Abstract
The objectives of this review are twofold. Our first objective is to evaluate the evidence supporting a role for genetics in irritable bowel syndrome (IBS). Specific examples of the associations of genetic variation and symptoms, syndromes, and intermediate phenotypes, including neurotransmitter (serotonergic, α2-adrenergic, and cannabinoid) mechanisms, inflammatory pathways (IL-10, TNFα, GNβ3, and susceptibility loci involved in Crohn's disease), and bile acid metabolism, are explored. The second objective is to review pharmacogenetics in IBS, with the focus on cytochrome P-450 metabolism of drugs used in IBS, modulation of motor and sensory responses to serotonergic agents based on the 5-hydroxytryptamine (5-HT) transporter-linked polymorphic region (5-HTTLPR) and 5-HT3 genetic variants, responses to a nonselective cannabinoid agonist (dronabinol) based on cannabinoid receptor (CNR1) and fatty acid amide hydrolase (FAAH) variation, and responses to a bile acid (sodium chenodeoxycholate) and bile acid binding (colesevelam) based on klothoβ (KLB) and fibroblast growth factor receptor 4 (FGFR4) variation. Overall, there is limited evidence of a genetic association with IBS; the most frequently studied association is with 5-HTTLPR, and the most replicated association is with TNF superfamily member 15. Most of the pharmacogenetic associations are reported with intermediate phenotypes in relatively small trials, and confirmation in large clinical trials using validated clinical end points is still required. No published genome-wide association studies in functional gastrointestinal or motility disorders have been published.
Keywords: adrenergic, serotonergic, solute carrier 6A4, 5-hydroxytryptamine transporter-linked polymorphic region, inflammation, susceptibility, klothoβ, bile acid malabsorption
epidemiological studies of familial aggregation (57, 99) and twins (7, 70, 72, 82, 83) suggest a genetic contribution to irritable bowel syndrome (IBS). While the data are conflicting, they are generally consistent with the hypothesis that IBS may be a complex genetic disorder. Understanding of genetic variation and its influence on gut motility, secretion, sensation, and inflammation, as it is applied to IBS, has the potential to help elucidate mechanisms of control of a poorly understood syndrome. Similarly, the impact of genetic variation on the targets of therapy in IBS may enhance the efficacy of pharmacological therapies. These targets include receptors and regulators of gut function and variations in drug metabolism.
This review addresses the available literature on the role of genetics in IBS. There are no published genome-wide association studies in functional gastrointestinal (GI) or motility disorders specifically focusing on genetic epidemiology and pharmacogenetics.
Genetic Epidemiology of IBS: Candidate Gene Approach
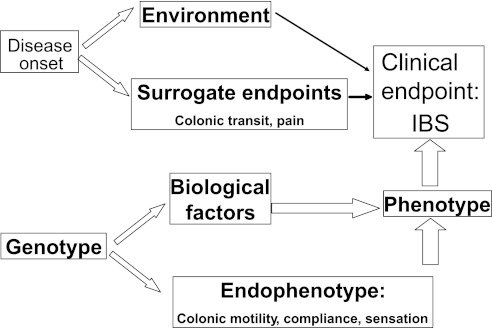
As the general approach followed in genotype association studies (Fig. 1), investigators have sought the relationship between genotype and clinical phenotype, while appreciating that intermediaries, such as the effect of the environment and other unknown biological factors, are largely unexplored. With this degree of imprecise control or knowledge of potential contributing factors, the identification of significant associations requires large numbers of participants and, importantly, replication of findings in different cohorts or ethnic groups. A second approach explores genotype-endophenotype association studies, that is, the relationship between candidate genes and quantitative traits (biomarkers, intermediate phenotypes, or endophenotypes) of interest. Endophenotypes are defined here as phenotypes that bear a closer relationship to the biological processes that give rise to the illness, have a hereditary component and cosegregate with illness within families, and are independent of whether or not the illness is active (46). Such biomarkers or quantitative traits typically are measurable, have a defined coefficient of variation, usually have a known relationship with the main manifestations of the clinical phenotype, and provide opportunities to assess genetic associations with much smaller sample sizes.
Fig. 1.
Genetic associations between candidate genes and intermediate phenotypes that are associated with manifestations of the clinical phenotype. One can also evaluate the role of the candidate mechanism. IBS, irritable bowel syndrome. [Adapted from Huang et al. (53).]
By studying the genetic associations between candidate genes and intermediate phenotypes that are associated with manifestations of the clinical phenotype, one can also evaluate the role of the candidate mechanism in IBS (see Fig. 1 adapted from Ref. 53). The intermediate phenotypes most commonly used in IBS are colonic transit, colonic motility and compliance, and sensation thresholds and ratings. Details regarding the published gene polymorphisms associated with IBS, the magnitude and significance of the disease association, the sample size, and the replication of findings are summarized in an extensive review by Saito et al. (97).
Neurotransmitter Mechanisms
Serotonin.
Serotonin [5-hydroxytryptamine (5-HT)] is involved in controlling GI secretion, motility, and visceral perception (41, 63). It is, therefore, not surprising that, in patients with IBS, studies have documented alterations in 5-HT levels (6) and 5-HT signaling (4); serotonin and serotonin receptor-modulating drugs are vigorously pursued in IBS, despite poor consensus on the optimal end points for trials and the regulatory agency requirement of rigorous safety standards for serotonergic agents because of potential vascular effects (14, 15, 24). As a result of the widespread interest in serotonin, genetic alterations in serotonergic mechanisms are the most extensively studied genetic associations with IBS.
SEROTONIN TRANSPORTER PROMOTER VARIANTS.
Serotonin transporter, or solute carrier 6A4 (SLC6A4), is the protein involved in the reuptake of 5-HT after it has interacted with the downstream receptor. Associations have been explored between IBS and two polymorphic regions within the promoter, as well as an intronic region of the gene near exon 2, with a variable number of tandem repeats (VNTR). The main polymorphic region in the promoter region is the 5-HT transporter long polymorphic region (5HTTLPR), which is located on chromosome 17q11.1–17q12 and organized into 14 exons spanning ∼38 and ∼1.4 kb upstream from the first exon (84). The long polymorphic region consists of a 44-bp insertion (71, 84), although this insertion is 43 bp in certain ethnic groups. The long and short variants of the polymorphic region (71) impact the function of SLC6A4, and the short allele results in a protein that is associated with reduced reuptake of 5-HT.
SLC6A4 is central to fine-tuning brain 5-HT neurotransmission; it is abundant in cortical and limbic areas, affects emotional aspects of behavior, and is associated with anxiety, psychiatric disease, and response to treatment. However, SLC6A4 is also expressed in the GI tract, and there is controversy as to whether it is underexpressed in mucosal biopsies from patients with IBS (17, 28).
Several studies have explored the association of any 5-HTTLPR allele and IBS, including one study from our center (64). Studies from Western countries and Asia on the association of the 5-HTTLPR genotype and IBS provide somewhat contradictory results (110), and a meta-analysis in 2007 suggests that, among Caucasians and Asians, odds ratios (OR) for IBS in subjects for SS or LS vs. LL [OR = 1.0, 95% confidence interval (CI) = 0.8–1.2] and for SS vs. LL or LS (OR = 1.0, 95% CI = 0.7–1.4) are not significant (110). Data from IBS patients from India (74% men) suggest an association of the homozygous S genotype (SS) with constipation-predominant IBS (C-IBS) (101). While the prevalence of IBS in India appears to be higher in men than in women (43), it is unclear why sex would explain the association of the SS genotype (for a gene located on chromosome 17, rather than an X or a Y chromosome) with C-IBS. A study that included patients with IBS from Germany and the United Kingdom [where prevalence is ∼1.3–1.5:1 in women compared with men (38, 47)] showed a lower prevalence of the SS genotype in IBS and, particularly, in diarrhea-predominant IBS (D-IBS), but this was only observed in male patients (34 male IBS patients and 30 male controls) (88).
The biological basis of these genotype-phenotype associations of the 5-HTTLPR SS genotype and C-IBS in India and protection from D-IBS in Germany and the United Kingdom is unclear. Thus, on the basis of the observations of Lesch et al. (71), the SS genotype would be expected to result in increased 5-HT (e.g., in a synaptic cleft), as it is associated with reduced 5-HT reuptake. It is unclear how increased 5-HT (which promotes secretion and motility in the gut) would be associated with C-IBS in patients from India. Similarly, the “protection” from D-IBS in male patients from the United Kingdom and Germany is difficult to understand. These observations emphasize the importance of balancing ethnicity among disease and control conditions, especially in increasingly multiethnic countries. The frequency of the short allele is higher among people of Asian ancestry, and ethnic imbalance in the disease and control groups may account for some of these associations. In summary, the association of 5-HTTLPR and IBS symptoms remains unproven.
Other studies show association with features that are related to IBS, such as psychological distress, rectal sensorimotor functions, and cerebral blood flow during rectal distensions.
Jarrett et al. (56) reported on 5-HTTLPR polymorphisms in 21 men and 117 women with IBS: the 5-HTTLPR genotype was not associated with GI symptom severity score or current levels of depression, anxiety, or general psychological distress. On the other hand, participants with the 5-HTTLPR SS genotype or those carrying a STin2.9 VNTR allele (see below) were more likely to have a history of depression, and participants with the 5-HTTLPR LL genotype had a reduced level of social functioning (56). It was concluded that the 5-HTTLPR genotype may modify risk for depressive episodes in IBS.
The 5-HTTLPR genotype (S allele) is associated with higher pain sensory ratings during rectal distension studies in health and IBS, and the increased sensation ratings in carriers of the S allele are not caused by lower rectal compliance (19).
The 5-HTTLPR SS genotype is also associated with greater regional cerebral blood flow in response to colorectal distension in patients with IBS. The most pronounced regional increases were in the left anterior cingulate cortex, right parahippocampal gyrus, and left orbitofrontal cortex (40). The anterior cingulate cortex and orbitofrontal region have been shown to be preferentially activated in patients with IBS during or in anticipation of rectosigmoid stimulation (87). Taken together, these two observations converge on the increased pain sensitivity in IBS patients, owing to increased activity in the emotional motor system of the brain.
Another genetic variation related to 5-HTTLPR is STin2 VNTR, which is located in intron 2; the biological function of this polymorphism is unclear. It consists of a variable number (e.g., 9, 10, or 12) of identical 17-bp segments (65); STin2.12 has been reported to possess greater transcriptional activity than STin2.10 (76). The 10/12 genotype has been associated with IBS in one study (114). However, at least five other studies found no association between STin2 VNTR and IBS (65, 73, 88, 90, 121).
A single-nucleotide polymorphism (SNP), rs25531, located immediately upstream of 5-HTTLPR, is strongly linked with the latter. The rs25531 G allele lowers SLC6A4 transcription compared with the A allele and occurs most frequently with the 5-HTTLPR L allele. In a study of 186 patients with IBS and 50 healthy control subjects, the odds of having IBS were increased (OR = 3.3, 95% CI = 1.1–9.6) in carriers of the G allele of rs25531 compared with healthy controls (65). However, the minor allele frequency is only 10–18% (52); therefore, these results require replication.
SEROTONIN RECEPTOR VARIANTS.
The 5-HT2A receptor gene is expressed in the brain, vagal nuclei, and the gut (11, 74). The −1438 (G/A) polymorphism has been associated with anorexia nervosa, alcohol dependence, obesity, obsessive compulsive disorder, and seasonal affective disorder (33, 86). The AA genotype is associated with higher 5-HT2A gene expression in cell lines endogenously expressing 5-HT2A (91). The −1438 polymorphism is in linkage disequilibrium with the 102 T/C polymorphism, which appears to be functionally silent (85).
In a study from Turkey of 54 patients with IBS based on Rome I criteria and 107 healthy controls, increased risk of IBS was associated with the homozygous C allele of the 102 T/C polymorphism (OR = 7.89, 95% CI = 1.079–57.76) and the homozygous A allele of the −1438 G/A polymorphism (OR = 11.14, 95% CI = 1.59–78.06) of the 5-HT2A receptor gene. In addition, the T/T genotype of the 102 T/C polymorphism may be associated with more severe pain in patients with IBS (92).
More recently, a study from Greece (78) of 124 patients with Rome III-positive IBS criteria and 238 healthy individuals replicated the association with the A allele (OR = 1.59, 95% CI = 1.17–2.18) and AA genotype (OR = 2.41, 95% CI = 1.30–4.44) of the −1438 G/A polymorphism and IBS. However, associations with the −102 C/T polymorphism with IBS and with more severe pain were not replicated (78).
Expression of 5-HT receptors (HTR3A and HTR3E) and micro-RNA (miRNA)-510 has been detected in human colonic epithelial cells by in situ hybridization (58). miRNAs are short (20- to 24-nucleotide) noncoding RNAs that are involved in posttranscriptional regulation of gene expression. No HTR3E expression was detectable within the lamina propria; a weak expression of miRNA-510 was shown within the lamina propria; HTR3E and miRNA-510 colocalize in enterocytes (58). 5-HT3C, 5-HT3D, and 5-HT3E subunits are coexpressed with 5-HT3A in cell bodies of human colonic myenteric neurons. Furthermore, 5-HT3A and 5-HT3D are expressed in the submucosal plexus of the human large intestine (59).
In a study of genetic variation of 5-HT3 receptors in IBS patients in the United Kingdom and Germany (58), the novel HTR3E 3′-untranslated region variant c.*76G>A (rs62625044) was associated with female D-IBS (OR = 8.53, 95% CI = 1.04–70.28) in the United Kingdom and replicated in D-IBS patients in Germany (OR = 4.92, 95% CI = 1.49–16.30). Using a reporter assay, Kapeller et al. (58) showed that c.*76G>A affected the binding of miRNA-510 to the HTR3E 3′-untranslated region and caused elevated protein expression in two different cell lines. While these data suggest an association between rs62625044 and IBS, it is worth noting that the minor allele frequency of rs62625044 is only 0.03 (95% CI = 0.011–0.064), and the difference in prevalence of the c.*76G>A genotype (11% in D-IBS patients and 4% in controls) in the pooled populations may represent a chance finding that requires replication.
The HTR3A polymorphism c.−42C>T (C178T; rs1062613), which causes increased 5-HT3A subunit expression in vitro (58), is associated with amygdala responsiveness in IBS patients (62); thus the C/C genotype, compared with T carrier status, is associated with increased anxiety and amygdala responsiveness during emotional and nonemotional tasks.
α2-Adrenergic receptors.
Several studies have also emphasized the role of the autonomic nervous system in GI motility and, consequently, alteration of this part of the nervous system in IBS (32, 48, 106). In general, adrenergic function is integral to regulation of the autonomic nervous system; more specifically, adrenergic function directly modulates gut and colon function and sensation (8, 112). The α2-adrenergic receptor has also been proposed as a mechanism whose genetic variation may modify motor and sensory functions in IBS patients. In 274 IBS patients (90 of whom had C-IBS) and 120 controls, there was a significant association between C-IBS and the α2C Del 322–325 deletion, which alters the coding region of the gene and encodes a receptor with a loss-of-function phenotype (64). This variation results in a receptor that has markedly decreased agonist-mediated responses in vitro (103). There is also evidence that the same variation results in altered cold pain perception without affecting cognition (66).
A nonsignificant (P = 0.08) association of C-IBS with the α2A −1291 C>G genotype (rs1800544, located in the promoter region of the α2A-adrenoceptor gene) provides further support for association between genetic variations in α2A- and α2C-adrenergic receptors and C-IBS. From a study in Northern India, a genotypic association between the α2A −1291 C>G polymorphism and D-IBS (OR = 2.08, 95% CI = 1.06–4.07, P < 0.05) but no significant association with C-IBS or mixed-bowel-function IBS (M-IBS) was observed (100).
In our study, however, we did not detect associations between the α2A genotype and a wide range of GI sensory or motor functions (19). Further work is required to assess the potential genetic role of α2-adrenergic mechanisms in IBS.
Cathechol-O-methyltransferase.
Cathechol-O-methyltransferase (COMT) is one of the enzymes that metabolizes catecholamines, thereby acting as a key modulator of dopaminergic and adrenergic/noradrenergic neurotransmission. The COMT Val158Met polymorphism is associated with a difference in thermostability, leading to a three- to fourfold reduction in the activity of the COMT enzyme (75). The Met/Met genotype results in reduced COMT enzymatic activity (60). The COMT Val158Met polymorphism influences the human experience of pain and may underlie interindividual differences in the adaptation and responses to pain and other stressful stimuli (124). There is evidence of association between COMT Val158Met genetic variation and reduced presynaptic phasic dopamine release in Met carriers that may result in impaired ability of the prefrontal cortex to downregulate startle responses (and amygdala complex activity) during threat and increased attention to a prepulse (108). The authors suggest that altered early response mechanisms to potential and symptom-related threats may play a role in central pain amplification and hypervigilance and may partially explain the greater reported prevalence of the Met allele in chronic pain syndromes (31).
Cannabinoid receptors and metabolism.
Cannabinoid receptors modulate a variety of GI functions, including pain modulation, inflammation, and gastric and colonic motility (55), in healthy humans (34, 35). A recent study has further demonstrated that use of a cannabinoid agonist may reduce fasting colonic motility in patients with nonconstipated IBS and in patients with IBS (116) and, therefore, represents a therapeutic target. As a result, a search for genetic variations in cannabinoid receptors and signaling is warranted.
The main endogenous cannabinoid agonists (endocannabinoids) are anandamide (AEA) and 2-arachidonyl glycerol, and there are two classes of cannabinoid receptors: CB1 receptors (located in the central nervous system, GI tract, enteric nervous system, liver, fat, and muscle) and CB2 receptors (found predominantly in immune cells) encoded by the CNR1 and CNR2 genes, respectively. The proteins involved in endocannabinoid metabolism are fatty acid amide hydrolase (FAAH) for AEA and monoacylglycerol lipase for 2-arachidonyl glycerol. The endocannabinoids are synthesized on demand in postsynaptic neurons (unlike classic neurotransmitters, which are presynthesized and stored in synaptic vesicles), and they signal as retrograde messengers binding to presynaptic CB1 receptors on cholinergic neurons, activating intracellular signaling cascades, inhibiting cAMP production, and, thereby, modulating protein kinases and ion channels, leading to inhibition of presynaptic neurotransmitter release (specifically, ACh in the enteric nervous system). AEA is then inactivated by FAAH; reduced activity of this enzyme leads to more AEA reaching the presynaptic membrane and a greater effect on ACh release from the presynaptic neuron. A common SNP is FAAH C385A, which causes reduced cellular expression and activity of FAAH enzyme (27). FAAH C385A was significantly associated with D-IBS, M-IBS, and rapid colon transit in a Mayo Clinic study (21).
In a study of 162 Rome II-positive IBS patients and 423 healthy controls from Korea, Park et al. (90) explored the association of IBS with the CNR1 gene. The gene that codes for CNR1 contains a polymorphic (AAT)n triplet repeat in the 3-flanking region of the CNR1 gene: a higher number of AAT triplets may induce a Z-shaped conformation in the DNA, thereby altering gene transcription. Thus, expression of the gene can be inversely proportional to the number of AAT repeats. This study showed significant associations between the CNR1 >10/>10 genotype and IBS overall (C-IBS, D-IBS, and M-IBS) and with severity of abdominal pain in IBS (90).
Association of the CNR1 genotype with colonic motor response to a nonselective cannabinoid agonist, dronabinol, is described in Pharmacogenetics in IBS.
SCN5A.
The SCN5A-encoded Na+ channel, Nav1.5, is expressed in interstitial cells of Cajal and smooth muscles in the circular muscle layer of the human intestine. Among 49 patients with IBS associated with at least moderately severe abdominal pain, 1 had a loss-of-function missense mutation, G298S (98). This mutation was not observed in 1,500 healthy controls. This G298S SCN5A missense mutation is functionally relevant, as it resulted in marked reduction of whole cell Na+ current and loss of function of Nav1.5 in transfected human embryonic kidney (HEK-293) cells (98).
Inflammatory Mechanisms
There is mounting evidence suggesting that, in some patients, IBS may be associated with inflammation or immune activation. The evidence includes the occurrence of IBS as a postinfectious phenomenon (79, 104), an increase in gut inflammatory cells noted on mucosal biopsy (reviewed in Ref. 89) or full-thickness biopsy (107), and experimental data that demonstrate alterations in gut motility and sensation after naturally acquired or experimentally administered infection (54, 69). As with celiac disease and inflammatory bowel disease, the pauci-inflammatory IBS may result from the interaction between environmental influences, such as nutrients, antibiotics, and the microflora, and the host genetics (95). As in inflammatory bowel disease, genetic alterations in inflammatory signaling and regulation may predispose to IBS.
IL-10, TNFα, and G proteins.
IL-10 and transforming growth factor-β1 have anti-inflammatory properties; genotype variation in the production of these cytokines would be consistent with the hypothesis that some patients are genetically predisposed to produce smaller amounts of the anti-inflammatory cytokine (e.g., IL-10) and potentially develop the mild inflammatory manifestations of IBS. In a first study from the United Kingdom (45), frequencies of the high IL-10 producer genotype for IL-10 (IL-10 G-1082A) were significantly reduced in patients with IBS compared with controls (21% vs. 32%, P = 0.003). Polymorphisms in the 5′-flanking region of IL-10 genetically affect interindividual differences in IL-10 production (36). A subsequent study from the Netherlands (109) did not replicate these results for IL-10 G-1082A. On the other hand, in the study from the Netherlands, TNFα G/A (G-308A, high producer) was more prevalent in IBS patients than in controls (41% vs. 26%, P = 0.02), and more patients than controls (41% vs. 30%) were positive for the A allele (P = 0.044, OR = 1.68, 95% CI = 1.01–2.79).
G protein-coupled receptors are present on excitable cells and cells that are susceptible to regulation (81). Activation of these receptors leads to production (catalyzed by GNβ3) of the βγ heterodimer from the heterotrimeric G protein. First studies suggested an association between GNβ3 C825T and dyspepsia (13, 50); however, this finding was not replicated in patients with IBS (3, 96).
A study from Korea replicated none of these three associations (68).
Susceptibility genes for Crohn's and inflammatory bowel diseases.
The main susceptibility genes for Crohn's disease affect several etiopathogenetic mechanisms, including bacterial recognition, epithelial transport and barrier function, prostaglandins, autophagy, and Th17 differentiation (10). Three genes, Toll-like receptor 9 [TLR9, rs5743836 (OR = 1.536, 95% CI = 1.080–2.182)], interleukin-6 [IL6, rs206986 (OR = 1.509, 95% CI = 1.031–2.209)], and E-cadherin 1 [CDH1, rs16260 (OR = 1.398, 95% CI = 1.069–1.829)], have been reported in association with postinfectious IBS in a Walkerton, Ontario, Canada cohort (111).
In a study involving patients from Sweden and the United States (125), associations between 30 Crohn's disease candidate genes and IBS identified a significant association of TNF superfamily member 15 [TNFSF15, rs4263839 (G/a)] with IBS (P = 2.2 × 10−5, OR = 1.37) and C-IBS (P = 8.7 × 10−7, OR = 1.79). Importantly, the association was detected independently in patients in Sweden and in the Upper Midwest in the United States (125), many of whom are of Scandinavian and Northern European ethnicity. More recently, the association of IBS with TNFSF15 was also reported in a cohort of patients in England (105). TNFSF15 is strongly associated with Crohn's disease in Japan and more modestly in European populations. TNFSF15 is upregulated in Crohn's disease and has a variety of actions, including induction of nuclear factor-κB activation, potentiation of IL-2 signaling, and secretion of γ-interferon by T lymphocytes (122).
In the same US cohort of IBS patients and controls, we observed univariate associations of IBS with IL23R [rs11465804 (T/g), P = 0.006], positive regulatory domain I-binding factor [PRDM1, rs7746082 (G/c), P = 0.031], and immunity-related GTPase family M protein [IRGM, rs11747270 (A/g), P = 0.050] and D-IBS with Toll-like receptor 9 [TLR9, rs5743836 (A/g), P = 0.02]; the latter confirmed the observations in postinfectious IBS patients in Ontario, Canada (111). The association with IL23R and the other genetic variants was not significant, as P = 0.0015 was required by Bonferroni's correction for 34 genes (22). Also, the association with IL23R was not observed in the Swedish cohort (125).
Univariate associations (not corrected for multiple comparisons) of inflammation susceptibility loci with the intermediate phenotype (quantitative trait) of scintigraphic colonic transit were also reported in the Mayo Clinic study of 172 IBS patients (22). On the basis of the dominant inheritance model, rs7746082 (PRDM1, P = 0.01) is associated with colonic transit at 24 and 48 h, rs5743836 (TLR9, P = 0.01) with colonic transit at 48 h, and rs7927894 [chromosome 11 open reading frame 30 (c11orf30), P = 0.007] and rs2872507 (ORMDL3, P = 0.014) with colonic transit at 24 h. The association with TNFSF15 and colonic transit was not significant, even at the univariate level (P = 0.174 and P = 0.256 for colonic transit at 24 and 48 h, respectively).
The biological plausibility of these associations is illustrated by the biological functions of the susceptibility genes. PRDM1 [also called B lymphocyte-induced maturation protein 1 (BLIMP-1)] is a zinc finger transcriptional repressor that silences β-interferon gene expression, regulates plasma cell differentiation, and controls gene expression in T lymphocytes, macrophages, sebaceous gland, and skin epidermis (9). However, the equivalent molecule in zebrafish is essential for neural crest and sensory neuron specification (49), suggesting that the PRDM1 polymorphism may impact colorectal functions by influencing neural function. TLR9 is involved in barrier function and pattern-recognition receptors that elicit innate/adaptive immune response and inflammation, and c11orf30 is involved in epithelial immunity, growth, and/or differentiation (42, 77). The orosomucoid (ORM) gene family encodes transmembrane proteins localized in the endoplasmic reticulum; ORMDL3 is involved in control of sphingolipids (12), which are structural components of membranes, including those in neural tissues.
Neuropeptide S receptor 1.
The neuropeptide S (NPS) receptor 1 (NPSR1) gene on chromosome 7 is associated with asthma and inflammatory bowel disease (30). NPSR1 is expressed on the intestinal epithelium and is upregulated in inflammation; NPS-NPSR1 signaling induced increased expression of cholecystokinin, vasoactive intestinal peptide, peptide YY, and somatostatin in HEK-293 cells (23). In a study with 699 participants (approximately two-thirds patients and one-third healthy controls) of 18 NPSR1 polymorphisms (23) that span the gene, rs1419793 was significantly associated with colonic transit (P < 0.003, with false discovery rate correction). The mechanisms whereby NPSR1 SNPs might result in altered motor or secretory functions are unclear.
Genetic Predisposition to Bile Acid Malabsorption in IBS
Bile acids have potent secretory effects on the colonic mucosa. As a result, patients with predispositions to bile acid malabsorption (BAM), such as those who have undergone extensive ileal resection, commonly suffer from diarrhea. More recent data, however, have demonstrated that, in subjects and patients with intact small bowel, alterations in BAM may contribute to gut symptoms. For example, among patients with chronic functional diarrhea (which overlaps with D-IBS), a systematic review estimated that BAM, shown by 75Se-homocholic acid taurine scanning, affects ∼30% of patients (115). A surrogate serological test indicating increased bile acid synthesis is serum 7αC4, which was elevated in 20% of D-IBS patients in a small study (25). Elevation of fecal deoxycholate has been documented in patients with IBS (67). Walters et al. (113) provided evidence that impaired feedback inhibition of bile acid synthesis by the ileal hormone FGF-19 plays a role in bile acid-associated diarrhea. Conversely, patients with C-IBS and functional constipation had altered diurnal rhythm with no serum 7αC4 peak at lunchtime (1). These data might suggest that alterations in genes that regulate bile acid metabolism and feedback may contribute to abnormal bowel pattern and symptoms in IBS.
Genetic variation in bile acid metabolism.
Wong et al. (117) explored the hypothesis that variants of genes regulating hepatic bile acid synthesis play a role in D-IBS. In 435 IBS patients and 279 healthy subjects, we tested individual associations of 15 common SNPs from 7 genes critical to bile acid homeostasis. S rs17618244 [Arg728Gln in klothoβ (KLB)] was associated with colonic transit at 24 h. The G allele (Arg728) compared with the A allele (Gln728) was associated with accelerated colonic transit (P = 0.0007) in the overall cohort. This association was restricted to D-IBS (P = 0.0018); both were significant after Bonferroni's and false discovery rate corrections. Interaction tests of KLB rs17618244 with 3 nonsynonymous SNPs of FGF receptor 4 (FGFR4) revealed that rs1966265 (Val10Ile) and rs351855 (Gly388Arg) modulate the association of rs1768244 with colonic transit in D-IBS (P = 0.0025 and P = 0.0023, respectively). The associated SNP in KLB (rs17618244) was functionally significant. KLB Arg728 significantly reduced protein stability compared with KLB Gln728 (117).
The diminished stability of the KLB protein resulting from KLB Arg728 (G allele) is expected to weaken the FGFR4-KLB receptor complex on the hepatocyte plasma membrane, leading to decreased negative feedback by endogenous FGF-19, increased hepatocyte CYP7A1 expression, and increased hepatic bile acid synthesis. Excess bile acid reaching the small bowel may overwhelm the ileal capacity for bile acid absorption, spill over into the colonic lumen, and accelerate colonic transit by stimulating motility and secretion (5, 80).
Genetic variation in the bile acid receptor TGR5.
The membrane-bound bile acid receptor TGR5 is located on myenteric, cholinergic, and nitrergic neurons in colon and proximal small intestine. In humans, TGR5 is colocalized with the CFTR in gallbladder epithelium, and stimulation of TGR5 in gallbladder cells activates CFTR (61). In a study of 230 healthy controls and 414 patients with lower functional GI disorders, we tested the association between TGR5 SNP rs11554825 (minor allele frequency of 41%), located in the untranslated TGR5 exon 1, which is associated with reduced expression of TGR5 mRNA in a lymphoblastoid cell line (51) with symptom phenotype, and the intermediate phenotypes of small bowel and colonic transit by radioscintigraphy, which was available in 213 people (26). There was no significant association with symptom phenotype. We observed a potential association with overall small bowel (colonic filling at 6 h, P = 0.061) and colonic transit at 24 h (P = 0.083). The association of the SNP with colonic filling at 6 h in the D-IBS subgroup (P = 0.017) indicated an average 50% faster small bowel transit in the TC/CC than the TT subgroup. No association was observed in D-IBS patients for colonic transit at 24 h (26). Further studies are required to characterize the potential role of the bile acid receptor TGR5 and genetic variation in TGR5 in the mechanism of D-IBS.
Pharmacogenetics in IBS
In addition to understanding the pathophysiology of IBS, insight into pharmacogenetics and, specifically, the manner in which genetic abnormalities influence drug activity, binding, and metabolism has become central to proposing control mechanisms in IBS and may impact the management of individual IBS patients.
Drug metabolism and pharmacogenetics.
Overall effects of medications result from metabolism, disposition, and effects of the agent. The enzymatic metabolism of drugs involves modifications of functional groups (phase I reactions, such as oxidation, dehydrogenation, and esterification) or conjugation with endogenous substituents (phase II reactions) (37). The commonest and most relevant drug modifications in IBS result from cytochrome P-450 (CYP) 2D6 (CYP2D6) metabolism, which is an example of phase I drug metabolism. There are distinct geographic variations of the CYP genes, suggesting that population substructure can strongly affect the variation in pharmacogenetic loci (102).
The number of functional alleles (>3, 2, 1, and 0) determines whether CYP2D6 metabolism is ultrarapid, extensive, intermediate, or poor. About 1% of Asians and 5–10% of Caucasians are poor metabolizers (120). Gene multiplication may result in ≥3 functional alleles, and ethnic groups with >10 functional alleles (29) are infrequent among Northern Europeans but frequent (as high as 29%) in East African populations (2). The most common nonfunctional alleles are CYP2D6*3, CYP2D6*4, CYP2D6*5, and CYP2D6*6, which constitute ∼98% of nonfunctional alleles in Caucasians (44).
CYP2D6 metabolism impacts the extensively used tricyclic antidepressants and selective serotonin reuptake inhibitors in functional GI disorders and visceral hypersensitivity (39). Although not yet fully appreciated in IBS practice, a multitude of drugs are metabolized by CYP2D6 and many drugs that inhibit or activate CYP2D6, setting up the possibility of significant drug interactions. In some ethnic groups, it may be advisable to use CYP2D6 testing if antidepressants are being prescribed (123).
The second category of pharmacogenetic modulation of drug effects in IBS reflects variations in receptors, transporters, or function of rate-limiting enzymes.
Serotonergic pharmacogenetics.
The most informative studies of pharmacogenetics in IBS revolve around 5-HTTLPR genetics and the efficacy of alosetron in normalizing colonic transit in D-IBS (18) and the efficacy of tegaserod in the treatment of bowel dysfunction in C-IBS (73). Thus the 5-HT3 antagonist alosetron is more effective when the 5-HTTLPR LL genotype results in greater efficacy of the 5-HT reuptake process, presumably because less 5-HT needs to be competitively inhibited at the 5-HT3 receptor. Conversely, the 5-HT4 agonist tegaserod results in lower efficacy in carriers of the LL genotype (73), since there is less endogenous 5-HT to complement the effects of the exogenous tegaserod in activating the 5-HT4 receptor.
Adrenergic pharmacogenetics.
In pharmacodynamic studies of low doses of the α2-adrenergic agonist clonidine, there were significant associations between post-clonidine responses and α2A 1291C>G SNPs for gastric accommodation and rectal sensations of gas and urgency (20).
Bile acids and pharmacogenetics in C-IBS and D-IBS.
In addition to the observations that genetic variations in KLB and, possibly, FGFR4 are associated with accelerated colonic transit in patients with D-IBS (see Fig. 2 in Ref. 117), variations in the same genes influence the colonic transit response to chenodeoxycholic acid in C-IBS (94) and to colesevelam in D-IBS patients (118).
Dronabinol pharmacogenetics, CNR1, and colonic motility in nonconstipated IBS.
Wong et al. (116) found that, in 75 IBS patients (35 with C-IBS, 35 with D-IBS, and 5 with M-IBS), dronabinol (a synthetic tetrahydrocannabinol derivative, CB1 agonist) decreased fasting colonic phasic motility and increased colonic compliance, and the effects were greatest in patients with D-IBS or M-IBS. CNR1 rs806378 (CC vs. CT/TT) appeared to affect fasting proximal colonic motility (prior to dronabinol treatment) in all patients with IBS (P = 0.075), and dronabinol affected fasting proximal colonic motility in patients with D-IBS or M-IBS with the variant FAAH rs324420 CA/AA (P = 0.013).
In a second study of 36 D-IBS patients (119), differential treatment effects of 5 mg (but not 2.5 mg) of dronabinol compared with placebo were observed for CNR1 rs806378 (CC vs. CT/TT), with the CT/TT group demonstrating a trend to slowing of overall colonic transit (P = 0.13); this underpowered study requires replication, preferably with peripherally restricted and CB1-selective cannabinoid agents.
Conclusion
Within the past few decades, the combination of major breakthroughs in genetic analysis and understanding the pathophysiology of IBS has led to a more unified approach to understanding mechanisms and potential therapy in IBS. Specifically, the understanding of genetic variation, its influence on gut motility, secretion, sensation, and inflammation, and its application to IBS have provided further elucidation of a syndrome that is so little understood. The most convincing association of the IBS phenotype is with TNFSF15, which has been observed in three independent cohorts in Sweden, the United States, and England. Genes influencing neural, barrier, mast cell, or immune function are univariately associated with colonic transit in IBS, supporting the hypothesis that local immune activation and, possibly, altered barrier function, such as increased permeability, and reflex motor responses may contribute to development of IBS. Study of the genetic abnormalities associated with IBS may enhance pharmacological therapies by targeting key controlling receptors and regulators of gut function and by understanding the influence of variations in drug metabolism, potentially leading to individualized medicine in IBS.
GRANTS
This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-067071, DK-092179, and DK-079866 to M. Camilleri.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.C. and D.A.K. prepared the figures; M.C. and D.A.K. drafted the manuscript; M.C. and D.A.K. edited and revised the manuscript; M.C. and D.A.K. approved the final version of the manuscript.
REFERENCES
- 1.Abrahamsson H, Ostlund-Lindqvist AM, Nilsson R, Simrén M, Gillberg PG. Altered bile acid metabolism in patients with constipation-predominant irritable bowel syndrome and functional constipation. Scand J Gastroenterol 43: 1483–1488, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP 2D6 alleles. J Pharmacol Exp Ther 278: 441–446, 1996 [PubMed] [Google Scholar]
- 3.Andresen V, Camilleri M, Kim HJ, Stephens DA, Carlson PJ, Talley NJ, Saito YA, Urrutia R, Zinsmeister AR. Is there an association between GNβ3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology 130: 1985–1994, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology 130: 34–43, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Bampton PA, Dinning PG, Kennedy ML, Lubowski DZ, Cook IJ. The proximal colonic motor response to rectal mechanical and chemical stimulation. Am J Physiol Gastrointest Liver Physiol 282: G443–G449, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Bearcroft CP, Perrett D, Farthing MJ. Postprandial 5-hydroxytryptamine in diarrhea predominant irritable bowel syndrome: a pilot study. Gut 42: 42–46, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengtson MB, Ronning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: genes and environment. Gut 55: 1754–1759, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharucha AE, Camilleri M, Zinsmeister AR, Hanson RB. Adrenergic modulation of human colonic motor and sensory function. Am J Physiol Gastrointest Liver Physiol 273: G997–G1006, 1997 [DOI] [PubMed] [Google Scholar]
- 9.Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev 19: 379–385, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Brand S. Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut 58: 1152–1167, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Breijer MR, Mathis C, Schuurkes JA. 5-HT receptor types in the rat ileum longitudinal muscle: focus on 5-HT2 receptors mediating contraction. Neurogastroenterol Motil 9: 231–237, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camilleri CE, Carlson PJ, Camilleri M, Castillo EJ, Locke GR, 3rd, Geno DM, Stephens DA, Zinsmeister AR, Urrutia R. A study of candidate genotypes associated with dyspepsia in a US community. Am J Gastroenterol 101: 581–592, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Camilleri M. Pharmacology of the new treatments for lower gastrointestinal motility disorders and IBS. Clin Pharmacol Ther 91: 44–59, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Camilleri M. LX-1031, a tryptophan 5-hydroxylase inhibitor, and its potential in chronic diarrhea associated with increased serotonin. Neurogastroenterol Motil 23: 193–200, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camilleri M. Non-malignant gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, Smyrk TC, Urrutia R, Aerssens J, Thielemans L, Göhlmann H, van den Wyngaert I, Coulie B. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology 132: 17–25, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilleri M, Atanasova E, Carlson PJ, Ahmad U, Kim HJ, Viramontes BE, McKinzie S, Urrutia R. Serotonin transporter polymorphism pharmacogenetics in diarrhea-predominant irritable bowel syndrome. Gastroenterology 123: 425–432, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Camilleri M, Busciglio I, Carlson P, McKinzie S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Candidate genes and sensory functions in health and irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 295: G219–G225, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camilleri M, Busciglio I, Carlson P, McKinzie S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Pharmacogenetics of low dose clonidine in irritable bowel syndrome. Neurogastroenterol Motil 21: 399–410, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camilleri M, Carlson P, McKinzie S, Grudell A, Busciglio I, Burton D, Baxter K, Ryks M, Zinsmeister AR. Genetic variation in endocannabinoid metabolism, gastrointestinal motility, and sensation. Am J Physiol Gastrointest Liver Physiol 294: G13–G19, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Camilleri M, Carlson P, McKinzie S, Zucchelli M, D'Amato M, Busciglio I, Burton D, Zinsmeister AR. Genetic susceptibility to inflammation and colonic transit in lower functional gastrointestinal disorders: preliminary analysis. Neurogastroenterol Motil 23: 935–943, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camilleri M, Carlson P, Zinsmeister AR, McKinzie S, Busciglio I, Burton D, Zucchelli M, D'Amato M. Neuropeptide S receptor induces neuropeptide expression and associates with intermediate phenotypes of functional gastrointestinal disorders. Gastroenterology 138: 98–107, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camilleri M, Chang L. Challenges to the therapeutic pipeline for IBS: end points and regulatory hurdles. Gastroenterology 135: 1877–1891, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camilleri M, Nadeau A, Tremaine WJ, Lamsam J, Burton D, Odunsi S, Sweetser S, Singh R. Measurement of serum 7α-hydroxy-4-cholesten-3-one (or 7α4), a surrogate test for bile acid malabsorption in health, ileal disease and irritable bowel syndrome using liquid chromatography-tandem mass spectrometry. Neurogastroenterol Motil 21: 734–e43, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camilleri M, Vazquez-Roque MI, Carlson P, Burton D, Wong BS, Zinsmeister AR. Association of bile acid receptor TGR5 variation and transit in health and lower functional gastrointestinal disorders. Neurogastroenterol Motil 23: 995–e458, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang KP, Gerber AL, Sipe JC, Cravatt BF. Reduced cellular expression and activity of the P129T mutant of human fatty acid amide hydrolase: evidence for a link between defects in the endocannabinoid system and problem drug use. Hum Mol Genet 13: 2113–2119, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Coates MD. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology 126: 1657–1664, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Dalén P, Dahl ML, Bernal Ruiz ML, Nordin J, Bertilsson L. 10-Hydroxylation of nortriptyline in Caucasians with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin Pharmacol Ther 63: 444–452, 1998 [DOI] [PubMed] [Google Scholar]
- 30.D'Amato M, Bruce S, Bresso F, Zucchelli M, Ezer S, Pulkkinen V, Lindgren C, Astegiano M, Rizzetto M, Gionchetti P, Riegler G, Sostegni R, Daperno M, D'Alfonso S, Momigliano-Richiardi P, Torkvist L, Puolakkainen P, Lappalainen M, Paavola-Sakki P, Halme L, Farkkila M, Turunen U, Kontula K, Lofberg R, Pettersson S, Kere J. Neuropeptide S receptor 1 gene polymorphism is associated with susceptibility to inflammatory bowel disease. Gastroenterology 133: 808–817, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Diatchenko L, Nackley AG, Slade GD, Bhalang K, Belfer I, Max MB, Goldman D, Maixner W. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain 125: 216–224, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Elsenbruch S, Orr WC. Diarrhea- and constipation-predominant IBS patients differ in postprandial autonomic and cortisol responses. Am J Gastroenterol 96: 460–466, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Enoch MA, Goldman D, Barnett R, Sher L, Mazzanti CM, Rosenthal NE. Association between seasonal affective disorder and the 5-HT2A promoter polymorphism, −1438G/A. Mol Psychiatry 4: 89–92, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Esfandyari T, Camilleri M, Busciglio I, Burton D, Baxter K, Zinsmeister AR. Effects of a cannabinoid receptor agonist on colonic motor and sensory functions in humans: a randomized, placebo-controlled study. Am J Physiol Gastrointest Liver Physiol 293: G137–G145, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Esfandyari T, Camilleri M, Ferber I, Burton D, Baxter K, Zinsmeister AR. Effect of a cannabinoid agonist on gastrointestinal transit and postprandial satiation in healthy human subjects: a randomized, placebo-controlled study. Neurogastroenterol Motil 18: 831–838, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Eskdale J, Gallagher G, Verweij C, Keijsers V, Westendorp R, Huizinga T. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA 95: 9465–9470, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286: 487–491, 1999 [DOI] [PubMed] [Google Scholar]
- 38.Ford AC, Forman D, Bailey AG, Axon AT, Moayyedi P. Irritable bowel syndrome: a 10-year natural history of symptoms and factors that influence consultation behavior. Am J Gastroenterol 103: 1229–1239, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Ford AC, Talley NJ, Schoenfeld PS, Quigley EM, Moayyedi P. Efficacy of antidepressants and psychological therapies in irritable bowel syndrome: systematic review and meta-analysis. Gut 58: 367–378, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Fukudo S, Kanazawa M, Mizuno T, Hamaguchi T, Kano M, Watanabe S, Sagami Y, Shoji T, Endo Y, Hongo M, Itoyama Y, Yanai K, Tashiro M, Aoki M. Impact of serotonin transporter gene polymorphism on brain activation by colorectal distention. Neuroimage 47: 946–951, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology 132: 397–414, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Ghadimi D, Vrese M, Heller KJ, Schrezenmeir J. Effect of natural commensal-origin DNA on Toll-like receptor 9 (TLR9) signaling cascade, chemokine IL-8 expression, and barrier integrity of polarized intestinal epithelial cells. Inflamm Bowel Dis 16: 410–427, 2010 [DOI] [PubMed] [Google Scholar]
- 43.Ghoshal UC, Abraham P, Bhatt C, Choudhuri G, Bhatia SJ, Shenoy KT, Banka NH, Bose K, Bohidar NP, Chakravartty K, Shekhar NC, Desai N, Dutta U, Das G, Dutta S, Dixit VK, Goswami BD, Jain RK, Jain S, Jayanthi V, Kochhar R, Kumar A, Makharia G, Mukewar SV, Mohan Prasad VG, Mohanty A, Mohan AT, Sathyaprakash BS, Prabhakar B, Philip M, Veerraju EP, Ray G, Rai RR, Seth AK, Sachdeva A, Singh SP, Sood A, Thomas V, Tiwari S, Tandan M, Upadhyay R, Vij JC. Epidemiological and clinical profile of irritable bowel syndrome in India: report of the Indian Society of Gastroenterology Task Force. Indian J Gastroenterol 27: 22–28, 2008 [PubMed] [Google Scholar]
- 44.Givens RC, Watkins PB. Pharmacogenetics and clinical gastroenterology. Gastroenterology 125: 240–248, 2003 [DOI] [PubMed] [Google Scholar]
- 45.Gonsalkorale WM, Perrey C, Pravica V, Whorwell PJ, Hutchinson IV. Interleukin 10 genotypes in irritable bowel syndrome: evidence for an inflammatory component? Gut 52: 91–93, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160: 636–645, 2003 [DOI] [PubMed] [Google Scholar]
- 47.Gulewitsch MD, Enck P, Hautzinger M, Schlarb AA. Irritable bowel syndrome symptoms among German students: prevalence, characteristics, and associations to somatic complaints, sleep, quality of life, and childhood abdominal pain. Eur J Gastroenterol Hepatol 23: 311–316, 2011 [DOI] [PubMed] [Google Scholar]
- 48.Gupta V, Sheffield D, Verne GN. Evidence for autonomic dysregulation in the irritable bowel syndrome. Dig Dis Sci 47: 1716–1722, 2002 [DOI] [PubMed] [Google Scholar]
- 49.Hernandez-Lagunas L, Choi IF, Kaji T, Simpson P, Hershey C, Zhou Y, Zon L, Mercola M, Artinger KB. Zebrafish narrow minded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev Biol 278: 347–357, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holtmann G, Siffert W, Haag S, Mueller N, Langkafel M, Senf W, Zotz R, Talley NJ. G protein β3-subunit 825 CC genotype is associated with unexplained (functional) dyspepsia. Gastroenterology 126: 971–979, 2004 [DOI] [PubMed] [Google Scholar]
- 51.Hov JR, Keitel V, Laerdahl JK, Spomer L, Ellinghaus E, ElSharawy A, Melum E, Boberg KM, Manke T, Balschun T, Schramm C, Bergquist A, Weismüller T, Gotthardt D, Rust C, Henckaerts L, Onnie CM, Weersma RK, Sterneck M, Teufel A, Runz H, Stiehl A, Ponsioen CY, Wijmenga C, Vatn MH, Study Group IBSEN, Stokkers PC, Vermeire S, Mathew CG, Lie BA, Beuers U, Manns MP, Schreiber S, Schrumpf E, Häussinger D, Franke A, Karlsen TH. Mutational characterization of the bile acid receptor TGR5 in primary sclerosing cholangitis. PLos One 5: e12403, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Center for Biotechnology Information dbSNP Short Genetic Variations (Online). http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=25531 [2011].
- 53.Huang GH, Hsieh CC, Chen CH, Chen WJ. Statistical validation of endophenotypes using a surrogate endpoint analytic analogue. Genet Epidemiol 33: 549–558, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Hughes PA, Brierley SM, Martin CM, Brookes SJ, Linden DR, Blackshaw LA. Post-inflammatory colonic afferent sensitization: different subtypes, different pathways, and different time-courses. Gut 58: 1333–1341, 2009 [DOI] [PubMed] [Google Scholar]
- 55.Izzo AA, Camilleri M. Emerging role of cannabinoids in gastrointestinal and liver diseases: basic and clinical aspects. Gut 57: 1140–1155, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Jarrett ME, Kohen R, Cain KC, Burr RL, Poppe A, Navaja GP, Heitkemper MM. Relationship of SERT polymorphisms to depressive and anxiety symptoms in irritable bowel syndrome. Biol Res Nurs 9: 161–169, 2007 [DOI] [PubMed] [Google Scholar]
- 57.Kalantar JS, Locke GR, 3rd, Zinsmeister AR, Beighley CM, Talley NJ. Familial aggregation of irritable bowel syndrome: a prospective study. Gut 52: 1703–1707, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Büchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet 17: 2967–2977, 2008 [DOI] [PubMed] [Google Scholar]
- 59.Kapeller J, Möller D, Lasitschka F, Autschbach F, Hovius R, Rappold G, Brüss M, Gershon MD, Niesler B. Serotonin receptor diversity in the human colon: expression of serotonin type 3 receptor subunits 5-HT3C, 5-HT3D, and 5-HT3E. J Comp Neurol 519: 420–432, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karling P, Danielsson Å, Wikgren M, Söderström I, Del-Favero J, Adolfsson R, Norrback KF. The relationship between the Val158Met catechol-O-methyltransferase (COMT) polymorphism and irritable bowel syndrome. PLos One 6: e18035, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Häussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology 50: 861–870, 2009 [DOI] [PubMed] [Google Scholar]
- 62.Kilpatrick LA, Labus JS, Coveleskie K, Hammer C, Rappold G, Tillisch K, Bueller JA, Suyenobu B, Jarcho JM, McRoberts JA, Niesler B, Mayer EA. The HTR3A polymorphism c.−42C>T is associated with amygdala responsiveness in patients with irritable bowel syndrome. Gastroenterology 140: 1943–1951, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim DY, Camilleri M. Serotonin: a mediator of the brain-gut connection. Am J Gastroenterol 95: 2698–2709, 2000 [DOI] [PubMed] [Google Scholar]
- 64.Kim HJ, Camilleri M, Carlson PJ, Cremonini F, Ferber I, Stephens D, McKinzie S, Zinsmeister AR, Urrutia R. Association of distinct α2-adrenoceptor and serotonin-transporter polymorphisms associated with constipation and somatic symptoms in functional gastrointestinal disorders. Gut 53: 829–837, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kohen R, Jarrett ME, Cain KC, Jun SE, Navaja GP, Symonds S, Heitkemper MM. The serotonin transporter polymorphism rs25531 is associated with irritable bowel syndrome. Dig Dis Sci 54: 2663–2670, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohli U, Muszkat M, Sofowora GG, Harris PA, Friedman EA, Dupont WD, Scheinin M, Wood AJ, Stein CM, Kurnik D. Effects of variation in the human α2A- and α2C-adrenoceptor genes on cognitive tasks and pain perception. Eur J Pain 14: 154–159, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 10: 4208–4218, 2011 [DOI] [PubMed] [Google Scholar]
- 68.Lee HJ, Lee SY, Choi JE, Kim JH, Sung IK, Park HS, Jin CJ. G protein β3-subunit, interleukin-10, and tumor necrosis factor-α gene polymorphisms in Koreans with irritable bowel syndrome. Neurogastroenterol Motil 22: 758–763, 2010 [DOI] [PubMed] [Google Scholar]
- 69.Lee KJ, Kim YB, Kim JH, Kwon HC, Kim DK, Cho SW. The alteration of enterochromaffin cell, mast cell, and lamina propria T lymphocyte numbers in irritable bowel syndrome and its relationship with psychological factors. J Gastroenterol Hepatol 23: 1689–1694, 2008 [DOI] [PubMed] [Google Scholar]
- 70.Lembo A, Zaman M, Jones M, Talley NJ. Influence of genetics on irritable bowel syndrome, gastro-oesophageal reflux and dyspepsia: a twin study. Aliment Pharmacol Ther 25: 1343–1350, 2007 [DOI] [PubMed] [Google Scholar]
- 71.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Müller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274: 1527–1531, 1996 [DOI] [PubMed] [Google Scholar]
- 72.Levy RL, Jones KR, Whitehead WE, Feld SI, Talley NJ, Corey LA. Irritable bowel syndrome in twins: heredity and social learning both contribute to etiology. Gastroenterology 121: 799–804, 2001 [DOI] [PubMed] [Google Scholar]
- 73.Li Y, Nie Y, Xie J, Tang W, Liang P, Sha W, Yang H, Zhou Y. The association of serotonin transporter genetic polymorphisms and irritable bowel syndrome and its influence on tegaserod treatment in Chinese patients. Dig Dis Sci 52: 2942–2949, 2000 [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Wei X, Zhao C, Hu S, Duan J, Ju G, Wong-Riley MT, Liu Y. 5-HT induces enhanced phrenic nerve activity via 5-HT2A receptor/PKC mechanism in anesthetized rats. Eur J Pharmacol 657: 67–75, 2011 [DOI] [PubMed] [Google Scholar]
- 75.Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melén K, Julkunen I, Taskinen J. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry 34: 4202–4210, 1995 [DOI] [PubMed] [Google Scholar]
- 76.MacKenzie A, Quinn J. A serotonin transporter gene intron 2 polymorphic region, correlated with affective disorders, has allele-dependent differential enhancer-like properties in the mouse embryo. Proc Natl Acad Sci USA 96: 15251–15255, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Magnúsdóttir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci USA 104: 14988–14993, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Markoutsaki T, Karantanos T, Gazouli M, Anagnou NP, Karamanolis DG. 5-HT2A receptor gene polymorphisms and irritable bowel syndrome. J Clin Gastroenterol 45: 514–517, 2011 [DOI] [PubMed] [Google Scholar]
- 79.Marshall JK, Thabane M, Garg AX, Clark WF, Salvadori M, Collins SM, Walkerton Health Study Investigators Incidence and epidemiology of irritable bowel syndrome after a large waterborne outbreak of bacterial dysentery. Gastroenterology 131: 445–450, 2006 [DOI] [PubMed] [Google Scholar]
- 80.Mekjian HS, Phillips SF, Hofmann AF. Colonic secretion of water and electrolytes induced by bile acids: perfusion studies in man. J Clin Invest 50: 1569–1577, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller LJ. G protein-coupled receptor structures, molecular associations, and modes of regulation. Ann NY Acad Sci 1144: 1–5, 2008 [DOI] [PubMed] [Google Scholar]
- 82.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in irritable bowel syndrome: a twin study. Am J Gastroenterol 100: 1340–1344, 2005 [DOI] [PubMed] [Google Scholar]
- 83.Morris-Yates A, Talley NJ, Boyce PM, Nandurkar S, Andrews G. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol 93: 1311–1317, 1998 [DOI] [PubMed] [Google Scholar]
- 84.Murphy DL, Lerner A, Rudnick G, Lesch KP. Serotonin transporter: gene, genetic disorders, pharmacogenetics. Mol Interv 4: 109–123, 2004 [DOI] [PubMed] [Google Scholar]
- 85.Murphy GM, Jr, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry 160: 1830–1835, 2003 [DOI] [PubMed] [Google Scholar]
- 86.Nakamura T, Matsushita S, Nishiguchi N, Kimura M, Yoshino A, Higuchi S. Association of a polymorphism of the 5HT2A receptor gene promoter region with alcohol dependence. Mol Psychiatry 4: 85–88, 1999 [DOI] [PubMed] [Google Scholar]
- 87.Naliboff BD, Derbyshire SW, Munakata J, Berman S, Mandelkern M, Chang L, Mayer EA. Cerebral activation in patients with irritable bowel syndrome and control subjects during rectosigmoid stimulation. Psychosom Med 63: 365–375, 2001 [DOI] [PubMed] [Google Scholar]
- 88.Niesler B, Kapeller J, Fell C, Atkinson W, Möller D, Fischer C, Whorwell P, Houghton LA. 5-HTTLPR and STin2 polymorphisms in the serotonin transporter gene and irritable bowel syndrome: effect of bowel habit and sex. Eur J Gastroenterol Hepatol 22: 856–861, 2010 [DOI] [PubMed] [Google Scholar]
- 89.Ohman L, Simrén M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol 7: 163–173, 2010 [DOI] [PubMed] [Google Scholar]
- 90.Park JM, Choi MG, Cho YK, Lee IS, Kim SW, Choi KY, Chung IS. Cannabinoid receptor 1 gene polymorphism and irritable bowel syndrome in the Korean population: a hypothesis-generating study. J Clin Gastroenterol 45: 45–49, 2011 [DOI] [PubMed] [Google Scholar]
- 91.Parsons MJ, D'Souza UM, Arranz MJ, Kerwin RW, Makoff AJ. The −1438A/G polymorphism in the 5-hydroxytryptamine type 2A receptor gene affects promoter activity. Biol Psychiatry 56: 406–410, 2004 [DOI] [PubMed] [Google Scholar]
- 92.Pata C, Erdal E, Yazc K, Camdeviren H, Ozkaya M, Ulu O. Association of the −1438 G/A and 102 T/C polymorphism of the 5-HT2A receptor gene with irritable bowel syndrome 5-HT2A gene polymorphism in irritable bowel syndrome. J Clin Gastroenterol 38: 561–566, 2004 [DOI] [PubMed] [Google Scholar]
- 93.Pata C, Erdal ME, Derici E, Yazar A, Kanik A, Ulu O. Serotonin transporter gene polymorphism in irritable bowel syndrome. Am J Gastroenterol 97: 1780–1784, 2002 [DOI] [PubMed] [Google Scholar]
- 94.Rao AS, Wong BS, Camilleri M, Odunsi-Shiyanbade ST, McKinzie S, Ryks M, Burton D, Carlson P, Lamsam J, Singh R, Zinsmeister AR. Chenodeoxycholate in females with IBS-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology 139: 1549–1558, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosenstiel P, Sina C, Franke A, Schreiber S. Towards a molecular risk map—recent advances on the etiology of inflammatory bowel disease. Semin Immunol 21: 334–345, 2009 [DOI] [PubMed] [Google Scholar]
- 96.Saito YA, Locke GR, 3rd, Zimmerman JM, Holtmann G, Slusser JP, de Andrade M, Petersen GM, Talley NJ. A genetic association study of 5-HTT LPR and GNβ3 C825T polymorphisms with irritable bowel syndrome. Neurogastroenterol Motil 19: 465–470, 2007 [DOI] [PubMed] [Google Scholar]
- 97.Saito YA, Mitra N, Mayer EA. Genetic approaches to functional gastrointestinal disorders. Gastroenterology 138: 1276–1285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saito YA, Strege PR, Tester DJ, Locke GR, 3rd, Talley NJ, Bernard CE, Rae JL, Makielski JC, Ackerman MJ, Farrugia G. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol 296: G211–G218, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saito YA, Zimmerman JM, Harmsen WS, De Andrade M, Locke GR, 3rd, Petersen GM, Talley NJ. Irritable bowel syndrome aggregates strongly in families: a family-based case control study. Neurogastroenterol Motil 7: 790–797, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sikander A, Rana SV, Sharma SK, Sinha SK, Arora SK, Prasad KK, Singh K. Association of α2A-adrenergic receptor gene (ADRAlpha2A) polymorphism with irritable bowel syndrome, microscopic and ulcerative colitis. Clin Chim Acta 411: 59–63, 2010 [DOI] [PubMed] [Google Scholar]
- 101.Sikander A, Rana SV, Sinha SK, Prasad KK, Arora SK, Sharma SK, Singh K. Serotonin transporter promoter variant: analysis in Indian IBS patients and control population. J Clin Gastroenterol 43: 957–961, 2009 [DOI] [PubMed] [Google Scholar]
- 102.Sistonen J, Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 19: 170–179, 2009 [DOI] [PubMed] [Google Scholar]
- 103.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of β1- and α2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med 347: 1135–1142, 2002 [DOI] [PubMed] [Google Scholar]
- 104.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology 136: 1979–1988, 2009 [DOI] [PubMed] [Google Scholar]
- 105.Swan C, Duroudier NN, Campbell E, Hastings M, Neal KR, Dukes GE, Whorwell PJ, Hall I, Spiller RC. Association of proinflammatory genetic polymorphisms with the irritable bowel syndrome (IBS): phenotype and genotype correlation (Abstract). Gastroenterology 140: S525, 2011 [Google Scholar]
- 106.Tillisch K, Mayer EA, Labus JS, Stains J, Chang L, Naliboff BD. Sex specific alterations in autonomic function among patients with irritable bowel syndrome. Gut 54: 1396–1401, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Törnblom H, Lindberg G, Nyberg B, Veress B. Full-thickness biopsy of the jejunum reveals inflammation and enteric neuropathy in IBS. Gastroenterology 123: 1972–1979, 2002 [DOI] [PubMed] [Google Scholar]
- 108.Truong TT, Kilpatrick L, Naliboff BD, Dandekar S, Papp JC, Jarcho J, Gilbert CD, Licudine A, Craske M, Ornitz EM, Mayer EA, Lin Chang L. COMT genetic polymorphism is associated with alterations in attentional processing in patients with IBS and other functional pain syndromes (Abstract). Gastroenterology 136: A74, 2009 [Google Scholar]
- 109.van der Veek PP, van den Berg M, de Kroon YE, Verspaget HW, Masclee AA. Role of tumor necrosis factor-α and interleukin-10 gene polymorphisms in irritable bowel syndrome. Am J Gastroenterol 100: 2510–2516, 2005 [DOI] [PubMed] [Google Scholar]
- 110.Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther 26: 979–986, 2007 [DOI] [PubMed] [Google Scholar]
- 111.Villani AC, Lemire M, Thabane M, Belisle A, Geneau G, Garg AX, Clark WF, Moayyedi P, Collins SM, Franchimont D, Marshall JK. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 138: 1502–1513, 2010 [DOI] [PubMed] [Google Scholar]
- 112.Viramontes BE, Malcolm A, Camilleri M, Szarka LA, McKinzie S, Burton DD, Zinsmeister AR. Effects of α2-adrenergic agonist on gastrointestinal transit, colonic motility and sensation in humans. Am J Physiol Gastrointest Liver Physiol 281: G1468–G147613, 2001 [DOI] [PubMed] [Google Scholar]
- 113.Walters JR, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol 7: 1189–1194, 2009 [DOI] [PubMed] [Google Scholar]
- 114.Wang BM, Wang YM, Zhang WM, Zhang QY, Liu WT, Jiang K, Zhang J. Serotonin transporter gene polymorphism in irritable bowel syndrome. Zhong-Hua Nei Ke Za Zhi Chin J Intern Med 43: 439–441, 2004 [PubMed] [Google Scholar]
- 115.Wedlake L, A'Hern R, Russell D, Thomas K, Walters JR, Andreyev HJ. The prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 30: 707–717, 2009 [DOI] [PubMed] [Google Scholar]
- 116.Wong BS, Camilleri M, Busciglio I, Carlson P, Szarka LA, Burton D, Zinsmeister AR. Pharmacogenetic trial of a cannabinoid agonist shows reduced fasting colonic motility in patients with nonconstipated irritable bowel syndrome. Gastroenterology 141: 1638–1647, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wong BS, Camilleri M, Carlson PJ, Guicciardi ME, Burton D, McKinzie S, Rao AS, Zinsmeister AR, Gores GJ. A klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology 140: 1934–1942, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wong BS, Camilleri M, Carlson PJ, Odunsi-Shiyanbade S, McKinzie S, Busciglio I, Burton D, Zinsmeister AR. Pharmacogenetics of the effects of colesevelam on colonic transit in IBS with diarrhea. Dig Dis Sci. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong BS, Camilleri M, Eckert D, Carlson P, Ryks M, Burton D, Zinsmeister AR. Randomized pharmacodynamic and pharmacogenetic trial of dronabinol effects on colon transit in IBS-diarrhea. Neurogastroenterol Motil 24: 358, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xie HG, Kim RB, Wood AJJ, Stein CM. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol 41: 815–850, 2001 [DOI] [PubMed] [Google Scholar]
- 121.Yeo A, Boyd P, Lumsden S, Saunders T, Handley A, Stubbins M, Knaggs A, Asquith S, Taylor I, Bahari B, Crocker N, Rallan R, Varsani S, Montgomery D, Alpers DH, Dukes GE, Purvis I, Hicks GA. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut 53: 1452–1458, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang H, Massey D, Tremelling M, Parkes M. Genetics of inflammatory bowel disease: clues to pathogenesis. Br Med Bull 87: 17–30, 2008 [DOI] [PubMed] [Google Scholar]
- 123.Zhang JP, Malhotra AK. Pharmacogenetics and antipsychotics: therapeutic efficacy and side effects prediction. Expert Opin Drug Metab Toxicol 7: 9–37, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, Koeppe RA, Stohler CS, Goldman D. COMT Val158Met genotype affects μ-opioid neurotransmitter responses to a pain stressor. Science 299: 1240–1243, 2003 [DOI] [PubMed] [Google Scholar]
- 125.Zucchelli M, Camilleri M, Nixon Andreasson A, Bresso F, Dlugosz A, Halfvarson J, Torkvist L, Schmidt PT, Karling P, Ohlsson B, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister A, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 60:1671–1677, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]