Abstract
The regulation of cardiomyocyte hypertrophy is a complex interplay among many known and unknown processes. One specific pathway involves the phosphatase calcineurin, which regulates nuclear translocation of the essential cardiac hypertrophy transcription factor, nuclear factor of activated T-cells (NFAT). Although metabolic dysregulation is frequently described during cardiac hypertrophy, limited insights exist regarding various accessory pathways. One metabolically derived signal, beta-O-linked N-acetylglucosamine (O-GlcNAc), has emerged as a highly dynamic posttranslational modification of serine and threonine residues regulating physiological and stress processes. Given the metabolic dysregulation during hypertrophy, we hypothesized that NFAT activation is dependent on O-GlcNAc signaling. Pressure overload-induced hypertrophy (via transverse aortic constriction) in mice or treatment of neonatal rat cardiac myocytes with phenylephrine significantly enhanced global O-GlcNAc signaling. NFAT-luciferase reporter activity revealed O-GlcNAc-dependent NFAT activation during hypertrophy. Reversal of enhanced O-GlcNAc signaling blunted cardiomyocyte NFAT-induced changes during hypertrophy. Taken together, these results demonstrate a critical role of O-GlcNAc signaling in NFAT activation during hypertrophy and provide evidence that O-GlcNAc signaling is coordinated with the onset and progression of cardiac hypertrophy. This represents a potentially significant and novel mechanism of cardiac hypertrophy, which may be of particular interest in future in vivo studies of hypertrophy.
Keywords: nuclear factor of activated t cells, transverse aortic constriction
cardiac hypertrophy reflects a compensatory response to pressure or volume overload, sarcomeric abnormalities, or loss of contractile ability through partial death of the myocardium (6, 14). The development of pathological pressure overload-induced hypertrophy has deleterious consequences on the heart and often progresses to decompensation and overt heart failure, the leading cause of death in the industrialized world (30). The molecular mechanisms contributing to cardiac hypertrophy are common to several stressors (25), involve the activation of the nuclear factor of activated T-cells (NFAT), and induce the fetal gene program (17). Intracellular Ca2+ participates as an obligatory signaling molecule during hypertrophy after an increase in cardiac workload or in response to myocyte stretch (2). Elevated Ca2+ levels activate the phosphatase calcineurin culminating in NFAT dephosphorylation and nuclear translocation of NFAT (17). The final macromolecular response involves an increase in cell size and protein synthesis (25) and is accompanied by an eventual energetic defect (7, 14, 30).
Although several groups have investigated the metabolic defects accompanying cardiac hypertrophy, little attention has been given to the contribution of accessory pathways of glucose metabolism, such as the hexosamine biosynthetic pathway (see Refs. 20, 21 for review). The hexosamine biosynthetic pathway converts cellular glucose to uridine diphosphate-β-N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc serves as the monosaccharide donor for the enzyme uridine diphospho- (OGT), which catalyzes the addition of the sugar to serine/threonine residues on proteins. This sugar modification remains on the target protein until removed by the enzyme β-N-acetylglucosaminidase (OGA) (35). O-GlcNAcylation modulates the function of many target proteins (9). One remarkable and relevant example is the activation of the transcription factor NFAT in T-cells (8). In fact, the O-GlcNAcylating enzyme, OGT, may interact with NFAT and directly modify this protein and activate immune response genes in T-cells (8). Yet, there is no known relationship between NFAT activation and O-GlcNAc signaling during cardiac hypertrophy. Here, we demonstrate that O-GlcNAc signaling is essential for NFAT transcriptional activity during cardiac hypertrophy.
MATERIALS AND METHODS
Transverse aortic constriction surgery.
The transverse aortic constriction (TAC) surgery was conducted in 3-mo-old, male C57BL/6J mice by constriction of the transverse aorta as described (1) and in accordance with the University of Louisville Animal Care and Use Committee. Briefly, C57BL/6J mice were anesthetized with ketamine (50 mg/kg, intraperitoneal) and pentobarbital (50 mg/kg, intraperitoneal), orally intubated with a polyethylene-60 tubing, and ventilated (Harvard Apparatus Rodent Ventilator, model 845) with oxygen supplementation. Mice were maintained under anesthesia with an isoflurane vaporizer (1%) supplemented with 100% oxygen. Tidal volumes and breathing rates were set based on standard allometric equations. The aorta was visualized through an intercostal incision. A 7-0 nylon suture was looped around the aorta between the brachiocephalic and left common carotid arteries. The suture was tied around a 27-gauge needle (put adjacent to the aorta) to constrict the aorta to a reproducible diameter. Then the needle was removed, leaving a discrete region of stenosis (TAC mice), and the chest was closed. Mice were extubated upon recovery of spontaneous breathing and were allowed to recover in warm, clean cages supplemented with oxygen. Analgesia (ketoprofen, 5 mg/kg, subcutaneous) was given before mice recovered from anesthesia (and by 24 and 48 h later). Sham age-matched mice were subjected to the same procedure except the suture was only passed underneath the aorta and not tied off. At the end of the study, TAC or sham-operated mice were euthanized, and the hearts were rapidly excised and weighed. The hearts were then immediately frozen in liquid nitrogen and stored at −80°C or perfused and fixed for immunohistochemical analysis.
Neonatal rat cardiac myocyte isolation and culture.
Neonatal rat cardiac myocytes (NRCMs) were isolated from 1- to 2-day-old Sprague-Dawley rats and cultured according to a well characterized protocol (11, 19, 23, 30). Briefly, the first 4 days of DMEM culture medium contained the antimitotic BrdU (0.1 mM) to inhibit fibroblast growth in addition to 5% fetal bovine serum, penicillin/streptomycin, and vitamin B12. Cells were maintained at 37°C in the presence of 5% CO2 in a humidified incubator. Where indicated, cells were treated with 100 μM phenylephrine (Phe) in serum-free DMEM plus insulin (10 μg/ml) for 48 h. Where indicated, NRCMs were pre-incubated with 40 μM 6-diazo-5-oxonorleucine (DON; an inhibitor of glutamine:fructose-6-phosphate amidotransferase) for 24 h before any additional reagents.
Adenoviral infection.
Replication-deficient adenovirus was used to infect NRCMs as described previously (19, 23, 30). Functional expression was confirmed by appropriate immunoblot analysis or real-time PCR. Ad-GFP or Ad-null was used as a control virus. Cells were infected with Ad-OGA, Ad-OGT, Ad-Null, or Ad-GFP at a multiplicity of infection (MOI) of 100 (19). Adenoviruses were delivered to the cells for 2 h (NRCMs) or 6 h (H9c2), and then the medium was changed to fresh DMEM. The cells were infected with adenovirus 24 h before treatment with phenylephrine at the indicated concentration.
NFAT-tagged GFP nuclear translocation.
For the NFATc3-GFP nuclear translocation studies, the cells were infected with Ad-OGA, Ad-OGT, or Ad-null for 24 h. The cells were then exposed to NFAT-GFP expressing adenovirus (kindly donated by Dr. Jeffery Molkentin's laboratory) incubated for 24 h, serum starved for 6 h, and then imaged. The cells were treated with DON for the same period of time as the adenoviruses. The effect of calcium treatment was imaged after 90 min of Ca2+ (4 mM) in DMEM lacking FBS. The NRCMs were grown in 35-mm glass-bottom dishes, and images were acquired with a Nikon A1 confocal at ×60/1.4 magnification Plan Apochromat oil immersion objective (Nikon, Japan). NFATc3-GFP nuclear translocation was analyzed with NIS elements software (Nikon Instruments).
Stretch procedures.
H9c2 myoblasts (ATCC, catalog no. CRL1446) were grown in six-well flexible surface Flexcell culture plates coated with laminin. The myoblasts were then subjected to repetitive cycles of stretch and relaxation for 24 h (45 cycles/min) using a computer-driven, vacuum-operated instrument (Flexcell Strain Unit FX-4000 Tension Plus, Flexcell International). The elongation in the diameter of the flexible surface was maintained at 10–12%. The temperature was maintained at 37°C in a 5% CO2 and 95% O2 atmosphere. Flexcell culture plates exposed to the same conditions but not submitted to stretch and relaxation procedures served as the control.
Heart and NRCM lysates.
Following isolation and respective treatments, the NRCM cellular protein content was harvested using a cell scraper in buffer containing (in mM) 5 Hepes, 1 EDTA, 1 EGTA, 50 KCl, 200 mannitol, and 68 sucrose (pH = 7.4 with KOH). DTT (1 mM), protease inhibitor (0.0001%), Triton X-100 (0.4%), NP-40 (0.4%), sodium orthovanadate (1 mM), sodium fluoride (1 mM), alloxan (OGT inhibitor, 1 mM), and O-(2-acetamido-2-deoxy-d-glucopyranosylidenamino) N-phenylcarbamate (i.e., PUGNAc, which is an OGA inhibitor, 1 μM) were added to the buffer to avoid artificial O-GlcNAc addition or removal, respectively, to the proteins in vitro. Hearts were homogenized with buffer containing (in mM) 50 Tris·HCl (pH 7.4), 150 NaCl, 0.01 deoxycholic sodium salt, 1 EDTA, 1 sodium orthovanadate, 1 sodium fluoride, 0.001 PUGNAc, and 0.001 alloxan monohydrate. Protease inhibitor (556 μl/l; Sigma P8340) and NP-40 (10%) were freshly added to the buffer. Heart and NRCM lysates were sonicated twice at 4°C for 25 s each, with 30 min of time separating the sonications. After the second sonication, the lysates were centrifuged 15,000 g (NRCMs) or 12,500 g (hearts) at 4°C for 5 min. The protein content was determined and normalized using Bio-Rad protein assay (Bio-Rad), and bovine albumin served as a control. Heart and cell lysates were frozen in liquid nitrogen immediately and stored at −80°C until used.
Western blotting.
The proteins harvested from NRCM or whole hearts were submitted to electrophoresis in SDS-PAGE (4–10%) and transferred to nitrocellulose membrane. For the O-GlcNAc antibody samples, whole hearts were first precleared with sepharose G (GE Healthcare) to limit the interaction of the secondary antibody (anti-mouse) with endogenous immunoglobulins. The membrane blot was blocked (room temperature) using Tris-buffered saline pH 7.5 (TBS) containing nonfat milk (0.5%). After that, the blot was probed with primary antibody against O-GlcNAc: RL2 (1:1,000, Affinity Bioreagents) or CTD 110.6 (1:1,000, Covance), OGT (SQ-17, 1:2,000, Sigma-Aldrich), alpha-tubulin (1:2,000, Sigma-Aldrich) in TBS containing nonfat milk (1%). After overnight incubation at 4°C, the blot was washed in TBS containing Tween-20 (TBS-T; 0.1%). The blot was again blocked for 15 min in TBS-T plus nonfat milk (1%) and incubated with the horseradish peroxidase-labeled secondary antibody goat anti-mouse IgG-HRP (Santa Cruz Biotechnology), goat anti-mouse IgM-HRP (Santa Cruz Biotechnology), goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology) in dilutions from 1:2,000 to 1:4,000, depending on the antibody, for 1 h. After washing four times with TBS-T, the blot was detected with an enhanced chemiluminescent detection system (Pierce). Densitometry was executed using nonsaturated chemiluminescent membranes exposed and quantified using Fuji LAS-3000 bio-imaging analyzer. To confirm the linear range of the signal, multiple exposures from every experiment were performed. Levels of proteins in each lane were normalized to loading protein content (tubulin) or to Ponceau stain and expressed as relative to control (set as 100%).
NFAT-luciferase assay.
NRCMs were infected with adenovirus containing firefly luciferase under the transcriptional control of four repeats of the NFAT consensus sequence binding site ggaaaa (NFAT-Luciferase, 10 MOI; Vector Biolabs). For an adenoviral loading control, we treated cells with adenovirus to overexpress the enzyme beta-galactosidase (Ad-betagal, 10 MOI; Vector Biolabs). All other adenoviruses when used in the luciferase experiment were added at 80 MOI. The control adenovirus Ad-null was added to apply the same virus load per condition. The cells were treated with phenylephrine for 6 h without serum and subsequently lysed for 20 min at room temperature using passive lysis buffer (Promega), followed by centrifugation at 2,500 g for 2 min to sediment the debris. A total of 20 μl of the cell lysate was mixed in 100 μl of the luciferase assay solution (Promega). The relative luminescence was measured with a single-tube multimode reader from Turner Biosystems.
β-Galactosidase assay.
The normalization of the NFAT-Luciferase activity was performed by β-galactosidase assay in 10 μl of NRCMs lysate in 90 μl of buffer containing 2-nitrophenyl β-d-galactopyranoside (1 mg/ml), 2-mercaptoethanol (50 mM), magnesium chloride (1 mM), and sodium phosphate (200 mM, pH 7.5). All were all purchased from Sigma (St. Louis, MO). The plate was covered and incubated for 30 min at 37°C, and absorbance at 405 nm was determined with a Thermo Electro Multiscan Spectrum plate reader. β-Galactosidase activity was expressed as A405 U/mg total protein.
Subcellular fractionation assay.
NRCMs were fractionated using Thermo Scientific Subcellular Protein Fractionation Kit, according to the manufacturer's protocol.
Protein-to-DNA ratio.
Cells were washed with PBS, then 200 μl of perchloric acid (0.2 N) were added to each well. Plates were placed on a rocker for 5 min, after which cells were scraped and collected in 1-ml tubes. Samples were then centrifuged for 10 min at 10,000 g at 4°C. Samples were then incubated at 60°C with 30–40 μl of KOH for 20 min, and protein was analyzed using standard Bradford technique. DNA content was determined by using 1 mM Hoechst solution in Tris·NaCl. Diluted Hoechst solution (200 μl) was placed in each well on a 96-well plate along with 10–20 μl of cell homogenate. Fluorescence was measured at 350-nm excitation (slit 2.5) and 460-nm emission (slit 2.5) at 200 scan speed.
Tritiated leucine assay.
The rate of protein synthesis in NRCMs was determined by [3H]leucine incorporation. Briefly, NRCMs were infected with vehicle + Ad-null, phenylephrine + Ad-null, or phenylephrine + Ad-OGA for 48 h. In the last 8 h, the cells were incubated with [3H]leucine (5 μCi/ml). After incubation, cells were rinsed with PBS, and protein was harvested in 10% TCA. The precipitate was resuspended in 0.5 N NaOH and then measured by scintillation. Values were normalized to DNA concentration measured by Hoechst fluorescence using salmon sperm DNA as a standard.
Enzymatic labeling of O-GlcNAc-modified proteins.
O-GlcNAc modified proteins were labeled using Invitrogen's Click-iT enzymatic labeling kit according to manufacturer's instructions. Briefly, chloroform/methanol was added to the sample to precipitate the detergents followed by centrifugation at 14,000 g for 5 min at 4°C. The interface layer containing the protein precipitate was washed twice with methanol. The resulting pellet was allowed to dry for 15 min in the fume hood. The dried pellet was resuspended in 40 μl of SDS (1%) and HEPES buffer (20 nM, pH 7.9), boiled at 90°C for 5 min, vortexed briefly, and allowed to cool on ice for 3 min. Labeling buffer and MgCl2 were added followed by UDP-GalNAz (azide-modified UDP-N-aceltylgalactosamine) and mutant β-1-4-galactosyltransferase. The next day, the GalNAz-labeled O-GlcNAc-modified protein mixture was precipitated using the chloroform/methanol precipitation method and resuspended in 50 μl of buffer containing SDS (1%) and Tris (50 mM, pH 8). The azide-labeled proteins were tagged with a fluorescent dye by adding ×2 Click-iT (with TAMRA) reaction buffer. The mixture was vortexed for 5 s after the addition of each component. The mixture was then rotated for 1 h at 4°C for the conversion of the azide group to a stable triazole conjugate. DTT (25 mM) was added and incubated at 4°C for 15 min to stop the reaction. Precipitation was via the chloroform/methanol precipitation method. The dried-labeled protein sample was resuspended in SDS-PAGE buffer for electrophoresis.
Reverse transcriptase PCR and real-time PCR.
The total RNA from NRCMs or from hearts was extracted with Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA levels were quantified using the ratio of absorbance at 260–280 nm (A260/A280 ratio) with the NanoDrop 1000 Spectrophotometer (Thermo Scientific). To check for organic contaminants like phenol and other aromatic compounds (Trizol, for example), the total RNA was verified by the absorbance ratio of 260 to 230 nm (A260/A230). We limited the use of RNA to samples with 260/230 ratio >1.8. Total RNA (1 μg) was then subjected to reverse transcriptase in a 20-μl final volume reaction for 30 min to synthesize the cDNA using IScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The relative levels of mRNA transcripts were quantified by real-time PCR using SYBR Green (Bio-Rad). The data generated was normalized to 18s ribosomal RNA (rRNA) or β-actin threshold cycle (CT) values by using the ΔΔCT comparative method (13). rRNA was diluted 1:32 with nuclease-free water. In each well, the final concentration of all other primers was 0.1 μg/μl. The expected size of DNA for each primer was confirmed by electrophoresing the DNA product of the qRT-PCR in agarose gel (2%). The absence of extra bands was verified. All the primers were designed using Primer 3.0 software (25).
Statistical analysis.
Results are shown as means ± SE. The statistical analysis (GraphPad 5.0) was conducted using Student's t-test or one-way ANOVA followed by Newman-Keuls multiple comparison test when appropriate. Differences were considered statistically significant at P ≤ 0.05.
RESULTS
Hypertrophy reactivates fetal gene program concomitantly with O-GlcNAc signaling.
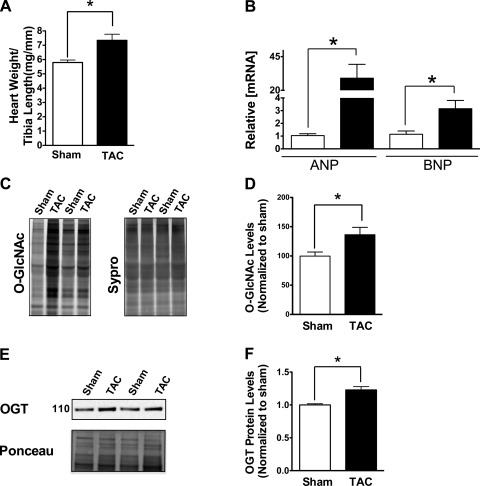
Several signaling pathways are implicated in the pathogenesis of hypertrophy (5). Here, we used the TAC model to induce cardiac hypertrophy. After 1 wk, hypertrophy was indicated by significantly elevated heart weight/tibia length (HW/TL) and ANP/BNP mRNA levels (Fig. 1, A and B, respectively). To determine whether O-GlcNAc signaling is involved in the development of hypertrophy in vivo, we analyzed total O-GlcNAc levels using a non-immune technique (Click chemistry-based approach) in hearts from sham or TAC mice and found an enhancement of O-GlcNAc signaling 1 wk after TAC (Fig. 1, C and D). These results were confirmed by evaluating the O-GlcNAc content of cardiac lysates by Western blot analyses (not shown). Interestingly, we also found an upregulation in OGT protein in TAC mice compared with sham (Fig. 1, E and F).
Fig. 1.
Cardiac hypertrophy stimulates O-GlcNAc signaling and reactivation of the fetal gene program. A: induction of hypertrophy after transverse aortic constriction (TAC), heart weight/tibia length (HW/TL) from sham and TAC-operated mice 1 wk after surgery. B: mRNA expression of ANP and BNP analyzed by real-time qPCR. C: enzymatically labeled O-GlcNAc proteins (TAMRA O-GlcNAc) show total O-GlcNAc from control hearts (sham operated) of hearts submitted to TAC for 1 wk. D: quantification of Fig. 1C. E: OGT protein is increased after 1 wk of TAC. Western blot for OGT protein and Ponceau loading control are shown. F: results are expressed as means ± SE, n ≥ 3/group. *Significant difference (P < 0.05).
Reversal of O-GlcNAc signaling reverses hypertrophic transcriptional reprogramming.
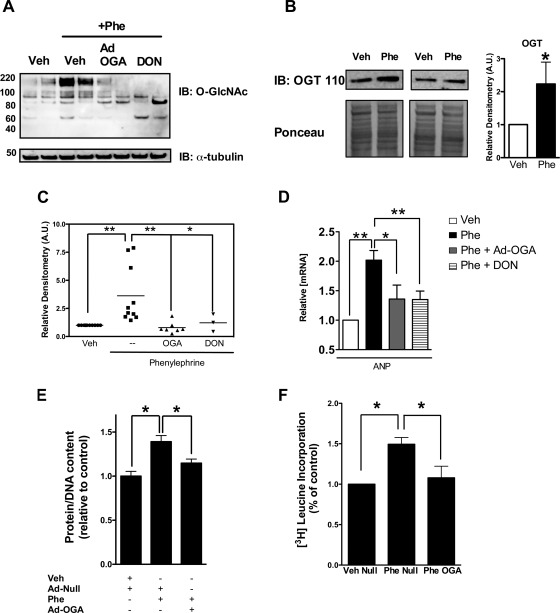
The observation that pressure overload (TAC) induced O-GlcNAc signaling prompted us to evaluate the effect of reduced O-GlcNAc modification during cardiac hypertrophy. To better understand this process, we exposed NRCMs to the alpha adrenergic receptor agonist phenylephrine, which produces cellular hypertrophy (1, 26) through activation of NFAT signaling and consequent resurrection of the fetal gene program (17). Similar to the in vivo hypertrophy model, we observed that O-GlcNAc signaling was upregulated by phenylephrine treatment of NRCMs (Fig. 2, A and C). Similarly, we found that the treatment of NRCMs with phenylephrine increased OGT protein (Fig. 2B) and significantly elevated ANP mRNA (Fig. 2D). Interestingly, exposing NRCMs to OGA adenovirus (Ad-OGA; 100 MOI) or inhibiting the rate-limiting enzyme (i.e., GFAT) of the hexosamine biosynthetic pathway with 6-diazo-5-oxo-l-norleucine (DON; 40 μM) significantly reversed the phenylephrine-induced increase in O-GlcNAc (Fig. 2, A and C) and minimized the induction of ANP during phenylephrine treatment (Fig. 2D). We also showed that O-GlcNAc is necessary for the increased protein production that accommodates hypertrophy (Fig. 2, E and F).
Fig. 2.
Modulation of O-GlcNAcylation reverses the changes in ANP in response to phenylephrine. A: effect of phenylephrine (Phe, 0.1 mM for 48 h) on total protein O-GlcNAcylation. The cardiomyocytes were exposed to phenylephrine either alone or in the presence of Ad-OGA (Ad-OGA; 100 MOI) or the GFAT inhibitor 6-diazo-5-oxonorleucine (DON; 0.04 mM), as indicated. B: Western blot showing OGT protein upregulation after 48 h of treatment with phenylephrine as in A. C: densitometric analysis of Western blot on A. D: OGA (Ad-OGA; 100 MOI) or 6-diazo-5-oxonorleucine (DON; 0.04 mM) treatment reverses the induction of ANP expression by phenylephrine. Ponceau stain or tubulin served as a loading control. The mRNA levels are expressed relative to 18S ribosomal RNA. E: increased total protein-to-DNA ratio due to phenylephrine treatment is significantly reduced with Ad-OGA treatment. F: increased de novo protein synthesis with phenylephrine treatment is significantly attenuated in the presence of Ad-OGA. Results are expressed as means ± SE, n ≥ 3 per group. Significant difference: *P < 0.05; **P < 0.01.
Inhibition of O-GlcNAc signaling blocks NFAT activation during hypertrophy.
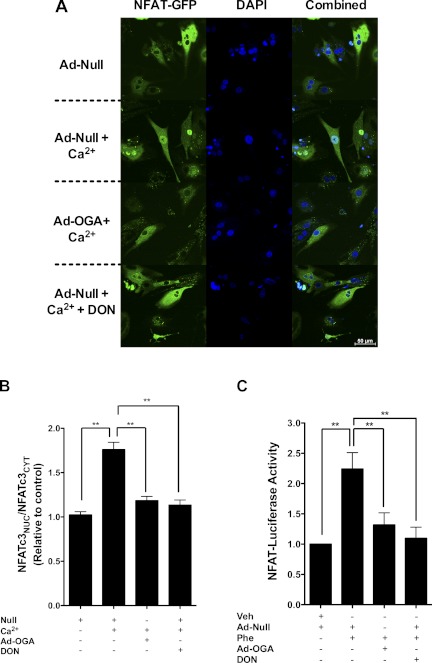
ANP mRNA expression is largely regulated by the activity of the hypertrophy transcription factor NFAT (17). To evaluate whether changes in O-GlcNAc signaling influenced NFAT activation during hypertrophy, we infected NRCMs with Ad-NFAT-luciferase reporter and exposed them to phenylephrine (Fig. 3C). Luciferase reporter activity was stimulated by phenylephrine and blunted by reversal of enhanced O-GlcNAc signaling using Ad-OGA (Fig. 3C). To confirm these findings, we used another adenoviral vector containing full-length NFATc3 fused to GFP (AdNFATc3-GFP), which indicates nuclear translocation of NFAT (i.e., another surrogate of activation). In Fig. 3, A and B, supraphysiologic calcium (4 mM) induced nuclear translocation of NFAT within 90 min. In parallel experiments, we co-infected AdNFATc3-GFP NRCMs with Ad-OGA (or Ad-null) and found that reduction in O-GlcNAc signaling blocked translocation of NFAT to the nucleus. DON, an inhibitor of the hexosamine biosynthetic pathway, exerted similar effects.
Fig. 3.
O-GlcNAcylation is essential for calcium-activated NFATc3 activation and nuclear translocation. A: NRCMs expressing NFATc3-GFP protein were co-infected with Ad-OGA or Ad-null in the presence or absence of the calcium. The HBP inhibitor DON was given to the cells 24 h before treatment with calcium. Ad-null was added to samples not containing any adenovirus treatment to serve as a control. DAPI was used to mark the nucleus. B: quantification data of NFATc3-GFP translocation. C: the effect of phenylephrine on Ad-NFAT uciferase reporter activity. Tubulin served as a loading control. The Ad-null was added to samples not containing any adenovirus treatment to serve as a control. Results were normalized to beta-galactosidase activity. Values are expressed as means ± SE, n ≥ 3 per group. **Significant difference (P < 0.01).
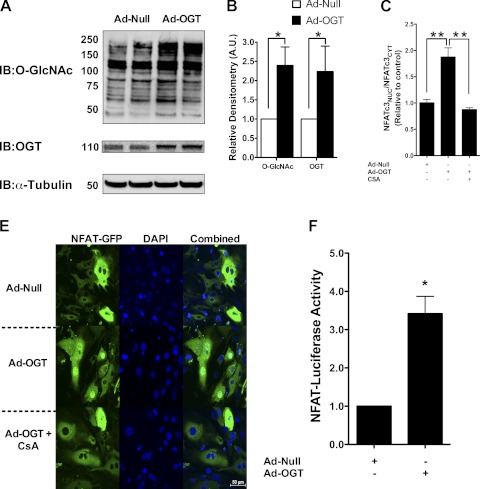
Such experiments indicate that enhanced O-GlcNAc signaling was necessary for hypertrophic reprogramming but did not answer the question of sufficiency. Next, we augmented O-GlcNAc signaling (in the absence of high calcium) by adenoviral overexpression of OGT (Ad-OGT; Fig. 4, A and B). Here, we found augmented NFAT activation (according to the Ad-NFAT-luciferase driven gene reporter) and nuclear translocation of NFAT (Fig. 4C). We also confirmed that cyclosporin A (CsA) could inhibit the Ad-OGT-induced nuclear translocation of NFAT.
Fig. 4.
Activation and nuclear translocation of NFATc3 by O-GlcNAcylation. A: effect of O-GlcNAcylating enzyme OGT (Ad-OGT, 100 MOI) on total protein O-GlcNAcylation. B: quantification of Fig. 5A. C: the nuclear-to-cytoplasmic ratio relative to control shows increased nuclear presence of NFATc3 with Ad-OGT. This elevation is abrogated with the addition of the calcineurin inhibitor cyclosporin A (CsA; 2.5 μM). D: confocal microscope images showing effect of Ad-OGT and Ad-OGT + CsA on NFAT translocation. E: Ad-NFAT Luciferase reporter activity in NRCMs in the presence of Ad-null and Ad-OGT. Luciferase data was normalized to β-galactosidase activity. Values are expressed as means ± SE, n ≥ 3 per group. *Significant difference (P < 0.05). **Significant difference (P < 0.01).
Hypertrophic NFAT activation requires O-GlcNAc signaling.
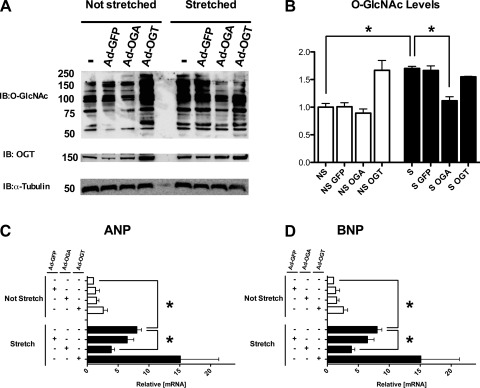
Both ANP and BNP promoters have binding sites for NFAT (17). We tested whether activation of NFAT by O-GlcNAc signaling during hypertrophy is of functional significance by focusing on the transcription of ANP and BNP (NFAT-dependent genes). We addressed this question by subjecting H9c2 cardiomyoblasts to stretch-induced hypertrophy (12). H9c2 cells express low levels of ANP and BNP under basal conditions, making them a suitable model to study the modulation of NFAT activation (at the level of ANP/BNP) by O-GlcNAc signaling, whereas ANP/BNP expression in NRCMs is relatively higher. Thus using H9c2 cells will address the potential influence of baseline elevations of ANP/BNP. As shown in Fig. 5, A and B, overexpression of OGT increased O-GlcNAc signaling, and, conversely, the overexpression of OGA reduced it. Stretching the H9c2 myoblasts for 24 h significantly augmented total O-GlcNAcylation, and such augmentation was sensitive to Ad-OGA. Both ANP and BNP mRNA were upregulated after stretch (Fig. 5, C and D). Strikingly, BNP mRNA was upregulated >100-fold during hypertrophy. Interestingly, the 1.8-kb BNP promoter has three binding sites for NFAT and is activated >100-fold in the presence of NFAT, GATA4, and calcineurin (17). In agreement with Fig. 4, Ad-OGT enhanced the levels of ANP and BNP in the absence of stretch. Conversely, Ad-OGT had no additive effects on these parameters when coupled with stretch (Fig. 5, A–D). Also, the control adenovirus (Ad-GFP) had no effect on any parameter analyzed (data not shown).
Fig. 5.
O-GlcNAc mediates the fetal gene program reactivation during stretch-induced hypertrophy. A: representative immunoblotting showing the effect of stretch (24 h) on total protein O-GlcNAcylation and OGT levels in H9c2 cells. The stretch was conducted, as described in materials and methods, alone or in the presence of Ad-OGA (100 MOI), Ad-OGT (100 MOI), or Ad-GFP (100 MOI). Tubulin serves as a loading control. B: quantification of blot in Fig. 5A. Mechanical stretch induces hypertrophy according to ANP (C) and BNP (D) levels. The mRNA levels are expressed relative to 18s ribosomal RNA. Results are expressed as means ± SE, n ≥ 4 per group. *Significant difference (P < 0.05).
To determine whether reducing O-GlcNAc signaling during hypertrophy is sufficient to reduce ANP/BNP upregulation, we overexpressed OGA and submitted the cells to stretch for 24 h. Ad-OGA prevented the increase in O-GlcNAc signaling and also minimized the reactivation of the fetal gene program (according to qPCR for ANP and BNP; Fig. 5) during stretch. These results confirm that O-GlcNAc signaling is enhanced during hypertrophy and that reducing O-GlcNAc signaling could block NFAT function during hypertrophy.
We also found that hypertrophic O-GlcNAc signaling coincides with a dysregulation of GLUT1 and GLUT4 expression, the latter of which is regulated by PGC-1α. TAC (in vivo) or phenylephrine treatment in cardiomyocytes increased the ratio of GLUT1:GLUT4 due to elevated GLUT1 expression and reduced GLUT4 expression. Interestingly, Ad-OGA, which reversed the hypertrophic transcriptional program in NRCMs, also blocked the increase in GLUT1:GLUT4 ratio in cardiomyocytes (data not shown). These findings confirm the observation that myocardial hypertrophy involves dysregulation of basal glucose uptake and utilization (18), wherein insulin-independent glucose transport (GLUT1) is relatively more favored compared with insulin-dependent glucose transport (GLUT4).
DISCUSSION
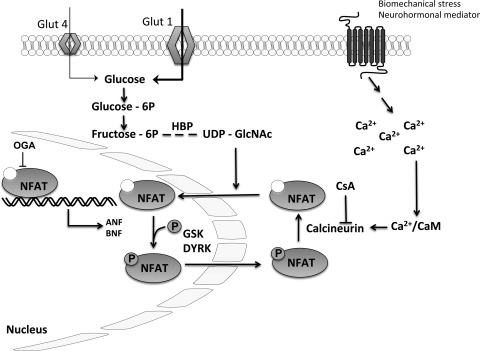
We have established a new paradigm in which we show that O-GlcNAc levels not only control NFAT nuclear translocation but also NFAT activation. Following a hypertrophic stimulus, the phosphatase calcineurin is activated and dephosphorylates NFAT. Dephosphorylated NFAT translocates to the nucleus where it promotes the transcription of several hypertrophic genes, including ANP. This is a well established process, and several nuclear kinases, such as GSK and DYRK, phosphorylate NFAT and promote its relocation back to the cytoplasm (10, 16). Here, we have established a novel paradigm in the regulation of cardiomyocyte hypertrophy whereby a metabolic signal (O-GlcNAc) represents an essential element of NFAT translocation to the nucleus. Moreover, augmentation of this signal, per se, is sufficient to induce NFAT activation in the absence of classical stimuli for hypertrophy.
O-GlcNAc signaling dynamically increases in response to cellular stress (36, 37) and protects against acute myocardial infarction, oxidative stress, and hypoxia (11, 19, 21, 22). Interestingly, activation of NFAT by calcineurin improves cardioprotection against ischemia/reperfusion injury both in vivo and in vitro (4). The concept of alterations in hexosamine signaling contributing to hypertrophy was first mentioned by Young et al. (34), in which they found elevation in UDP-GlcNAc levels following aortic banding in rats, although they were unable to draw specific conclusions related to changes in O-GlcNAc signaling or to demonstrate causality. Chatham's group recently reported that protein O-GlcNAcylation may also be important in the diabetic heart (15). Nevertheless, their findings are consistent with the present results, both of which emphasize the importance of metabolism and metabolic signaling during hypertrophy and heart failure.
Although we provided new insights regarding the emerging role of protein O-GlcNAcylation in hypertrophy, the role of NFAT/calcineurin signaling has been well established as a regulator of hypertrophic growth. Genetic disruption/inhibition of NFAT inhibits cardiomyocyte hypertrophy, albeit to varying extents with different NFAT isoforms (31, 33). Moreover, targeted inhibition of calcineurin can exert similar results in terms of limiting hypertrophy (17, 23, 27, 28). More recently, novel, indirect approaches indicate that hypertrophic signaling via calcineurin/NFAT can be indirectly regulated by miRNA (3). With continued investigation, the present findings can be extended into more detailed molecular mechanistic insights.
Despite the novelty of this study, there are several limitations. The use of NRCM (or H9c2) may not mimic all of the changes observed in the intact adult heart. This is why we confirmed that O-GlcNAc signaling was induced during TAC in vivo. More importantly, we needed to use the NRCM system to transfer genes of interest and induce metabolic re-programming during phenylephrine treatment; this is currently not feasible in multi-day studies of isolated adult cardiac myocytes. Furthermore, had we performed these studies in vivo, we would lack the cardiomyocyte-specific insights gathered from the present data. Nevertheless, we are developing strategies to translate the present findings to in vivo systems in future studies, which represents an important question. This is particularly important given our recent findings that protein O-GlcNAcylation during infarct-induced heart failure appears to be important for the endogenous, although insufficient, compensatory response during heart failure (32).
In summary, this study provides novel insights into mechanisms of metabolic reprogramming in the hypertrophic heart (summarized in Fig. 6). In addition, this work provides potential insights into metabolic dysregulation that could be extended to studies of diabetic tissues since O-GlcNAc is produced in the hexosamine biosynthetic pathway (one accessory pathway of glycolysis). Undoubtedly, the interaction of O-GlcNAc signaling with NFAT translocation/activation should spark additional studies designed to identify the specific signaling modules and site-specific modification of O-GlcNAc in the hypertrophic cardiomyocyte. Many of the critical targets of O-GlcNAc remain elusive, but the significance of O-GlcNAc signaling in primary disease is becoming unquestionable. As our understanding of the influence of metabolic signaling in the form of O-GlcNAc continues to grow, the likelihood escalates that novel therapeutics might arise from such discoveries.
Fig. 6.
Hypothetical scheme of the interplay between protein O-GlcNAcylation and hypertrophic reprogramming.
GRANTS
This work was supported by grants to S. P. Jones from the National Heart, Lung, and Blood Institute (NHLBI; R01 HL-083320, R01 HL-094419, and P01 HL-078825), American Heart Association National Center Scientist Development Grant (0535270N), National Center for Research Resources (NCRR; P20 RR-024489), and Kentucky Science and Engineering Foundation grant (KSEF-1677-RDE-011). S. D. Prabhu was supported by the NHLBI (P01 HL-078825, R01 HL-099014), NCRR (P20 RR-024489), and a Veterans' Administration Merit Award. T. Hamid was supported by American Heart Association Scientist Development Grant (0835456N). H. T. Facundo was an American Heart Association Postdoctoral Fellow, Great Rivers Affiliate (0825643D). R. E. Brainard (09PRE2390017), L. J. Watson (0815502D), and G. A. Ngoh (0815502D) were American Heart Association Predoctoral Fellows, Great Rivers Affiliate.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: H.T.F., R.E.B., L.J.W., G.A.N., and T.H. performed experiments; H.T.F., R.E.B., L.J.W., G.A.N., and T.H. analyzed data; H.T.F., R.E.B., G.A.N., T.H., S.D.P., and S.P.J. interpreted results of experiments; H.T.F. and R.E.B. prepared figures; H.T.F. and S.P.J. drafted the manuscript; R.E.B. and S.P.J. edited and revised the manuscript; R.E.B. and S.P.J. approved the final version of the manuscript; S.P.J. conception and design of research.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Allison Aird, Elizabeth Fine, Bethany Long, and Linda Harrison (University of Louisville).
REFERENCES
- 1.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci USA 103: 10086–10091, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustamante JO, Ruknudin A, Sachs F. Stretch-activated channels in heart cells: relevance to cardiac hypertrophy. J Cardiovasc Pharmacol 17, Suppl 2: S110–S113, 1991 [DOI] [PubMed] [Google Scholar]
- 3.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, Hansen A, Coenen-de Roo CJ, Bierhuizen MF, van der Nagel R, van Kuik J, de Weger R, de Bruin A, Condorelli G, Arbones ML, Eschenhagen T, De Windt LJ. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 12: 1220–1227, 2010 [DOI] [PubMed] [Google Scholar]
- 4.De Windt LJ, Lim HW, Taigen T, Wencker D, Condorelli G, Dorn GW, 2nd, Kitsis RN, Molkentin JD. Calcineurin-mediated hypertrophy protects cardiomyocytes from apoptosis in vitro and in vivo: an apoptosis-independent model of dilated heart failure. Circ Res 86: 255–263, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Dorn GW, 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest 115: 527–537, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation 109: 1580–1589, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Gaasch WH, Zile MR, Hoshino PK, Weinberg EO, Rhodes DR, Apstein CS. Tolerance of the hypertrophic heart to ischemia. Studies in compensated and failing dog hearts with pressure overload hypertrophy. Circulation 81: 1644–1653, 1990 [DOI] [PubMed] [Google Scholar]
- 8.Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J 26: 4368–4379, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart GW, Housley MP, Slawson C. Cycling of O-linked [beta]-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 7: 589–600, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marban E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation 117: 1172–1182, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Liang F, Wu J, Garami M, Gardner DG. Mechanical strain increases expression of the brain natriuretic peptide gene in rat cardiac myocytes. J Biol Chem 272: 28050–28056, 1997 [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Lorell BH, Grossman W. Cardiac hypertrophy: the consequences for diastole. J Am Coll Cardiol 9: 1189–1193, 1987 [DOI] [PubMed] [Google Scholar]
- 15.Marsh SA, Dell'Italia LJ, Chatham JC. Activation of the hexosamine biosynthesis pathway and protein O-GlcNAcylation modulate hypertrophic and cell signaling pathways in cardiomyocytes from diabetic mice. Amino Acids 40: 819–828, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res 63: 467–475, 2004 [DOI] [PubMed] [Google Scholar]
- 17.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montessuit C, Thorburn A. Transcriptional activation of the glucose transporter GLUT1 in ventricular cardiac myocytes by hypertrophic agonists. J Biol Chem 274: 9006–9012, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Ngoh GA, Facundo HT, Hamid T, Dillmann W, Zachara NE, Jones SP. Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury. Circ Res 104: 41–49, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngoh GA, Facundo HT, Zafir A, Jones SP. O-GlcNAc signaling in the cardiovascular system. Circ Res 107: 171–185, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngoh GA, Jones SP. New insights into metabolic signaling and cell survival: the role of O-GlcNAc. J Pharmacol Exp Ther 327: 602–609, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngoh GA, Watson LJ, Facundo HT, Dillmann W, Jones SP. Non-canonical glycosyltransferase modulates post-hypoxic cardiac myocyte death and mitochondrial permeability transition. J Mol Cell Cardiol 45: 313–325, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson EN, Molkentin JD. Prevention of cardiac hypertrophy by calcineurin inhibition: hope or hype? Circ Res 84: 623–632, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Sadoshima J, Izumo S. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu Rev Physiol 59: 551–571, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Simpson P. Stimulation of hypertrophy of cultured neonatal rat heart cells through an alpha 1-adrenergic receptor and induction of beating through an alpha 1- and beta 1-adrenergic receptor interaction. Evidence for independent regulation of growth and beating. Circ Res 56: 884–894, 1985 [DOI] [PubMed] [Google Scholar]
- 27.Sussman MA, Lim HW, Gude N, Taigen T, Olson EN, Robbins J, Colbert MC, Gualberto A, Wieczorek DF, Molkentin JD. Prevention of cardiac hypertrophy in mice by calcineurin inhibition. Science 281: 1690–1693, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Taigen T, De Windt LJ, Lim HW, Molkentin JD. Targeted inhibition of calcineurin prevents agonist-induced cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 97: 1196–1201, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teshima Y, Akao M, Jones SP, Marban E. Uncoupling protein-2 overexpression inhibits mitochondrial death pathway in cardiomyocytes. Circ Res 93: 192–200, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O'Donnell C, Kittner S, Lloyd-Jones D, Goff DC, Jr, Hong Y, Adams R, Friday G, Furie K, Gorelick P, Kissela B, Marler J, Meigs J, Roger V, Sidney S, Sorlie P, Steinberger J, Wasserthiel-Smoller S, Wilson M, Wolf P. Heart disease and stroke statistics: 2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 113: e85–e151, 2006 [DOI] [PubMed] [Google Scholar]
- 31.van Rooij E, Doevendans PA, de Theije CC, Babiker FA, Molkentin JD, de Windt LJ. Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J Biol Chem 277: 48617–48626, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM, Long BW, Prabhu SD, Xuan YT, Jones SP. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci USA 107: 17797–17802, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkins BJ, De Windt LJ, Bueno OF, Braz JC, Glascock BJ, Kimball TF, Molkentin JD. Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol Cell Biol 22: 7603–7613, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young ME, Yan J, Razeghi P, Cooksey RC, Guthrie PH, Stepkowski SM, McClain DA, Tian R, Taegtmeyer H. Proposed regulation of gene expression by glucose in rodent heart. Gene Regul Syst Biol 1: 251–262, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc. Biochim Biophys Acta 1761: 599–617, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta 1673: 13–28, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Zachara NE, O'Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem 279: 30133–30142, 2004 [DOI] [PubMed] [Google Scholar]