Abstract
Activating mutations in the KRAS oncogene are common in colorectal cancer. However, the complete spectrum of KRAS targets that mediate its tumorigenic effect has not yet been fully delineated. We identified bone morphogenetic protein 4 (Bmp4), a transforming growth factor-β family member that regulates development and tissue homeostasis, as a new target of KRAS. In SW480, Hela, and 293 cells, oncogenic KRASV12 downregulated BMP4 RNA levels, a BMP4 promoter luciferase construct, and Bmp4 protein levels. The MEK inhibitor PD98059 but not the phosphatidylinositol 3-kinase inhibitor LY294002 blocked this downregulation of BMP4. To identify the region of the BMP4 promoter that mediated this regulation by KRAS, serial 5′-deletions of the promoter were generated. An inhibitory region was identified between −3,285 and −3,258 bp in the Bmp4 promoter. In summary, oncogenic KRAS can downregulate Bmp4 through a transcriptional pathway that depends on ERK. These findings point to a unique link between two pathways that are frequently altered in colon cancer.
Keywords: bone morphogenetic protein 4, GATA2, colon cancer, extracellular signal-regulated kinase, phosphatidylinositol 3-kinase
kras, a gdp/gtp binding protein that functions as a key intracellular signal transducer, is one of the most frequently activated oncogenes in human cancer. As many as 17–25% of all tumors harbor a KRAS mutation (18). It plays a particularly important role in the pathogenesis of colon cancer, where mutations are identified in up to 50% of cases. In its active, GTP-bound state, KRAS signals through multiple downstream pathways including the RAF/MEK, MAPK, JNK, and phosphatidylinositol 3-kinase pathways (4, 6, 21, 23) and thereby regulates target genes that promote cell growth and survival. However, the complete spectrum of targets that mediate the oncogenic effects of KRAS has not yet been fully delineated.
In an attempt to identify novel genes regulated by oncogenic KRAS in colon cancer, we utilized a cDNA microarray approach and previously identified bone morphogenetic protein 4 (Bmp4), a member of the transforming growth factor-β (TGF-β) family. Alterations in TGF-β signaling are known to play an important role in the pathogenesis of many human cancers, including colon cancer (28, 32). Although most studies have focused on the role of TGF-β in malignancy, there is growing evidence that the BMP subfamily also plays an important tumor suppressive role (17). Bmp4 binds to its type II receptor and recruits a type I receptor (BMPR1A and BMPR1B). After heterodimerization, the type II receptor phosphorylates and activates the type I receptor, resulting in serine-threonine kinase activity and concomitant phosphorylation of the signal transducer molecules Smads-1/5/8. The phosphorylated Smads-1/5/8 then heterodimerize with Smad4 and translocate to the nucleus, where they regulate the expression of Bmp4 target genes.
Recently, it has been demonstrated that germline mutations in either BMPR1A or SMAD4 result in the inherited cancer predisposition syndrome juvenile polyposis (11–12, 37), indicating a role for the BMP pathway in the pathogenesis of intestinal polyps and tumors. This was confirmed in a mouse model, where inhibition of BMP signaling resulted in the formation of numerous ectopic crypts, mimicking the juvenile polyposis syndrome (7). In addition, BMP signaling controls duplication of intestinal stem cells through suppression of Wnt-β-catenin signaling, thereby preventing crypt fission and the subsequent increase in crypt number (9). Furthermore, BMPs exhibit tumor-suppressive properties in human colon cancer cells (1, 24). However, Bmp4 has not yet been linked to KRAS.
In the present study, we describe a novel interaction between KRAS and Bmp4 in colon cancer. Oncogenic KRAS can transcriptionally downregulate Bmp4 expression, and this is mediated through the ERK signaling pathway. In the BMP4 promoter, an inhibitory region was identified that was responsive to KRAS. These findings point to a unique link between two pathways that are frequently disrupted in colon cancer.
EXPERIMENTAL PROCEDURES
Cell culture.
293T and Hela cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin. The human colon cancer cell line SW480 stably transfected with an small interfering (si)RNA against mutant KRASV12 (SW480siKrasV12) or a mock siRNA (SW480sicontrol; Ref. 14) was maintained in DMEM supplemented with 10% FBS, penicillin/streptomycin, and 2 ug/ml puromycin. Cells were treated with 20 μM PD98059 (Calbiochem) and 50 μM LY290042 (Calbiochem) for 12–24 h.
DNA microarrays.
RNA was isolated from SW480sicontrol and SW480siKrasV12 cells utilizing the RNeasy mini prep kit per manufacturer's instructions (Qiagen, Valencia, CA). RNA quality control, target preparation, and array hybridization and scanning were performed per recommended specifications (Asuragen, Austin, TX). Genechip PrimeView human arrays were utilized (Affymetrix, Santa Clara, CA). CEL files were analyzed utilizing R/Bioconductor package, oneChannelGUI. Briefly, normalization of expression values was performed utilizing the “Expresso” option, selecting “RMA” as the background correction, “quantiles” as normalization, “pmonly” as the PM correction, and “MAS” as the expression. Probes were filtered with IQR settings of 0.25. P values were calculated using a two-tailed Student's t-test. Microarray data have been submitted to Gene Expression Omnibus (GSE35663).
Plasmids and constructs.
The phr-GFP-KRASV12, phr-GFP-KRASD12, phr-GFP-KRASD13, phr-GFP-KRASwt, and kinase-mutant ERK1/2 constructs have been described previously (14, 16, 31). Empty phr-GFP and pcDNA3 plasmids were used as controls, respectively. The human Bmp4-promoter luciferase plasmid 3.36-kb Bmp4-luc was generated by PCR amplifying the BMP4 promoter 1 region using the human RP112526II clone (Invitrogen) as a template and cloning the PCR product into pGL3basic vector using the unique restriction enzyme sites NHE1 and XHO1. The construct was confirmed by DNA sequencing. Deletion constructs were generated by digesting 3.36-kb Bmp4-luc with Psi and XHO1 to generate the 3.17-kb Bmp4-luc plasmid, with Kpn1 and Xho1 to generate the 2.1-kb Bmp4-luc plasmid, with AvrII and Nhe1 to generate the 1.7-kb Bmp4-luc plasmid and with SpeI and Nhe1 to generate the 0.46-kb Bmp4-luc plasmid. Site-directed mutants were generated using the Quick-Change protocol (Stratagene) and the mutagenic primers 5′-GGAATTAAGGGCTACTGCGCTTATAGGATTATCTTTTCAC and 5′-GTGAAAAGATAATCCTATAAGCGCAGTAGCCCTTAATTCC. The introduced mutations were confirmed by DNA sequencing. The human Bmp4-promoter 2-luciferase plasmid was generated by PCR amplifying the BMP4 promoter 2 region using the human RP112526II clone (Invitrogen) as a template and cloning the PCR product into pGL3basic vector using the unique restriction enzyme sites HindIII and XHO1. The construct was confirmed by direct sequencing. The full length BMP4 promoter luciferase plasmid, containing both promoter 1 and promoter 2, was generated by subcloning the 3.36-kb Bmp4 fragment into Bmp4-Prom2-luc. pRLnull plasmid was used as for assaying transfection efficiency.
Transfections.
Transient transfections of plasmid DNA was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. SW480siKrasV12 and SW480sicontrol cells were seeded in 12-well dishes to reach 50% confluency the day of transfection. The 0.8 ug of promoter plasmids were cotransfected with 50 ng pRL-null plasmid and harvested 24 h after transfection. Luciferase assays were performed using the Dual Luciferase Kit (Promega) on a luminometer (Monolight 3010; Pharmingen). 293T and Hela cells were seeded in 6 well dishes to reach 60% confluency the day of transfection. Two micrograms of expression plasmid were transfected, and RNA was harvested 24–48 h after transfection.
RNA analysis.
RNA was extracted using RNeasy mini kit (Qiagen) and quantitative reverse transcription PCR was performed using SuperScript III Platinum Two-Step qRT-PCR kit (Invitrogen). The 18S rRNA served as an endogenous control. Primer sequences for Bmp4 and 18S are available upon request. PCR cycles were 2 min at 95°C, followed by 40 cycles with annealing temperature of 55–58°C. A fluorogenic SYBR Green and MJ research detection system were used for real time quantification. Relative mRNA expression was calculated using the parameter threshold cycle (CT) values. ΔCT was the difference in the CT values derived from the specific gene being assayed and the 18S rRNA. ΔΔCT represented the difference between the paired samples, as calculated by the formula ΔCT of a sample −ΔCT of a reference. The amount of target was normalized to 18S, and the reference was calculated as 2−ΔΔCT.
ELISA.
Bmp4 protein concentration was assayed utilizing the Quantikine human Bmp4 kit (R&D Systems) following the manufacturer's recommendations. SW480sicontrol and SW480siKrasV12 cells were seeded into 6-cm dishes to reach 80% confluence the next day. Serum starvation was performed for 24 h. Supernatant was collected, centrifuged, and used undiluted for ELISA.
Western blot analysis.
Cells were lysed in chilled lysis buffer (Cell Signaling) supplemented with proteinase inhibitor (PSC; Roche). Then, 15–20 ug protein lysate were resolved on 4–12% NuPAGE Bis-Tris polyacrylamid gels (Invitrogen) and transferred to polyvinylidene difluoride membranes (Millipore). Nuclear extracts were isolated using NPER kit (Pierce). The blots were probed with KRAS (Santa Cruz; 1:1,000), pERK and total ERK (Cell Signaling; 1:1,000), pSMAD1/5/8 and SMAD1 (Cell Signaling; 1:1,000), and β-actin (Sigma; 1:3,000) antibodies. Immunoreactive proteins were visualized using Western Lighting Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences).
BMP4 mRNA decay assay.
SW480sicontrol and SW480siKrasV12 cells were treated with 5 ug/ml actinomycin D. RNA was harvested after 0, 1, 2, 4, 8, and 12 h and transcribed into cDNA. Quantitative (q)RT-PCR was performed with Bmp4 and 18S primers as described.
Methylation assays.
SW480sicontrol and SW480siKrasV12 cells were seeded in 60-mm dishes. Twenty-four hours later, 2 μM 5-azacytidine (Sigma) were added to the medium. Cells were treated for 4 days with daily replacement of 5-azacytidine. RNA was extracted 4 days after initiation of treatment and qRT-PCR performed as described.
Bisulfite sequencing.
DNA from SW480sicontrol and SW480siKrasV12 cells was extracted utilizing blood and cell culture kit (Qiagen). One microgram DNA of each cell line was diluted in 21 μl Tris-EDTA (TE) buffer. Four microliters of a 2-M NaOH solution were added and samples were incubated at 50°C for 20 min. After incubation, DNA was mixed with low-melting agarose, and beads were formed by dropping 10 μl of DNA mixture into cold mineral oil. DNA-containing agarose beads were incubated with 2.5 M sodium metabisulfite and 125 mM hydrochinone at 50°C for 4 h. Beads were washed four times with TE. After being washed, beads were desulphonated with 0.3 M NaOH for 15 min at room temperature. After being desulphonated, beads were again washed twice with TE and twice with water. Washed beads were used as template in a nested PCR. Primer sequences are available upon request. PCR products were purified and sequenced.
Acetylation assays.
SW480sicontrol and SW480siKrasV12 cells were seeded in sixwell dishes and treated with 50 nM trichostatin A for 48 h. RNA was extracted and qRT-PCR performed as described.
Cell growth assays.
Then, 1 × 105 SW480sicontrol and SW480siKrasV12 cells were seeded into sixwell dishes. Cells were treated for 5 days with 2 ug inhibitory anti-Bmp4 antibody (R&D Systems) or mock treated and counted each day using Trypan blue (Invitrogen) and a hemacytometer (Fisher Scientific).
EMSA.
Nuclear extracts were prepared utilizing NE-PER nuclear extraction reagent (Pierce). Sequences of the BMP4 promoter between 3,361 and 3,171 bp were divided into five fragments and utilized as oligonucleotide probes (Table 1). The 5′-ends of the oligonucleotides were labeled with biotin during synthesis, and complementary oligonucleotides were annealed to generate double-stranded fragments. EMSA was performed using LightShift chemiluminescent kit (Pierce) according to the manufacturer's protocol. Briefly, 5 μg of nuclear extracts were incubated with 20 fmol of biotinylated oligonucleotides in binding buffer including 50 ng/μl poly(dI-dC), 0.05% Nonidet P-40, 2.5% glycerol, 5 mM MgCl2, and 2 mM EDTA, and the reaction mix was loaded onto 6% DNA retardation gels (Invitrogen). DNA-protein complexes were transferred onto nylon membranes (Roche Applied Science), and the mobility shift was detected using a streptavidin-horseradish peroxidase conjugate and a chemiluminescent substrate. Specificity of shifts was confirmed by utilizing 200-fold molar excess of unbiotinylated oligonucleotides as a specific competitor. Mutagenesis was performed to further define the elements responsible for the specific shifts obtained, as described in Table 1.
Table 1.
Sequences of oligonucleotide probes used for gel-shift assays
| Oligonucleotides | Sequence |
|---|---|
| 1 | 5′-GAATTCCTTCCGTAGCTTCACCAGACACCTAATTGGCCAA |
| 2 | 5′-GGCCAAGAAGGTTTGAAGACCTGATGTGGTTCTTAATTGGGGATGG |
| 3 | 5′-GATGGGGAATTAAGGGCTACTGTATCTATAGGATTATCTTTTCACT |
| 3A | 5′-GATGGGGAATTAAGGGCTACTGTATCTA |
| 3B | 5′-TACTGTATCTATAGGATTATCTTTTCACT |
| 4 | 5′-CTTTTCACTTGCATAGACCTATTTGGTGTGTTCAGGGC |
| 5 | 5′-GGGCATAGTGATACTATAATTGCCATATTTAACAGTTTATAAAG |
| Mut1 | 5′-AGCAAAAGGTTAAGGGCTACTGTATCTA |
| Mut2 | 5′-GATGGGGAACCGGAAATCACTGTATCTA |
| Mut3 | 5′-GATGGGGAATTAAGGGCTGTCACGCTCG |
RESULTS
Downregulation of Bmp4 by mutant KRAS.
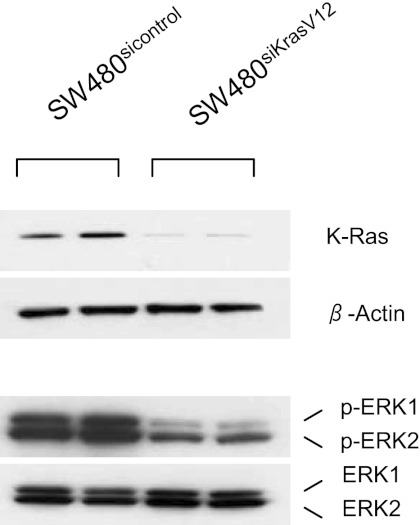
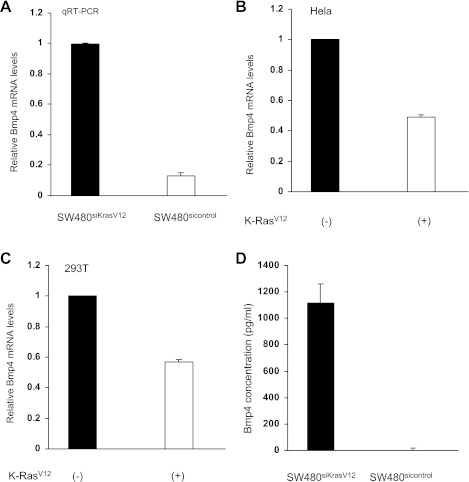
In SW480 colon cancer cells, the endogenous KRAS gene harbors an oncogenic G12V point mutation. Stable transfection of a specific siRNA against this mutant KRAS resulted in a 90% reduction of KRAS protein expression (Fig. 1). Accompanying this reduction in KRAS protein levels was a marked reduction of ERK1 and ERK2 phosphorylation (Fig. 1). cDNA microarray studies of RNA isolated from stably transfected SW480sicontrol and SW480siKrasV12 cells indicated that Bmp4 gene expression was twofold higher in cells in which mutant K-ras was knocked down (P < 0.05). qRT-PCR was performed to verify this result. There was a 7.7-fold increase in BMP4 mRNA expression in cells in which K-ras was knocked down (Fig. 2A). As a complementary study, we examined another colon cancer cell line with an endogenous wild-type K-ras gene (HT29). Introduction of a mutant K-ras expression suppressed BMP4 levels 52%. Of note, no such suppression was seen when a wild-type K-ras expression vector was transfected. To determine whether this was a cell-type-specific effect, 293T and Hela cells were transfected with a mutant KRASV12 expression plasmid and BMP4 mRNA levels were measured. qRT-PCR revealed an ∼50% reduction of BMP4 mRNA expression in these cells (Fig. 2, B and C), indicating that mutant KRAS can downregulate Bmp4 in a cell-type independent manner. In contrast, overexpression of wild-type KRAS in 293 cells led to only a 12% reduction in Bmp4 levels, indicating this was effect was KRAS genotype-specific. Finally, a significant downregulation of Bmp4 protein levels was confirmed by ELISA in SW480sicontrol cells (Fig. 2D). Thus mutant KRAS can downregulate Bmp4 expression.
Fig. 1.
Knockdown of K-ras blocks activation of ERK. Two different cell clones stably transfected with a small interfering (siRNA) against mutant K-rasV12 in SW480 colon cancer cells (SW480siKrasV12) or stably transfected with a control siRNA (SW480sicontrol) were analyzed for K-ras expression by Western blotting. Silencing of K-ras disrupts activation of ERK 1 and 2, shown as a reduction in phosphorylation (p-ERK1 and p-ERK2).
Fig. 2.
Mutant K-rasV12 can downregulate bone morphogenetic protein 4 (Bmp4). A: quantitative real-time PCR demonstrated a decrease in Bmp4 mRNA expression in SW480sicontrol cell lines. Bmp4 mRNA expression normalized to SW480siKrasV12. B and C: Hela and 293T cells were transiently transfected with phr-GFP-K-rasV12 or empty control vector. Bmp4 mRNA expression is downregulated ∼2-fold upon expression of mutant K-ras, as measured by quantitative RT-PCR. D: Bmp4 protein concentration is significantly lower in SW480sicontrol cells compared with SW480siKrasV12 cells, as assessed by ELISA.
Silencing of KRAS inhibits cell growth in a Bmp4-dependent manner.
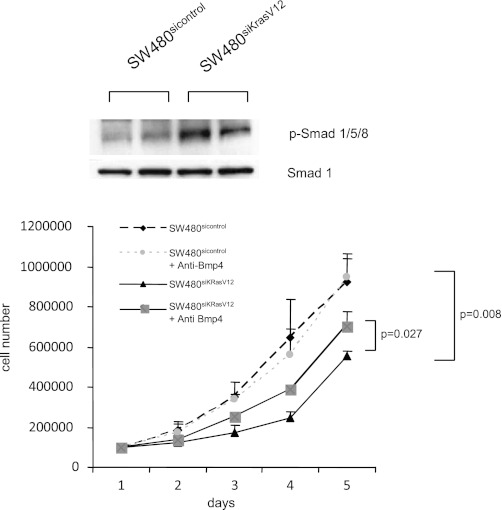
To verify that Bmp4 has a functional role in colon cancer cells, assays for Smad phosphorylation were performed. Downregulation of Bmp4 by mutant KRAS in SW480sicontrol cells reduced the phosphorylation levels of Smad1/5/8, which are key mediators of the intracellular Bmp4 signaling pathway (Fig. 3A). In vitro cell growth assays were then performed to define the effects of KRAS silencing. Stable knockdown of mutant KRAS was associated with a significant, twofold decrease in cell number after 5 days (Fig. 3B). Furthermore, treatment of SW480siKrasV12 cells with a neutralizing Bmp4 antibody over a 5-day period resulted in a marked increase in cell proliferation, whereas it did not affect the proliferation rate of SW480sicontrol cells (Fig. 3B). Silencing of KRAS can therefore reduce cellular proliferation in a Bmp4-dependent manner.
Fig. 3.
Bmp4 signaling is intact in SW480 cells and can negatively regulate cell growth. A: mutant K-rasV12 results in decreased phosphorylation of the Bmp4 effector proteins Smad1/5/8. B: Cell growth assays were performed on SW480sicontrol and SW480siKrasV12 cells that were treated with 2 μg/ml neutralizing Bmp4 antibody or mock treated. Cells were counted each day for 5 days. Shown are the mean cell numbers of 2 independent experiments.
KRAS downregulates Bmp4 through ERK.
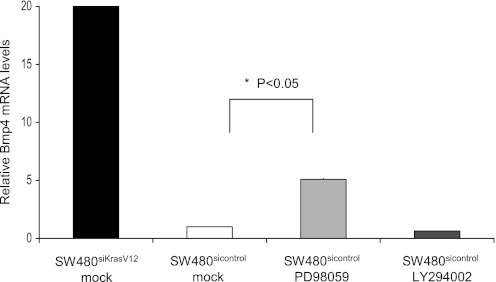
We next sought to identify which KRAS effector pathway may mediate this downregulation of Bmp4. The role of the ERK pathway was examined utilizing the specific inhibitor PD98059. Twenty micromoles of PD98059 increased BMP4 mRNA expression 5.1-fold in SW480sicontrol cells (P = 0.007; Fig. 4). However, inhibition of the phosphatidylinositol 3-kinase pathway with LY294002 failed to induce BMP4 mRNA levels in SW480sicontrol cells (Fig. 4). Of note, although inhibition of the ERK pathway strongly induced BMP4, it did not reach the peak induction observed with knockdown of KRAS, suggesting that the ERK pathway is a key but not sole mediator of the downregulation of Bmp4 by KRAS.
Fig. 4.
K-ras can downregulate Bmp4 through ERK activation. SW480sicontrol and SW480siKrasV12 cells were treated with an ERK inhibitor (20 μM PD98059), a phosphatidylinositol 3-kinase inhibitor (50 μM LY290042), or mock control (DMSO) for 24 h and Bmp4 mRNA levels were determined by quantitative RT-PCR. Inhibition of ERK resulted in a 5-fold induction of Bmp4 expression in SW480sicontrol cells.
KRAS regulates Bmp4 expression through transcriptional mechanisms.
To determine more specifically how KRAS regulates BMP4 mRNA levels, mRNA decay assays were performed to determine whether KRAS altered BMP4 mRNA stability. No difference in the half-life of Bmp4 was seen in SW480sicontrol and SW480siKrasV12 cells (data not shown). To determine whether KRAS regulated Bmp4 expression through epigenetic mechanisms, we examined the acetylation and methylation status of the BMP4 gene. The BMP4 promoter is GC-rich and harbors several CpG islands. However, treatment with either the histone deacetylase inhibitor trichostatin A or the demethylating agent 5-azacytidine did not result in any significant changes in BMP4 mRNA levels in SW480sicontrol or SW480siKrasV12 cells (data not shown). Furthermore, DNA bisulfite-sequencing of the CpG island closest to the transcriptional start site of the BMP4 promoter did not reveal any methylated CpG residues within this island (data not shown).
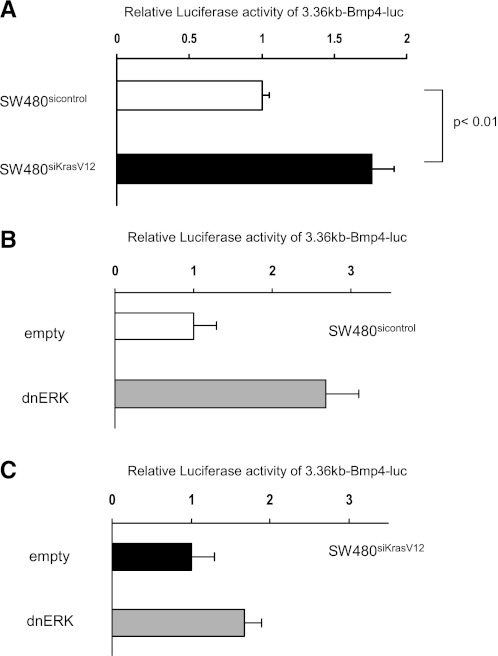
We then sought to determine whether KRAS regulates Bmp4 expression more directly through transcriptional mechanisms. The BMP4 gene contains two promoters that are separated by the first exon and are transcribed in a cell type- and differentiation-dependent manner (34). Because it is not known which promoter is active in SW480 cells, three different promoter constructs, spanning 3.36 kb of the first promoter (3.36-kb Bmp4-luc), 2.1 kb of the second promoter (2.1-kb Bmp4-luc), or spanning both promoters (5.5-kb Bmp4-luc), were introduced into SW480sicontrol and SW480siKrasV12 cells. Only the 3.36-kb Bmp4-luc construct showed activity in these cells, indicating that in SW480 cells BMP4 is under the control of the first promoter (data not shown). Furthermore, the levels of BMP4 were twofold higher in SW480siKrasV12 cells compared with SW480sicontrol cells that do not express mutant KRAS (Fig. 5A), suggesting that mutant KRAS regulates Bmp4 expression through transcriptional mechanisms.
Fig. 5.
K-ras downregulates Bmp4 through transcriptional mechanisms in an ERK dependent manner. A: SW480sicontrol and SW480siKrasV12 cells were transfected with the 3.36-kb Bmp4-luc reporter construct and luciferase activity was measured. Cotransfection with a dnERK plasmid or control vector (empty) was performed, and induction of Bmp4 luciferase activity was seen in SW480sicontrol (B) but not SW480siKrasV12 (C) cells. Mean values of 3 independent transfections are shown.
To confirm the role of ERK in the regulation of BMP4 gene transcription, the 3.36-kb Bmp4-luc promoter construct was cotransfected with a dominant-negative ERK plasmid (dnERK). Consistent with the effects of PD98059 on endogenous mRNA levels, dnERK enhanced BMP4 promoter activity 2.7-fold in SW480sicontrol cells but only 1.7-fold in SW480siKrasV12 cells (Fig. 5, B and C). Thus KRAS appears to downregulate BMP4 through transcriptional mechanisms and the ERK effector pathway is an important mediator.
Identification of a regulatory region of the BMP4 promoter responsive to mutant KRAS.
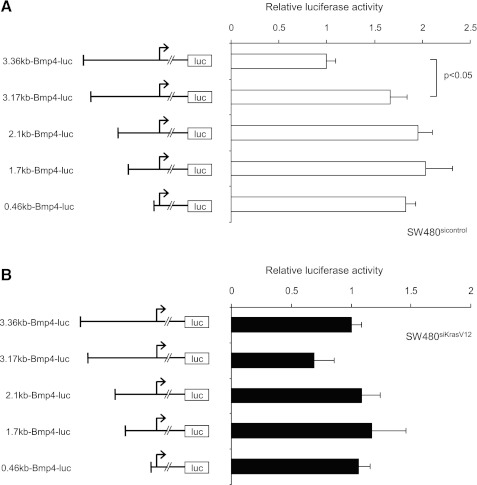
To identify regions of the BMP4 promoter that mediate its repression by mutant KRAS, we performed serial 5′-deletions. A significant induction of BMP4 promoter activity was observed in SW480sicontrol cells when the first 190 base pairs of the promoter construct were deleted (Fig. 6A), whereas there was only a slight reduction in promoter activity in SW480siKrasV12 cells (Fig. 6B). This 190-bp region appeared to contain a regulatory element that repressed transcription, as further deletions did not enhance this induction significantly.
Fig. 6.
K-ras-responsive element in the Bmp4 promoter is identified between 3.36 and 3.17 kb. The 5′-serial deletion constructs of the human Bmp4 promoter were generated and are schematically illustrated. Locations of the transcription start sites are indicated by the arrows. SW480sicontrol (white bars, A) and SW480siKrasV12 cells (black bars, B) were transiently transfected and luciferase activity measured 24 h later as described in materials and methods. Values were normalized to the activity of the full-length 3.36-kb reporter fragment in each cell line, respectively.
Identification of a critical regulatory element responsive to mutant KRAS.
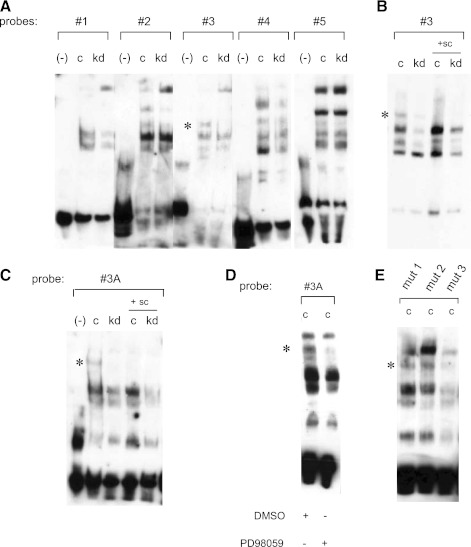
To further characterize the regulatory element in the BMP4 promoter between −3,361 and −3,171 bp that is responsive to KRAS, EMSAs were performed. The 190-bp region was divided into five fragments that were utilized as probes for EMSAs (Table 1). Nuclear extracts were isolated from SW480sicontrol and SW480siKrasV12 cells and incubated with the biotinylated probes. Several band shifts of differing intensity were observed in these experimental conditions, but we focused on a band shift that was almost completely absent using nuclear extracts from SW480siKrasV12 cells when incubated with oligonucleotide 3 (Fig. 7A). The specificity of the shift band was confirmed utilizing a 200-fold molar excess of unbiotinylated probe (Fig. 7B).
Fig. 7.
Identification of a critical regulatory element responsive to mutant K-ras. A: sequences between 3.36 and 3.17 kb in the Bmp4 promoter were divided into 5 fragments (probes 1–5) and EMSAs were performed. Nuclear extracts of SW480sicontrol (c) and SW480siKrasV12 (kd) were utilized. *Specific shift obtained in SW480sicontrol cells by probe 3. B: 200-fold molar excess of unlabeled probe 3 was used as a specific competitor (sc) to confirm specificity. C: specific shift was also observed in SW480sicontrol cells incubated with probe 3A, spanning the region 3,285–3258 bp of the Bmp4 promoter. D: treatment of SW480sicontrol cells with 20 μM PD98059 disrupts the binding of the critical element to the Bmp4 promoter. E: 3 different probes were generated that contain mutations of the first 10 bp (mut1), second 10 bp (mut2), and third 10 bp (mut3) of probe 3A. Specific shift (*) was lost in SW480sicontrol cells (c) incubated with probe mut3.
To further narrow the regulatory region, the 40-bp probe comprising oligonucleotide 3 was divided into two smaller oligonucleotides (probes 3A and 3B), and additional EMSAs were performed. A shifted band was readily detectable in nuclear extracts from SW480sicontrol cells that were incubated with oligonucleotide 3A (Fig. 7C) but was barely detectable when incubated with oligonucleotide 3B (data not shown). Treatment with the ERK inhibitor PD98059 resulted in a loss of the shifted band (Fig. 7D). Taken together, a regulatory element responsive to KRAS and ERK lies between −3,285 and −3,258 bp of the BMP4 promoter.
Transcription factor binding site analyses did not reveal any likely candidates for a transcriptional repressor that recognized DNA sequences within these 28 bp. Therefore, EMSAs were performed with 3 additional biotinylated oligonucleotides in which the first 9 bp, the second 9 bp, or the third 10 bp were mutated (“mut1,” “mut2,” and “mut3,” respectively). Only when nuclear extracts of SW480sicontrol cells were incubated with oligonucleotide “mut3” did the specific shifted band disappear (Fig. 7E), suggesting that the key regulatory element is located within these 10 bp. The bands obtained with the “full-length” probe 3 compared with the shorter probe 3A (and its mutants mut1, mut2, and mut3) differ, and this likely reflects differences in DNA-protein interactions due to differing flanking DNA sequences. There are three potential transcription factor-binding sites for GATA1, GATA2, and PLZF in this short promoter fragment. qRT-PCR and Western Blot analyses revealed that of these three transcription factors, only GATA2 was expressed in SW480 cells (data not shown). However, when a promoter construct that contained a specific mutation for the GATA2-binding sequence was tested, no difference was seen between SW480siKrasV12 and SW480sicontrol cells (data not shown). Furthermore, silencing of GATA2 with a siRNA did not significantly increase BMP4 mRNA in SW480sicontrol cells.
DISCUSSION
BMPs were first identified as regulators of bone formation in adults (33), but they also play an important role in the embryonic development of multiple organs, including the nervous system, musculature, skeleton, skin, hair, teeth, kidney, lung, and the intestinal tract (10, 20). Furthermore, there is growing evidence that BMP signaling is a key regulator of tumorigenesis. The role of BMP signaling in cancer is likely to be cell type and tissue specific. BMP signaling can inhibit proliferation of breast, prostate, gastric, and colon cancer cells (2–3, 30, 35). Specifically, signaling through Bmp2 in colon cancer results in growth inhibition (2, 8). Thus far Bmp4 has not been carefully evaluated in colon cancer pathogenesis, despite the fact that it is perhaps the best understood of all Bmp family members. The regulation of Bmp4 ligand activity has been described, and specific inhibitors such as noggin, chordin and follistatin have been identified (13, 27, 38). However, insights into the regulation of Bmp4 gene expression are lacking.
BMP4 is a novel target that is downregulated by oncogenic KRAS. A recent report using rat intestinal epithelial cells also demonstrated a dramatic reduction in BMP4 gene expression by oncogenic HRAS, consistent with the present results (15). This study suggested that HRAS regulation of BMP4 may be mediated through AU-rich element motifs that control mRNA stability. However, we did not observe any differences in mRNA stability of Bmp4 in cells that expressed oncogenic KRAS.
BMP4 mRNA levels may not always correlate with secreted Bmp4 protein levels, as there is evidence that production of TGF-β superfamily members can be controlled during posttranslational processing and secretion (20). However, the current studies demonstrate downregulation of Bmp4 both at the levels of mRNA and protein by KRAS. Furthermore, the functional activity of Bmp4 was verified through altered phosphorylation patterns of the downstream intracellular effectors Smad1/5/8. A limitation of the current study is that reduced levels of Bmp4 have not yet been demonstrated in human tumors with KRAS mutations.
KRAS can signal through various effector pathways, and we have demonstrated that oncogenic KRAS downregulates Bmp4 expression through MAPK/ERK signaling. This is particularly intriguing in light of previous reports that demonstrated an inhibitory effect on BMP signaling by ERK through the phosphorylation of SMAD1 in its linker region (19). In colon cancer cell lines, activation of ERK can prevent the growth inhibitory effects of BMP2/Smad signaling, potentially through this mechanism proposed by Kretzschmar et al. (19). Thus ERK may regulate BMP signaling on multiple levels. The effects of Bmp4 are classically mediated through SMAD4 pathways, but SMAD4-independent mechanisms have been described and these instead depend on ERK activation (29, 36). However, the role of these SMAD4-independent pathways in the suppression of tumor growth has yet to be defined.
Epigenetic processes including DNA methylation and histone modification are now recognized as critical events for regulation of gene expression in mammalian cells, and oncogenic KRAS has been implicated in both processes. It can repress RECK expression via histone deacetylation (5) and regulate DNA methylation via the expression of DNMT1 (DNA methyltransferase 1), resulting in decreased expression of uPA (urokinase-type plasminogen activator 1) and Fas (22, 25–26). In addition, the BMP4 promoter is GC-rich and a target for DNA methylation. However, we were unable to identify any epigenetic alterations that may mediate the downregulation of Bmp4 expression by KRAS.
Rather, oncogenic KRAS directly suppressed BMP4 promoter activity. A novel Ras-responsive region between −3,268 and −3,258 bp of the BMP4 promoter was identified containing a putative repressive element. Transcription factor binding site analysis revealed three potential transcription factors, namely PLZF, GATA1, and GATA2. Of these, only GATA2 is expressed in SW480 cells. However, further analysis demonstrated that GATA2 is not likely to be involved in the KRAS-mediated downregulation of Bmp4. Rather, there appears to be a new transcriptional repressor that has yet to be identified.
This relationship between KRAS and Bmp4 is likely to be broadly relevant. Oncogenic KRAS can downregulate BMP4 gene expression in many different tissues (colon, kidney, cervix), indicating that this effect is not tissue specific. Furthermore, this effect does not appear to be limited to the KRASV12 mutation, as overexpression of KRASD12 or KRASD13 mutants in 293T cells also downregulated BMP4 mRNA to the same extent as the KRASV12 mutation (data not shown).
Taken together, we present a novel mechanism for the regulation of BMP4 gene expression and provide evidence for cross talk between KRAS and BMP signaling in colon cancer cells that amplifies the effects on cellular proliferation. Oncogenic KRAS transcriptionally repressed BMP4 gene expression via the ERK pathway, and a novel transcriptional repressor appears to mediate this unique effect.
GRANTS
E.-M. Duerr was the recipient of a research grant from the German Research Foundation (Deutsche Forschungsgemeinschaft). This work was supported in part by National Cancer Institute Grant CA-92594 (to D. C. Chung).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: E.-M.D. and D.C.C. conception and design of research; E.-M.D., Y.M., K.M., W.-S.J., H.K., and R.J.X. performed experiments; E.-M.D. prepared figures; E.-M.D. drafted manuscript; Y.M., K.M., M.G., H.K., R.J.X., and D.C.C. analyzed data; K.M., M.G., R.J.X., and D.C.C. interpreted results of experiments; M.G. and D.C.C. approved final version of manuscript; D.C.C. edited and revised manuscript.
REFERENCES
- 1. Beck SE, Jung BH, Del Rosario E, Gomez J, Carethers JM. BMP-induced growth suppression in colon cancer cells is mediated by p21WAF1 stabilization and modulated by RAS/ERK. Cell Signal 19: 1465– 1472, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beck SE, Jung BH, Fiorino A, Gomez J, Rosario ED, Cabrera BL, Huang SC, Chow JY, Carethers JM. Bone morphogenetic protein signaling and growth suppression in colon cancer. Am J Physiol Gastrointest Liver Physiol 291: G135– G145, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brubaker KD, Corey E, Brown LG, Vessella RL. Bone morphogenetic protein signaling in prostate cancer cell lines. J Cell Biochem 91: 151– 160, 2004 [DOI] [PubMed] [Google Scholar]
- 4. Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2: 261– 274, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang HC, Liu LT, Hung WC. Involvement of histone deacetylation in ras-induced down-regulation of the metastasis suppressor RECK. Cell Signal 16: 675– 679, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Denhardt DT. Signal-transducing protein phosphorylation cascades mediated by Ras/Rho proteins in the mammalian cell: the potential for multiplex signalling. Biochem J 318: 729– 747, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303: 1684– 1686, 2004 [DOI] [PubMed] [Google Scholar]
- 8. Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, Offerhaus GJ, Van Deventer SJ, Peppelenbosch MP. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology 126: 111– 121, 2004 [DOI] [PubMed] [Google Scholar]
- 9. He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet 36: 1117– 1121, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev 6: 432– 438, 1996 [DOI] [PubMed] [Google Scholar]
- 11. Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 28: 184– 187, 2001 [DOI] [PubMed] [Google Scholar]
- 12. Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280: 1086– 1088, 1998 [DOI] [PubMed] [Google Scholar]
- 13. Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N. Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci USA 95: 9337– 9342, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jo WS, Mizukami Y, Duerr EM, Zukerberg LR, Chung DC. Wnt signaling can repress thrombospondin-1 expression in colonic tumorigenesis. Cancer Biol Ther 4: 1361– 1366, 2005 [DOI] [PubMed] [Google Scholar]
- 15. Kanies CL, Smith JJ, Kis C, Schmidt C, Levy S, Khabar KS, Morrow J, Deane N, Dixon DA, Beauchamp RD. Oncogenic Ras and transforming growth factor-beta synergistically regulate AU-rich element-containing mRNAs during epithelial to mesenchymal transition. Mol Cancer Res 6: 1124– 1136, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol 12: 253– 265, 2002 [DOI] [PubMed] [Google Scholar]
- 17. Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE 2007: cm1, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta 1756: 81– 82, 2005 [DOI] [PubMed] [Google Scholar]
- 19. Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature 389: 618– 622, 1997 [DOI] [PubMed] [Google Scholar]
- 20. Leong LM, Brickell PM. Bone morphogenic protein-4. Int J Biochem Cell Biol 28: 1293– 1296, 1996 [DOI] [PubMed] [Google Scholar]
- 21. Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways. Eur J Biochem 242: 171– 185, 1996 [DOI] [PubMed] [Google Scholar]
- 22. MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem 270: 11327– 11337, 1995 [DOI] [PubMed] [Google Scholar]
- 23. Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras. GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260: 1658– 1661, 1993 [DOI] [PubMed] [Google Scholar]
- 24. Nishanian TG, Kim JS, Foxworth A, Waldman T. Suppression of tumorigenesis and activation of Wnt signaling by bone morphogenetic protein 4 in human cancer cells. Cancer Biol Ther 3: 667– 675, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Pakneshan P, Szyf M, Rabbani SA. Methylation and inhibition of expression of uPA by the RAS oncogene: divergence of growth control and invasion in breast cancer cells. Carcinogenesis 26: 557– 564, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Peli J, Schroter M, Rudaz C, Hahne M, Meyer C, Reichmann E, Tschopp J. Oncogenic Ras inhibits Fas ligand-mediated apoptosis by downregulating the expression of Fas. EMBO J 18: 1824– 1831, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell 86: 589– 598, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piek E, Roberts AB. Suppressor and oncogenic roles of transforming growth factor-beta and its signaling pathways in tumorigenesis. Adv Cancer Res 83: 1– 54, 2001 [DOI] [PubMed] [Google Scholar]
- 29. Pino MS, Kikuchi H, Zeng M, Herraiz MT, Sperduti I, Berger D, Park DY, Iafrate AJ, Zukerberg LR, Chung DC. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology 138: 1406– 1417, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pouliot F, Labrie C. Role of Smad1 and Smad4 proteins in the induction of p21WAF1,Cip1 during bone morphogenetic protein-induced growth arrest in human breast cancer cells. J Endocrinol 172: 187– 198, 2002 [DOI] [PubMed] [Google Scholar]
- 31. Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem 268: 5097– 5106, 1993 [PubMed] [Google Scholar]
- 32. Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer 3: 807– 821, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Urist MR, Mikulski A, Lietze A. Solubilized and insolubilized bone morphogenetic protein. Proc Natl Acad Sci USA 76: 1828– 1832, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van den Wijngaard A, Pijpers MA, Joosten PH, Roelofs JM, Van zoelen EJ, Olijve W. Functional characterization of two promoters in the human bone morphogenetic protein-4 gene. J Bone Miner Res 14: 1432– 1441, 1999 [DOI] [PubMed] [Google Scholar]
- 35. Wen XZ, Miyake S, Akiyama Y, Yuasa Y. BMP-2 modulates the proliferation and differentiation of normal and cancerous gastric cells. Biochem Biophys Res Commun 316: 100– 106, 2004 [DOI] [PubMed] [Google Scholar]
- 36. Zhou Q, Heinke J, Vargas A, Winnik S, Krauss T, Bode C, Patterson C, Moser M. ERK signaling is a central regulator for BMP-4 dependent capillary sprouting. Cardiovasc Res 76: 390– 399, 2007 [DOI] [PubMed] [Google Scholar]
- 37. Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, Launonen V, Virta S, Pilarski R, Salovaara R, Bodmer WF, Conrad BA, Dunlop M, Hodgson SV, Iwama T, Jarvinen H, Kellokumpu I, Kim JC, Leggett B, Markie D, Mecklin JP, Neale K, Phillips R, Piris J, Rozen P, Houlston RS, Aaltonen LA, Tomlinson IP, Eng C. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet 69: 704– 711, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86: 599– 606, 1996 [DOI] [PubMed] [Google Scholar]