALS AND METHODS
Cells and cell culture.
The liver cell line Huh-7 and the monkey kidney line CV-1 were used. Huh-7 cells were cultured in RPMI 1640 with FBS and antibiotics. CV1 cells were cultured in DMEM with 10% FBS and antibiotics. All cells were grown in 5% CO2 in a humidified incubator maintained at 37°C. All cell lines were obtained from American Tissue Culture Collection.
Chemicals.
All chemicals were obtained from Sigma unless stated otherwise. siRNAs for Set7/9 was obtained from Dharmacon or Santa Cruz Biotechnology. Antibodies to Set7/9 were from Millipore. Pan methyl lysine (ab7315), and H3K4me1 (ab8895) antibodies were from Abcam. FXR (sc-25309) and GFP (sc-8334) antibodies were from Santa Cruz Biotechnology.
Plasmid construct.
The human bile salt export pump (BSEP) promoter sequence was generated by PCR and subcloned into the luciferase expression vector pSV0ALΔ5′ (p-1445/Luc), as described by us previously (1). Plasmids encoding FXR and RXRα were generously supplied by Dr. David Mangelsdorf, Dallas, TX. Set7/9 cDNA was subcloned into the mammalian expression vector pcDNA3 at the BamH1/Not1 sites. 3XFXRE-TK-Luc (FXRE sequence from the rat BSEP promoter) was constructed by cloning three copies of the FXRE upstream of the minimal thymidine kinase (TK) promoter and the luciferase opening reading frame.
For the mammalian two-hybrid (M2H) system, pBIND and pACT (Promega) are fusion vectors for the linkage of proteins to the Gal4 DNA binding domain and to the VP16 transactivation domain, respectively. The different domains of FXR or the full coding sequence of human FXR was amplified by PCR and cloned in-frame into the Xba1/NotI sites of pBIND fusion vector to produce the plasmid pBIND-FXR domains and pBIND-FXR. A complete coding sequence of the Set7/9 cDNA was inserted in-frame into the Xba1/NotI sites of the pACT fusion vector to produce the plasmid construct pACT-Set7/9.
The bacterial expression vector pGEX-6P-1 was used to produce glutathione S-transferase (GST)-fused recombinant proteins in Escherichia coli. The different domains of FXR or the full coding sequence of human FXR was amplified by PCR and cloned in-frame into the BamH1/XhoI sites of pGEX-6P-1 fusion vector to produce the plasmid pGEX-6P-1-FXR domains and pGEX-6P-1-FXR. A cDNA fragment encoding Set7/9 was also inserted in-frame into the BamHI/NotI sites of pGEX-6P-1vector to produce the GST tagged construct. All of the positive clones containing cDNA inserts were identified by restriction enzyme mapping and sequenced using the ABI automated DNA sequencer model 377. The short heterodimer partner (SHP) promoter plasmid generated from genomic DNA by PCR was further subcloned into the PGL3 and PXP2 vectors (Kpn1-Xho1).
Truncated FXR cDNAs were derived from full-length human FXR cDNAs by PCR, subcloned in pGEX-6P-1, and expressed in the E. coli BL21RP by isopropyl-β-d-thiogalactopyranoside induction (IPTG). Point mutations were introduced into FXR and Set7/9 cDNAs using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and appropriate mutant oligos.
For mutation of the Set7/9 active methyltransferase site (W352A, Y353A), an antisense primer (5′-CAGCTCCACCTGGgcCgcCTCAGGGGCTTCA-3′) and a sense primer (5′-TGAAGCCCCTGAGgcGgcCCAGGTGGAGCTG-3′) (mutated bases indicated in lowercase type) were used in a PCR according to the manufacturer's directions with the Set7/9 expression plasmids as a template.
Mutants were confirmed by nucleic acid sequencing. Upon transfection into mammalian cells, the mutagenized cDNAs produced FXR variants with desired AA substitutions. The Set7/9 full-length cDNA was cloned from Huh-7 cells by RT-PCR, sequence verified, and subcloned into pGEX-6P-1 or into pcDNA3.1. Recombinant proteins were expressed in E. coli BL21RP under IPTG induction and affinity purified on glutathione-S-Sepharose columns.
Chromatin immunoprecipitation analysis of cultured cell lines.
Chromatin immunoprecipitation (ChIP) analysis assays were conducted by a combination of previously described protocols (5, 6, 16) and manufacturer's instructions using EZ ChIP/MagnaChIP G kit from Upstate Biotechnology/Millipore (Millipore, MA). Briefly, cells from 3× 100-mm culture dishes were harvested after fixation with 1% formaldehyde. Following lysis, genomic DNA were sonicated in Diagenode Bioruptor Sonicator for 8× 30 s (twice, with 30 s on/30 s off cycles) resulting in DNA fragments of 200–1,000 bp. The fragmented DNA was diluted in ChIP dilution buffer and preadsorbed with Protein G Sepharose/salmon sperm DNA (Millipore, MA) for 1 h at 4°C. Then 5% of the chromatin was removed and saved as input. It was then incubated overnight at 4°C with 3–5 μg of the appropriate antibodies or normal mouse IgG (control). Antibody-chromatin complexes were captured by incubation with Protein G Sepharose and centrifuged. Protein G Sepharose beads were washed with low salt, high salt, lithium chloride, and finally with Tris-EDTA buffer. DNA from the beads was then eluted. Reversal of protein cross-linking and proteinase K digestion, followed by purification of the DNA, was then achieved. An aliquot of the DNA (2 μl) was used in a PCR ( standard and quantitative ) reaction using specific primers flanking the FXRE site of human BSEP and human SHP. Primers flanking a site distant from the FXRE site were used as negative controls. The positive control for the ChIP assay consisted of immunoprecipitation with CARM1 antibody. PCR products were run in a 2% agarose gel and stained with ethidium bromide to confirm the amplicon size.
Quantitative real-time PCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) in combination with primers for BSEP, SHP, or Set7/9 in a Step One Plus Real-time PCR system (ABI, CA), as previously described (3). Primers were designed using Primer Express analysis software (ABI) and dissociation curves after each set of primer use were checked to verify that a single PCR amplicon was obtained and no primer dimers were formed. PCR products were also run on agarose gels to further check the amplicon size. PCR reactions were monitored in real-time using SYBR Green dye detection. ChIP DNAs were quantified by real-time PCR and normalized to input DNA (5% input before ChIP). Differential Ct values from experimental and input DNAs (ΔCt) were used to calculate the amplified DNA yield for each experiment. ChIP with IgG (negative control) did not show a PCR signal within 40 amplification cycles. For each ChIP, the fold change was calculated relative to the value for DMSO-treated cells, which was set as 1. Each independent ChIP data point is the average of normalized values from PCR runs in duplicate wells. Each histogram is the average mean ± SE from independent ChIP experiments of n = 3. Chromatin Re-ChIP was performed according to Active Motif Re-ChIP-IT method.
EMSA.
Preparation of nuclear extracts from Huh-7 cells transfected with human FXR, RXRα, Set7/9wt, or the FXR K206Rmut (lysine changed to arginine), or the Set7/9mut (W352A, Y353A) was done using NE-PER. Expression plasmids (5 μg/dish) were transfected into Huh-7 cells (5×106 cells/100-mm dish) in three dishes using TransIT-LT at a DNA/TransIT-LT ratio of 1:3. Control untransfected cells were left in RPMI-1640 medium. Fugene-6 was replaced with RPMI-1640 medium 1 day later, and 3 days later, nuclear extracts were prepared using NE-PER from Pierce according to the manufacturer's directions. Nuclear extracts were stored in aliquots at −80°C until used. Oligonucleotide probes for the EMSAs were end-labeled with [γ-32P] ATP (3,000 or 6,000 mCi/mmol) by T4 polynucleotide kinase. The gel shift assay protocol was carried out as described previously (1). Briefly, 10 μg of nuclear extract together were added to 20 μl of binding reaction containing 12 mM HEPES (pH 7.9), 60 mM KCl, 4 mM Tris·HCl, 5% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 1 μg of polydeoxyinosinic-deoxycytidylic acid, and 5×104 cpm of probe on ice for 45 min. As a control, the probe was also incubated with the same amount of nuclear extract from the untransfected cells. In competition assays, unlabeled wild-type or mutant oligonucleotides were added to the reaction 15 min before the addition of the probe. DNA-protein complexes were resolved on 4% native polyacrylamide gel electrophoresis containing 0.53 TBE (0.89 M Tris, 0.89 M boric acid, 0.02 M disodium EDTA for 10× TBE). The gel was dried and exposed to X-ray film for varying lengths of time until a suitable image was obtained.
Methylation of FXR in vitro GST-FXR, and Set7/9.
A mixture of GST-Set7/9 and GST-FXR (wild-type or bearing K→R substitution) was incubated with 1 μM [3H-methyl] SAM (78 Ci/mmol, cat. no. NET155H; Perkin-Elmer, Wellesley, MA) at 30°C for 2 h in a reaction buffer containing 50 mM Tris·HCl (pH 8.5), 5 mM MgCl2, and 4 mM dithiothreitol, as described elsewhere (2). Reaction products were run on 10% SDS-PAGE, transferred onto PVDF membrane, which was sprayed with EN3HANCE (Perkin-Elmer) before autoradiography was done for 3 days. In some cases, the gel was incubated with EN3HANCE for 2 h before drying and autoradiography for 1 wk.
Glutathione S-transferase pull-down assay.
Full-length WT, K206Rmut, and different domain recombinant proteins of FXR were overexpressed as GST fusion proteins in E. coli BL21. Equal amounts of GST and GST fusion proteins were used for GST pull-down assays by incubating 5 μl of full length Set7/9 protein obtained by in vitro translation using TNT T7-coupled transcription and translation system (Promega). Methods exactly followed the recommended protocol from Pierce GST Pull-down assay kit. The bound proteins were washed with wash buffer and eluted into SDS-PAGE sample buffer by heat denaturation. The proteins were separated on 4–20% gradient gels. After transfer to PVDF membrane, the appropriate antibody was used to detect the signal.
Coimmunoprecipitation.
A pan-methyllysine-specific antibody (Abcam) was used to detect the methylated form of intracellular FXR. Cell lysates prepared in IP buffer [50 mM Tris (pH 8),150 mM NaCl, 0.5% NP-40, 5 mM EDTA plus protease inhibitor cocktail (Roche Applied Biosciences)] were precleared with protein A agarose beads for 30 min and incubated overnight with an anti-FXR antibody (D-3; Santa Cruz Biotechnology), anti-Set7/9 antibody (Millipore) or mouse IgG (Santa Cruz Biotechnology) at 4°C. The bead-bound immunoprecipitates were captured by centrifugation at 2,500 rpm, washed twice with the IP buffer, and then dissociated from the beads, after which the recovered supernatant (using 2× Laemmli's sample buffer at 98°C) was used for Western blot analysis after fractionation on 10% SDS-PAGE. Co-IP in FXRwt, K206Rmut, and Set7/9-transfected cells were also performed with total cell lysates.
Mammalian two-hybrid analysis.
The mammalian two-hybrid assay was performed by the modified Checkmate mammalian two hybrid system (Promega) following manufacturer's instructions, as used previously by us (29). Huh-7 cells were cotransfected with the pBIND/FXRs (containing WT, K204, 206Rmut, and different FXR domains), pACT/Set7/9 and pG5/luc plasmids employing Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommended protocol. pG5-Luc (Promega) contains the GAL4-binding site as five tandem repeats. Briefly, cells were seeded in 24-well plates at a density of 1× 105 cells/well and cultured to ∼ 90% confluency. Plasmid constructs were combined at a ratio of 2.4:1.4:0.2 (FXR/Set7/9/pG5) to a total of 1.0 μg/well; cells were transfected and cultured for 24 h followed by the addition of ligand GW4064 (1 μM) for another 24 h. Finally, luciferase activity was determined with luminometer (Promega) using a commercially available kit (Promega). The transfection efficiency was normalized using Renilla luciferase activity, which was simultaneously expressed from the pBIND plasmids.
Transient transfections and luciferase assays.
Huh-7 and CV1 cells were plated at a concentration of 1× 105 cells/well in 24-well plates 2 days earlier. They were transfected at day 0 with the human BSEP or SHP promoter at 0.5 μg/well (in triplicate/per group) and also cotransfected with 50 ng FXR/RXRα and various amounts of Set7/9 expression plasmids in OPTI-MEM (Invitrogen, CA) where indicated. Transfections were carried out using TransIT-LT (Mirus Bio, WI) at DNA:TransIT ratio of 1:3. On day 1, medium was changed to DMEM without Phenol Red and with charcoal-adsorbed FBS. FXR ligand GW4064 (1 μM) was added at this time and luciferase activities were measured 24 h later using Promega Kit (Promega, WI). Normalization of transfection efficiencies in the different wells was achieved by cotransfection with pCMV-β galactosidase and assay of galactosidase activity.
siRNA-mediated knockdown of Set7/9.
siRNAs against Set7/9 (targeting 4 separate regions of the Set7/9 sequence) were obtained from Dharmacon of Santa Cruz (siGenome pool). For knockdown experiments, Huh-7 cells were plated in six well plates (1× 106 cells/well) and incubated 2 days later with 50 nM siRNA using TransIT-TKO (MirusBio) at a ratio of 1:1 (μl/μl) according to manufacturer's instructions. Six hours later, medium was added to the wells, and 24 h later, spent medium was replaced with fresh RPMI-1640. One day after transfection cells were treated for 24 h with GW 4064 (1 μM) or vehicle (0.01% DMSO). Forty-eight hours later total RNA was prepared using Trizol kit (Invitrogen, Carlsbad, CA), and real-time PCR analysis was conducted following conversion of mRNA into cDNA (12).
Immunoblot analysis.
For the determination of proteins in cultured Huh-7 cells, cells were resuspended in gel loading buffer (50 mM Tris·HCl, pH 6.8, 2% SDS, 10% glycerol, 10% β-mercaptoethanol, and 0.1% bromphenol blue). Samples (20 μg, without boiling) were separated by 4–20% gradient SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked with Tris-buffered 5% nonfat milk overnight at 4°C and then incubated with the relevant anti-body (1:2,000 dilution) overnight at room temperature. The blots were washed three times (5 min each) in Tris-buffered saline/Tween 20 solution (25 mM Tris, pH 7.4, 150 mM NaCl, and 0.1% Tween 20) and incubated with a peroxidase-conjugated anti-rabbit secondary antibody (1:5,000 dilution) for 1 h at room temperature. The blots were further washed three times (5 min each) in Tris-buffered saline/Tween 20 solution and were visualized using the enhanced chemiluminescence detection system (ECL+Plus; Amersham Biosciences, Piscataway, NJ).
Statistics.
Calculation of mean ± SE and Student's t-test was done using Prism software. P values < 0.05 were considered statistically significant. All experiments were repeated at least three times with similar results.
RESULTS
Ligand-induced chromatin recruitment of the Set7/9 methytransferase to the promoters of FXR-target genes and associated histone H3 activation mark.
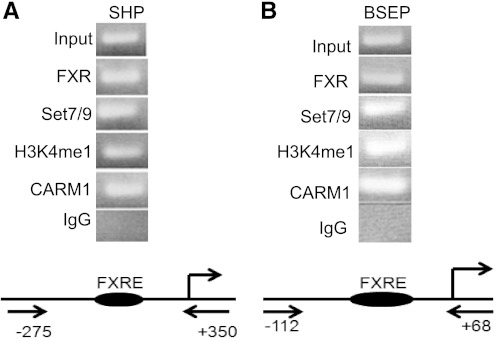
To determine whether Set7/9 could be detected at the promoters of FXR-target genes, chromatin immunoprecipitation (ChIP) was performed on the Huh-7 liver cell line (Fig. 1). Chromatin occupancy of Set7/9 at the FXRE of the endogenous promoters of the SHP and the BSEP was present in Huh-7 cells. The methylated histone H3K4 activation mark was also demonstrated. Coactivator arginine methyltransferase-1 (CARM1), previously shown by us to be involved ligand-dependent activation of FXR, was used as a positive control in the ChIP assay (20). It is well established that the H3K4 monomethylation is mediated by Set7/9. No reactivity was observed in ChIP assays employing anti-H3K4 di- and trimethyl antibodies (not shown).
Fig. 1.
Ligand-induced chromatin recruitment of Set7/9 to the promoters of FXR-target genes and the associated histone H3 activation mark. Chromatin immunoprecipitation (ChIP) analysis of hepatoma cells (Huh-7) showed recruitment of FXR, Set7/9, CARM1, and the H3K4 monomethylation mark of histone H3 to the endogenous short heterodimer partner (SHP; A) and human bile salt export pump (BSEP; B) promoters in the presence of the farnesoid X receptor (FXR)-ligand (GW4064). Schematic representation of the primers used for ChIP and the location of the sites on the SHP and BSEP promoters are shown below representative gels.
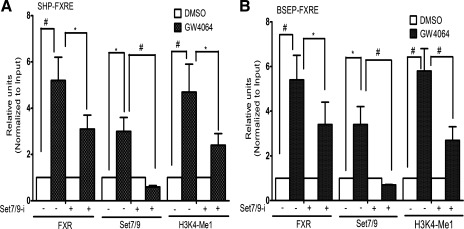
To address the question of whether Set7/9 is involved in regulation of FXR-target genes, we employed Set7/9-specific small-interfering RNA (siRNA) pools targeting four separate regions of the Set7/9 sequence in Huh-7 cells to reduce cellular Set7/9 levels compared with a nonsilencing siRNA control. In ChIP assays, treatment of Huh-7 cells with siRNA against Set7/9 markedly reduced ligand-induced occupancy of Set7/9 and FXR at the endogenous SHP (Fig. 2A) and BSEP (Fig. 2B) promoters, and decreased the H3K4 methylation mark, assessed by real-time quantitative PCR. The decrease in recruitment of FXR at the BSEP and SHP promoters after Set7/9 depletion suggests either that Set7/9 contributes to FXR expression or that methylation of FXR is important for its recruitment to the FXRE. These data indicate that Set7/9 contributes to the transactivation of FXR-target genes.
Fig. 2.
The effect of short interfering RNAs (siRNA) knockdown of Set7/9 expression on ligand-induced occupancy of Set7/9 and FXR and on H3K4 monomethylation at the endogenous BSEP and SHP promoters. siRNAs were used to reduce cellular Set7/9 levels with a nonsilencing siRNA used as control. Treatment of Huh-7 cells with siRNA against Set7/9 markedly reduced ligand-induced occupancy of Set7/9 and FXR at the SHP (A) and BSEP (B) promoters and decreased the H3K4 methylation mark, as assessed by real-time quantitative PCR. Each independent ChIP data point is the average of normalized values from PCR runs in duplicate wells. Each histogram is the average mean ± SE from independent ChIP experiments of n = 3. *P < 0.05, #P < 0.001.
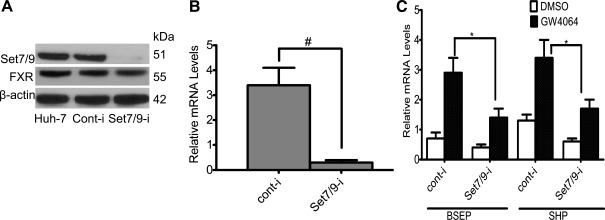
Figure 3A shows the results of additional experiments in which Set7/9 siRNAs were used in Huh-7 cells. Endogenous Set7/9 expression was markedly and specifically reduced compared with control RNAi-transfected cells at both the protein and mRNA level (Fig. 3, A and B). However, the amount of FXR protein was not significantly reduced by Set7/9 knockdown, indicating that Set7/9 does not regulate FXR expression. As a result of Set7/9 depletion, ligand-induced mRNA expression of the endogenous FXR target genes SHP and BSEP declined significantly (Fig. 3C). These findings further support the role of Set7/9 in enhancing the ligand-dependent transactivation of FXR-target genes.
Fig. 3.
Inhibition of ligand-induced FXR target gene expression in Set7/9-silenced Huh-7 cells. Set7/9 expression was markedly and specifically reduced compared with control in RNAi-transfected cells at both the protein (A) and mRNA level (B). C: SHP and BSEP mRNAs declined significantly, as assessed by real-time quantitative PCR. *P < 0.05, #P < 0.001.
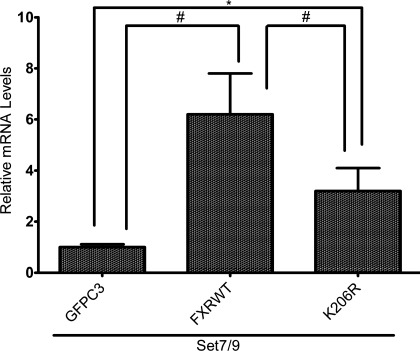
To further verify that Set7/9 activates the FXR-target gene, BSEP, endogenous BSEP mRNA levels were measured in Huh-7 cells after cotransfection with FXR/RXRα and Set7/9 by real-time PCR (Fig. 4). Overexpression of Set7/9 together with FXRwt led to a fourfold increase in BSEP mRNA levels in Huh-7 cells. In contrast, a K206R FXR mutant (lysine replaced with an arginine) was significantly less effective in inducing expression of BSEP mRNA.
Fig. 4.
Effect of Set7/9 overexpression on endogenous BSEP mRNA levels in Huh-7 cells. Huh-7 cells were cotransfected with FXR/RXRα and Set7/9. BSEP mRNA was measured by real-time PCR. Overexpression of Set7/9 together with FXRwt led to a significant increase in BSEP mRNA levels in Huh-7 cells. *P < 0.05, #P < 0.001.
FXR and Set7/9 occupy the same genomic locus at the SHP and BSEP promoters.
Figure 5A shows the results of an electromobility shift assay using nuclear extracts prepared from Huh-7 cells transfected with FXR, RXRα, and Set7/9 or FXR or Set7/9 mutants. Specific binding of FXR/RXRα to the FXRE was markedly enhanced by expression of Set7/9 (lanes 4 and 5). In contrast, specific binding was significantly reduced in cells expressing the FXR K206R mutant (lysine 206 is replaced with an arginine, lane 3) or a Set7/9 mutant (lane 2) that is deficient in methytransferase activity. In previous studies, mutagenesis identified two residues in the COOH terminus of Set7/9 (W352A, Y353A) that were essential for catalytic activity toward lysine-4 of histone H3 (34, 36). The residual binding can be explained by endogenous expression of wild-type FXR/RXRα and Set7/9 in Huh-7 cells. The specific binding of FXR/RXRα to the FXRE could be abrogated by the addition of excess amount of wild-type, but not mutated, FXRE (lanes 6 and 7). These data support the results shown in Fig. 2 in which siRNA knockdown of Set7/9 resulted in a decreased occupancy of FXR at the SHP and BSEP promoters in ChIP assays.
Fig. 5.
FXR and Set7/9 occupy the same genomic locus at the SHP and BSEP promoters. An electromobility shift assay was done using nuclear extracts prepared from Huh-7 cells transfected with FXR, RXRα, and Set7/9 (A). Specific binding of FXR/RXRα to the FXRE was enhanced by expression of Set7/9 (lanes 4 and 5). In contrast, specific binding was significantly reduced in cells expressing the FXR K206R mutant (lane 3) or a Set7/9 mutant (lane 2) that is deficient in methyltransferase activity. Competition analysis was performed with unlabeled 25-fold molar excess of wild-type (lane 6) or mutant FXRE (lane 7). Sp, specific complex; NSp, nonspecific complex. Sequential chromatin immunoprecipitation (ChIP Re-ChIP) confirms that FXR and Set7/9 occupy the same genomic site (FXRE) in vivo at both the SHP and BSEP loci (B and C). ChIP-ReChIP was done using chromatin prepared from Huh-7 cells transfected with FXR, RXRα, and Set7/9 or the FXR K206 mutant or the Set7/9 mutant. B: PCR results using the SHP primer that includes the FXR binding site. C: PCR results using the BSEP primer that include FXR binding site. Lane numbers are the same to indicate that the DNA is from the same chromatin sample. Top: results PCR performed on an aliquot of DNA removed from the experiment after the first ChIP assay. Bottom: PCR results on DNA from chromatin samples after both ChIP steps. Chromatin samples subjected to the first ChIP using FXR antibody (top, lane 2) were then subjected to a second ChIP with a Set7/9 antibody (bottom, lane 2). Mouse IgG and no antibody controls were also performed from the first ChIP using FXR antibody (lane 2). RXR, retinoic X receptor; IP, immunoprecipitation; Mu, mutant; WT, wild type.
Next , we performed a ChIP Re-ChIP assay that is a direct strategy to determine the in vivo colocalization of proteins interacting or in close contact in a chromatinized template on the basis of double and independent rounds of immunoprecipitations with antibodies against FXR and Set7/9. The ChIP-ReChIP assay was done using chromatin prepared from Huh-7 cells transfected with FXR, RXRα, and Set7/9 or the FXR K206 mutant or the Set7/9 mutant. Figure 5, B and C confirms that FXR and Set7/9 occupy the same genomic site (FXRE) in vivo at both the SHP and BSEP loci. Although this assay is not quantitative, it appears , that similar to the findings in the EMSA, less FXR and Set7/9 are recruited to the SHP and BSEP loci when mutant forms of FXR and SET7/9 are overexpressed in Huh-7 cells. The endogenous expression of FXR and SET7/9 contributes to the bands in each vertical lane.
Set7/9 methylation of FXR in vitro and in vivo by Set7/9 and identification of the lysine methylation site.
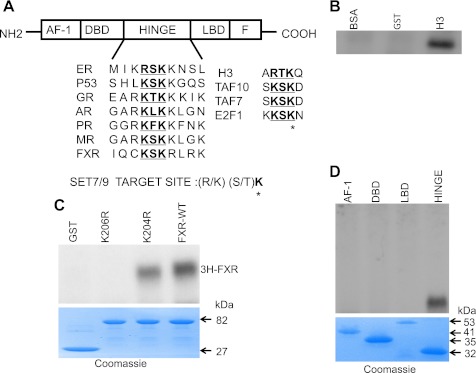
The schematic representation of FXR in Fig. 6A indicates domains conserved in the nuclear hormone receptor superfamily, namely, the AF-1 domain, DNA binding domain (DBD), hinge region (hinge), and LBD. The conserved methylation motif is highlighted in bold. The methylated sequence of various known substrates by Set7/9 is shown. The asterisk indicates the target lysine residue.
Fig. 6.
Methylation of FXR in vitro by Set7/9 and identification of the lysine methylation site. A: schematic representation of FXR indicating domains conserved in the nuclear hormone receptor superfamily: activation function-1 (AF-1), DNA binding domain (DBD), hinge region (hinge), and ligand-binding domain (LBD). The conserved methylation motif is underlined. The sequence methylated by Set7/9 in various known substrates is indicated to the right. *Target lysine residue. B: strong histone methyltransferase activity of a GST-Set7/9 preparation in vitro is indicated by methylation of recombinant histone H3. GST and BSA were negative control substrates. C: methylation of GST-FXR in an in vitro reaction. GST-Set7/9 (3 μg) was incubated with the GST-fused FXRwt and FXRmutants in the presence of [3H-methyl]-SAM. 3H signal of methylated FXR in the reaction mixture was detected by 10% SDS-PAGE and autoradiography. Recombinant Set7/9 methylated FXR. Mutation of the putative methylation site at K206 (K206R) to an arginine completely abrogated methylation of FXR by Set7/9. FXR was still methylated with mutation of an adjacent, irrelevant lysine (K204) to an arginine. GST lacked Set7/9 activity. bottom: Coomassie blue staining of the gel showing the protein substrates. D: methylation of various GST fused FXR fragments. The hinge domain but not the AF1, DBD, or LBD was methylated by Set7/9. Bottom: Coomassie blue staining of the gel shows the protein substrates.
Consistent with previous findings by other investigators, we found strong histone methyltransferase activity in vitro by a purified GST-Set7/9 preparation, indicated by methylation of recombinant histone H3. GST and BSA were used as negative control substrates (Fig. 6B).
Having determined that Set7/9 influences the expression of FXR-target genes, we next determined whether FXR itself was subject to methylation and if so, at which residue. The conserved lysine K206 and adjacent residues (KSKR) in the hinge domain of FXR closely resembles the Set7/9 consensus recognition sequence ([R/K]: [R/K] [S/T] K) (Fig. 6A). A mixture of GST-Set7/9 and GST-FXR (wild-type or bearing K→R substitution, K206R) was incubated with 1 μM [3H-methyl] SAM. Recombinant Set7/9 methylated FXR (Fig. 6C). Mutation of the putative methylation site at K206 (K206R) to an arginine completely abrogated methylation of FXR by Set7/9 (Fig. 6C). FXR was still methylated with mutation of an adjacent, irrelevant lysine (K204) to an arginine. Figure 6D shows that an FXR fragment containing the hinge domain but not fragments containing the AF1, DBD, or LBD was methylated by Set7/9.
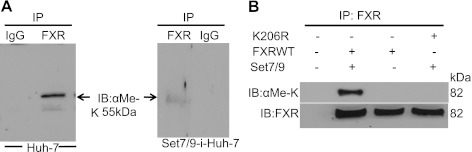
In Fig. 7A, left endogenous FXR from a lysate of Huh7 cells was found to be methylated when a Western blot analysis of immunoprecipitated FXR was probed with a pan-methyllysine lysine antibody. Knockdown of Set7/9 with siRNA markedly decreased the methylation of FXR (Fig. 7A, right). These findings indicated that FXR can be present in vivo in a methylated form.
Fig. 7.
Methylation of FXR in vivo by Set7/9. A: FXR immunoprecipitated (or IgG-incubated control) from Huh-7 cells total lysate was immunoblotted with the pan-methyllysine (αMe-K) antibody, and methylated FXR was detected by Western blot analysis. The methylation of FXR was abrogated in cells subjected to Set7/9 siRNA-mediated silencing. B: Huh-7 cells were cotransfected with plasmids encoding FXR (wild-type or K206Rmut) and Set7/9, and the Western blot FXR immunoprecipitate was probed with anti-αMe-K and anti-FXR antibodies. Immunoprecipitates from the transfected cells were then probed with the αMe-K antibody. Wild-type but not the K206R mutant was methylated when Set7/9 was expressed in these cells. Wild-type FXR transfected cells without Set7/9 cotransfection, or control, nontransfected cells were also analyzed. IB, immunoblotting.
Huh-7 cells were then transfected with FXR or the K206R FXR mutant and Set7/9. Immunoprecipitates from the transfected cells were then probed with the αMe-K antibody. Wild-type FXR but not the K206R mutant was methylated when Set7/9 was expressed in these cells (Fig. 7B). These results confirm the in vitro methylation data and show that the K206 residue is required for lysine methylation of intracellular FXR, as the αMe-K specific antibody recognized the transfected wild-type FXR but not the K206R mutant form in the Huh-7 cells.
Set7/9 associates with FXR in vivo.
After showing that FXR and Set7/9 are recruited to the same site in the genome and that Set7/9 methylates FXR, we then sought to identify a direct interaction between Set7/9 and FXR using several approaches.
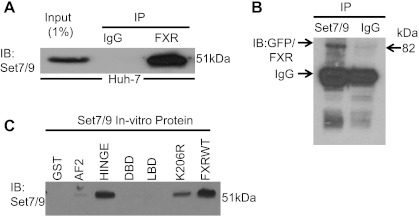
To demonstrate the interaction between FXR and Set7/9, reciprocal immunoprecipitations were performed. Figure 8A shows that when FXR was immunoprecipitated from a whole cell lysate of Huh-7 cells; association was demonstrated with Set7/9 on Western blot analysis with a Set7/9 antibody. To confirm the interaction, Huh-7 cells were transiently transfected with FXR-GFP. Set7/9 was then immunoprecipitated with a Set7/9 antibody and then probed with an GFP antibody on a Western blot (Fig. 8B). These data further support the association between Set7/9 and FXR.
Fig. 8.
Set7/9 associates with FXR in vivo. A: When FXR was immunoprecipitated from a whole cell lysate of Huh-7 cells, association was demonstrated with Set7/9 on Western blot analysis with a Set7/9 antibody. The input lysate was at 1% of the lysate used for immunoprecipitation. B: FXR-GFP also coimmunoprecipitated with Set7/9 from a Huh-7 cell lysate when Set7/9 was first immunoprecipitated, run on a Western blot analysis and probed with a GFP antibody. Huh-7 cells were transiently transfected with the FXR-GFP construct 1 day before collecting the cells. C: GST pull-down assays using various fragments of FXR and K206Rmut with in vitro translated Set7/9 protein was carried out as described in the materials and methods. FXR hinge domain bound to Set7/9, but binding was absent when the GST control and other domains were used in the assay. With use of the FXR K206R mut there was weak binding when compared with FXRwt. RLU, relative light unit.
Recombinant Set7/9 proteins and portions of the various domains of the FXR molecule linked to GST were then used in a GST pull down assay using a Set7/9 antibody. In this assay, Set7/9 bound strongly to wild-type FXR and a peptide corresponding to the hinge region, but not to the DBD, LBD, or the AF-1 peptides and minimally to a K206R FXR mutant (Fig. 8C). These data further support the direct association between Set7/9 and FXR-GST.
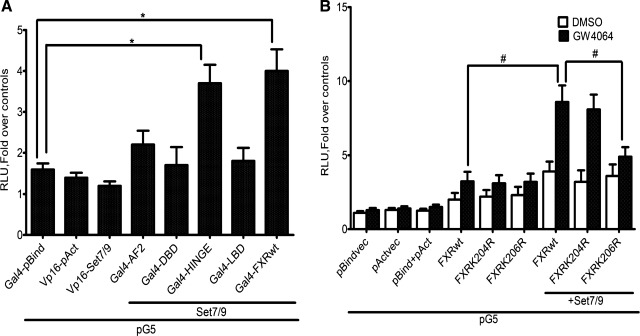
Mammalian two-hybrid analysis was used to further explore the interaction between FXR and Set7/9 (Fig. 9A). The results show that a construct consisting of the hinge region or a full-length FXR expressed in Huh-7 cells interacted with Set7/9, while the AF1, DBD, and LBD constructs did not. Additionally, the interaction between FXR and Set7/9 was enhanced in the presence of the FXR ligand GW4064 (Fig. 9B). The K206R FXR mutant was significantly less effective in this assay even in the presence of ligand. Mutation of an irrelevant, adjacent lysine residue to an arginine (K204R) had no effect on the interaction of FXR with Set7/9.
Fig. 9.
Mammalian two-hybrid analysis of the interaction between FXR and Set7/9. A: mammalian two hybrid analysis: The association between pBIND- FXRwt, different domains of the FXR and pACT-Set7/9, is shown. A 2- to 3-fold increase in reporter gene activity was observed with the FXR hinge region and full length constructs. No significant increase in activity was seen with the remaining constructs. B: mammalian two hybrid analysis between pBIND- FXRwt, K206Rmut, and pACT-Set7/9 in the absence/presence of 1 μM GW4064 and the ensuing increase in reporter gene activity over empty vector are shown. Note the increased reporter gene activity when Set7/9 was cotransfected with the FXRwt but not with expression the 206R FXRmut (*P < 0.05, #P < 0.001 compared with FXRwt only).
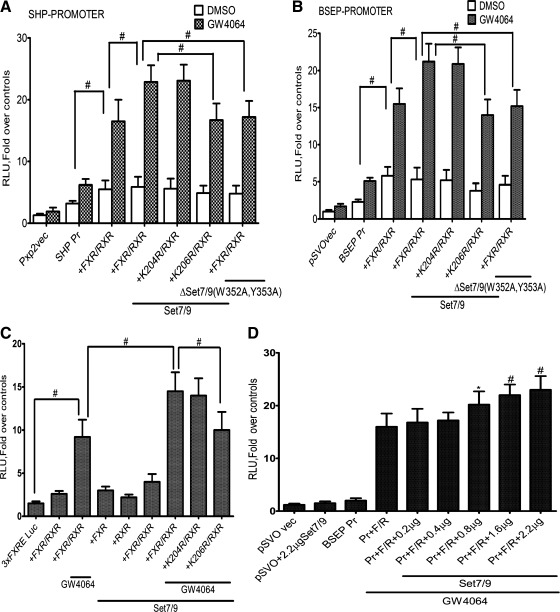
Synergistic activation of SHP and BSEP by FXR and Set79.
To test the functional and biological relevance of Set7/9 methylation of K206, we next assessed the effect of Set7/9-mediated methylation of FXR on ligand-induced transactivation of FXR target genes. Huh-7 cells were transiently transfected with plasmid vectors containing the SHP promoter-luciferase DNA or the BSEP promoter-luciferase DNA. Transfection with FXR/RXRα with the addition of the GW4064 ligand led to a significant increase in SHP (Fig. 10A) and BSEP (Fig. 10B) promoter-luciferase activity in Huh-7 cells. Transfection with Set7/9 but not with the Set7/9 (W352A, Y353A) mutant further increased luciferase activity. This effect was restricted to the wild-type FXR and not the K206R FXR mutant. In both cases mutation of an irrelevant, adjacent lysine residue to an arginine (K204R) had no effect on the transactivation of these FXR-target genes by Set7/9.
Fig. 10.
Synergistic activation of SHP and BSEP by FXR and Set7/9. A: Huh-7 cells transfected with indicated plasmids were treated with FXR ligand GW 4064 24 h after transfection, and reporter assays were performed. The mean and mean ± SE are shown (n = 3). Set7/9 expression significantly increased SHP reporter gene activity, which was not observed with expression of the K206R FXR mutant or the Set7/9 mutant deficient in methyltransferase activity. # P < 0.001. B: Huh-7 cells were cotransfected with indicated reporter plasmids and expression plasmids for Set7/9 or the Set7/9 mutant and FXR WT or the FXR mutant, as indicated. Set7/9 expression significantly increased BSEP reporter gene activity, which was not observed with expression of the K206R FXR mutant or the Set7/9 mutant deficient in methyltransferase activity. #P < 0.001. C: CV-1 cells in 12-well plates were transfected with 3XFXRE-TK-Luc along with wild-type FXR, RXRα, and Set7/9 or the K206R mutant (Fig. 9A). 3XFXRE-TK-Luc was constructed by cloning 3 copies of the BSEP FXRE upstream of the minimal thymidine kinase (TK) promoter and the luciferase opening reading frame. Expression of the FXR-RXRα heterodimer along with Set7/9 in CV-1 cells led to a marked increase in expression of the 3XFXRE-TK-LUC activity in the presence of GW4064. There was no activity of the promoter observed in the absence of FXR/RXRα. Set7/9 lysine methyltransferase failed to augment 3XFXRE-TK-LUC activity with expression of the K206R mutant FXR and RXRα in the presence of ligand. *P < 0.05 compared with transfection in the absence of FXR /RXRα or with mutant FXR. #P < 0.001. D: effect of increasing amount of Set7/9 in CV-1 cells. CV-1 cells were transfected with FXR, RXRα, and increasing amounts of full-length Set7/9 cDNA plasmid. FXR transactivation of BSEP promoter was monitored by luciferase activity. Significant (*P < 0.05 compared with activity in cells without Set7/9 plasmid cotransfection) dose-dependent stimulation of promoter activity was seen with 0.8 to 2.2 μg of Set7/9 cDNA. *P < 0.05 compared with transfection in the absence FXR /RXRα. #P < 0.001, Pr, BSEP promoter; F/R, FXR /RXRα.
Since Huh-7 cells synthesize bile acids and express low levels of FXR and RXRα, cotransfection assays were also done in CV-1 cells, derived from monkey kidney fibroblasts that do not express FXR. Similar to results seen with Huh-7 cells, expression of the FXR-RXRα heterodimer along with Set7/9 in CV-1 cells led to a marked increase in expression of the 3XFXRE-TK-Luc reporter in the presence of GW4064 (Fig. 10C). There was no activity of the promoter observed in the absence of FXR/RXRα or without the GW4064 ligand. CV-1 cells were also transfected with the BSEP promoter along with wild-type FXR, RXRα, and Set7/9. We observed a dose-dependent increase in the expression of the BSEP promoter with addition of increasing amounts of Set7/9 (Fig. 10D).
DISCUSSION
The FXR is a ligand-dependent transcription factor that is activated by bile acids and recruits an array of coactivator complexes that alter local chromatin structure and facilitate the assembly of the transcriptional machinery to activate target gene expression (8, 18). Many of these coactivators and associated proteins have intrinsic enzymatic activity that modifies histones or even components of the transcriptional complex including NRs themselves (15, 26). Lysine methyltransferases can regulate the activity of transcription factors and coregulators by methylating specific lysine residues (4, 17). The enzymatic activities of coregulators can either enhance or attenuate the activity of the FXR. Methylation of histone H3-K4 and demethylation of H3K9 at targeted promoters are thought to be activation marks (27).
In this study, we show that FXR is methylated by Set7/9 within its hinge region at lysine-206 in vitro and in vivo. The conserved lysine K206 and adjacent residues (KSKLR) in this hinge domain closely correspond to the Set7/9 recognition sequence. The methylation of FXR is catalyzed by Set7/9 which was originally defined as histone H3K4 monomethyltransferase, but more recently shown to also methylate the estrogen receptor-α, the androgen receptor, and several other nonhistone proteins (20, 21, 23, 28). ChIP assays demonstrated recruitment of Set7/9 and the presence of the associated H3K4 monomethylation activation mark at the SHP and BSEP loci. Interactions between FXR and Set7/9 were demonstrated by GST pull down, coimmunoprecipitation, and mammalian two-hybrid experiments. This ligand-dependent modification promoted binding of FXR/RXRα to the FXRE, as demonstrated on EMSA and ChIP assays. Set7/9 also significantly enhanced transactivation of the SHP and BSEP promoters by wild-type FXR but not a K206R mutant FXR. Depletion of Set7/9 by siRNA decreased the mRNA levels of two FXR target genes, SHP and BSEP. This effect is likely related to abrogation of FXR methylation, but monomethylation of histone H3K4, which is an activation mark, also mediated by Set7/9 would be affected.
These studies indicate that methylation of FXR by Set7/9 has a role in supporting the transactivation of FXR-target genes through methylation of histone H3K4 and FXR itself within the hinge domain, and adds to the complexity of coactivators and posttranslational modifications of NRs that contribute to gene regulation. Data gleaned from similar studies with the estrogen receptor-α and the AR would indicate that the FXR methylation by Set7/9 in the hinge region may serve to stabilize FXR and possibly promote its interaction with RXRα and the FXRE (20). The K206 methylation of FXR could be required for recruitment of a coactivator to the hinge region or could inhibit interaction with a corepressor. Methylation of the FXR on lysine 206 by Set7/9 could enhance transcriptional activity of the receptor by facilitating both interdomain communication between the NH2- and COOH-termini and recruitment to FXR-target genes, as described for the AR (20). In the AR receptor, the K630 site that is methylated by Set7/9 is also subject to acetylation by p/CAF and Tip60. The recruitment of p/CAF induced by androgens is impaired by siRNA depletion of Set7/9. Under these conditions there is an increase in histone H3K9-dimethylation, which is a repressive mark (20).
The hinge region of FXR has emerged as an important target for a growing number of posttranslational modifications. In addition to the methylation of lysine 206 by Set7/9 defined by our work, lysine 217 of FXR is the major acetylation/deacetylation site targeted by p300 and SIRT1 (19). Acetylation of FXR increases its stability but inhibits heterodimerization with RXRα, DNA binding, and transactivation activity. The level of FXR acetylation is reciprocally regulated by the acetylase p300 and the deacetylase SIRT1 (19). Set7/9-mediated histone methylation marks are potentially reversible through the action of LSD1, a lysine-specific demethylase that acts on di- or monomethylated H3K4 (30). Some histone demethylases, such as LSD1, have recently been found to demethylate nonhistone proteins, as occurs with methylated p53 and methylated DNMT1 (14, 32). It remains unknown whether methylation of FXR is subject to demethylation as another mechanism of dynamic regulation. Nuclear localization of FXR is facilitated by phosphorylation by PKCζ at Thr-442 (10). Phosphorylation of FXR by calcium-dependent PKCα at Ser-135, and Ser-154 in the DBD also contributes activation of FXR-target genes (11). Further studies are warranted to define the combinatorial nature and cross talk between these modifications in a manner similar to those defined for histones (31, 35).
The repertoire of coregulator proteins and associated posttranslational modifications continues to expand and provides mechanistic complexity required for transcription control of the many FXR-target genes. Although our work has focused on bile acid metabolism, FXR also plays an important and unexpected role in regulating lipid and glucose homeostasis (13, 38). There is some evidence to suggest that dysregulation of FXR signaling occurs in disease because of aberrant posttranslational modifications of histones. For example, we have recently reported that histone H3K4 trimethylation by the lysine methyl transferase MLL3 is important for supporting the transcriptional activation of FXR-target genes and is disrupted in obstructive cholestasis (3). In that study, Set7/9 was not one of the genes on a PCR array whose expression was altered, but it is certainly possible that it could be affected in other disorders.
GRANT
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-084434 (to F. J. Suchy).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
N.B., M.A., and F.J.S. conception and design of research; N.B. performed experiments; N.B. and F.J.S. analyzed data; N.B. and F.J.S. interpreted results of experiments; N.B. prepared figures; N.B. and F.J.S. drafted manuscript; N.B., M.A., and F.J.S. approved final version of manuscript.
REFERENCES
- 1. Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276: 28857–28865, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem 279: 54348–54357, 2004 [DOI] [PubMed] [Google Scholar]
- 3. Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 300: G771–G781, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell 109: 801–806, 2002 [DOI] [PubMed] [Google Scholar]
- 5. Chaya D, Zaret KS. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol 376: 361–372, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Das PM, Ramachandran K, vanWert J, Singal R. Chromatin immunoprecipitation assay. Biotechniques 37: 961–969, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol 18: 111–120, 2011 [DOI] [PubMed] [Google Scholar]
- 8. Eloranta JJ, Kullak-Ublick GA. The role of FXR in disorders of bile acid homeostasis. Physiology (Bethesda) 23: 286–295, 2008 [DOI] [PubMed] [Google Scholar]
- 9. Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem 283: 35086–35095, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun AQ, Balasubramaniyan N, Arias I, Setchell KD, Suchy FJ, Shneider BL. The membrane protein ATPase class I type 8B member 1 signals through protein kinase Cζ to activate the farnesoid X receptor. Hepatology 48: 1896–1905, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol 22: 2433–2447, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Gopalakrishnan B, Wolff J. siRNA and DNA transfer to cultured cells. Methods Mol Biol 480: 31–52, 2009 [DOI] [PubMed] [Google Scholar]
- 13. Hageman J, Herrema H, Groen AK, Kuipers F. A role of the bile salt receptor FXR in atherosclerosis. Arterioscler Thromb Vasc Biol 30: 1519–1528, 2010 [DOI] [PubMed] [Google Scholar]
- 14. Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Imhof A. Epigenetic regulators and histone modification. Brief Funct Genomic Proteomic 5: 222–227, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Jayani RS, Ramanujam PL, Galande S. Studying histone modifications and their genomic functions by employing chromatin immunoprecipitation and immunoblotting. Methods Cell Biol 98: 35–56, 2010 [DOI] [PubMed] [Google Scholar]
- 17. Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr Opin Struct Biol 20: 730–738, 2010 [DOI] [PubMed] [Google Scholar]
- 18. Kemper JK. Regulation of FXR transcriptional activity in health and disease: emerging roles of FXR cofactors and post-translational modifications. Biochim Biophys Acta 1812: 842–850, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab 10: 392–404, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Lysine methylation and functional modulation of androgen receptor by set9 methyltransferase. Mol Endocrinol 25: 433–444, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev 26: 147–170, 2005 [DOI] [PubMed] [Google Scholar]
- 22. Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci 31: 572–580, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, Wang H, Gu W, Roeder RG, Zhu WG. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc Natl Acad Sci USA 108: 1925–1930, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masatsugu T, Yamamoto K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys Res Commun 381: 22–26, 2009 [DOI] [PubMed] [Google Scholar]
- 25. Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pradhan S, Chin HG, Esteve PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 4: 383–387, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411, 2002 [DOI] [PubMed] [Google Scholar]
- 28. Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptorα by the SET7 lysine methyltransferase. Mol Cell 30: 336–347, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun AQ, Balasubramaniyan N, Liu CJ, Shahid M, Suchy FJ. Association of the 16-kDa subunit c of vacuolar proton pump with the ileal Na+-dependent bile acid transporter: protein-protein interaction and intracellular trafficking. J Biol Chem 279: 16295–16300, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metabol 12: 321–327, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villar-Garea A, Imhof A. The analysis of histone modifications. Biochim Biophys Acta 1764: 1932–1939, 2006 [DOI] [PubMed] [Google Scholar]
- 32. Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41: 125–129, 2009 [DOI] [PubMed] [Google Scholar]
- 33. Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111: 105–115, 2002 [DOI] [PubMed] [Google Scholar]
- 35. Winter S, Fischle W. Epigenetic markers and their cross-talk. Essays Biochem 48: 45–61, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421: 652–656, 2003 [DOI] [PubMed] [Google Scholar]
- 37. Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr Opin Struct Biol 13: 699–705, 2003 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA 103: 1006–1011, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18, 2011 [DOI] [PubMed] [Google Scholar]