Abstract
The causative agent of African sleeping sickness, Trypanosoma brucei, undergoes an unusual mitochondrial RNA editing process that is essential for its survival. RNA editing terminal uridylyl transferase 2 of T. brucei (TbRET2) is an indispensable component of the editosome machinery that performs this editing. TbRET2 is required to maintain the vitality of both the insect and bloodstream forms of the parasite, and, with its high-resolution crystal structure, it poses as a promising pharmaceutical target. Neither the exclusive requirement of UTP for catalysis, nor the RNA primer preference of TbRET2 is well understood. Using all-atom explicitly solvated molecular dynamics (MD) simulations, we investigated the effect of UTP binding on TbRET2 structure and dynamics, as well as the determinants governing TbRET2’s exclusive UTP preference. Through our investigations of various nucleoside triphosphate substrates (NTPs), we show that UTP pre-organizes the binding site through an extensive water-mediated H-bonding network, bringing Glu424 and Arg144 sidechains to an optimum position for RNA primer binding. In contrast, CTP and ATP cannot achieve this pre-organization and thus preclude productive RNA primer binding. Additionally, we have located ligand-binding “hot spots” of TbRET2 based on the MD conformational ensembles and computational fragment mapping. TbRET2 reveals different binding pockets in the apo and UTP-bound MD simulations, which could be targeted for inhibitor design.
Keywords: Molecular dynamics, terminal uridylyl transferase, TbRET2, computational solvent mapping, RNA editing, nucleotide specificity, Trypanosoma brucei
Introduction
Trypanosoma brucei is the causative agent of African sleeping sickness, a devastating disease threatening millions of people in Africa.1 The disease is uniformly fatal if untreated. Currently, pentamidine and suramin are used to treat the first stage, and melarsoprol and eflornithine are used to treat the second stage of the disease.2,3 The available drugs for the disease are inadequate and have many serious side effects including death, thus, there is an immediate need for novel drugs.
T. brucei is a member of the order Kinetoplastida, which has unusual mitochondrial DNA called “kDNA” in the form of maxicircles and minicircles.4,5 Maxicircle DNA encodes mitochondrial rRNAs as well as oxidative phosphorylation system proteins.4,5 However, most of the pre-edited mRNAs encoded by maxicircle DNA require post-transcriptional editing to become mature functional mRNAs that can successfully produce mitochondrial proteins suitable for the current life-cycle stage.4,5 The sequence of pre-edited mRNAs is modified by a multi-protein machinery called “the editosome” using guide RNAs (gRNA) encoded by the minicircles as a template.4,5 At the points of mismatch between the gRNAs and pre-mRNAs, uridine nucleotides (U) are either inserted to or deleted from the pre-mRNAs, and this process could be repeated multiple times using different gRNAs until a mature RNA forms.4,5 RNA editing is shown to be essential to sustain viability of both the insect and the bloodstream forms of T. brucei. 5
The T. brucei editosome is a dynamic, heterogenous assembly consisting of approximately 20 proteins. Although the individual components are characterized at different levels, much of their architecture and organization is not known. According to the current data available, the editosome is functionally and physically organized into two separate subcomplexes that are responsible for insertion editing and deletion editing.4,5,6 Within the editosome, editing starts with endonuclease cleavage of the pre-edited mRNA transcripts at the mismatch point between the RNA duplex of gRNA and pre-edited mRNA. This step is followed by either deletion by an exonuclease or U insertion by a terminal uridylyl transferase (TUTase). Finally, the cleavage products are joined by an RNA ligase.
RNA editing terminal uridylyl transferase (TUTase) 2 of T. brucei (TbRET2) is a 57 kDa monomeric protein and is one of the indispensable components of the editosome.7 Although there are multiple endonucleases, exonucleases and RNA ligases in the editosome, TbRET2 is the only enzyme that has the terminal uridylyl transferase function.8 The enzyme consists of three domains: the N-terminal domain (NTD), the C-terminal domain (CTD) and the middle domain. The binding site lies at the bottom of a deep cleft dividing NTD and CTD. The crystal structure of UTP-bound TbRET2 demonstrates a large binding site that interacts with the uridine portion of UTP mostly through water-mediated hydrogen bonds.9 Thus, it is still of interest how the enzyme exclusively prefers UTP among various nucleoside triphosphates (NTPs) for catalysis.
TbRET2 catalyzes the addition of a single U if the RNA substrate is single-stranded. In the case of a double-stranded RNA substrate, the enzyme adds as many Us as specified by the gRNAs.7 The mechanism of this U addition is thought to be a two-metal ion mechanism that is characteristic of all polymerases.9,10 In a two-metal ion mechanism, the 3′-hydroxyl group of the RNA primer attacks the α-phosphate of UTP and releases pyrophosphate without forming a covalent intermediate. A metal cation (metal cation A) will facilitate this reaction by lowering the affinity of 3′-hydroxyl for hydrogen, while the second metal cation (metal cation B) helps to stabilize the pyrophosphate leaving group. In the UTP-bound TbRET2 crystal structure, only one single metal cation that corresponds to metal cation B is unambiguously resolved.9 This is similar to the case of the minimal terminal uridylyl transferase, TbTUT4, crystal structures, in which only one single metal cation (metal cation B) is observed in UTP/CTP-bound TbTUT4. In contrast, two metal cations are observed in ATP/GTP-bound TbTUT4, as well as the ternary complex of the enzyme with RNA primer and UTP.11
TbRET2 is reported to be essential for the survival of both the insect12 and the bloodstream stages 9 of the pathogen. A family of seven putative TUTases with remote sequence similarity to TbRET2 is identified among the human host proteins 13,14,15,16,17,18. Only several of these, namely human TUTase4, U6 snRNA-specific TUTase (a.k.a. TUTase6), TUTase1 and TUTase7, are biochemically characterized, though further biochemical studies are needed for these enzymes and the rest of the family to achieve a detailed understanding of this enzyme family. It is not yet possible to assess how similar the human TUTases are structurally to TbRET2 due to the lack of experimental structural information.13,19,20 The only human enzyme that has TUTase activity and an available crystal structure is PAPD1 (a.k.a. human TUTase1) (PdbID:3PQ1).18 Despite less than 20% sequence identity18 PAPD1 shares the overall protein folds of the NTD and CTD domains of TbRET2 as well as the position and identity of the catalytic residues. However, there are still noticeable sequence and structural differences between the two proteins in the binding site; moreover, some parts of the PAPD1 binding site not resolved in the crystal structure may accommodate more differences.9 The recently elucidated high-resolution crystal structure of TbRET2 in apo and UTP-bound forms, and the lack of a closely homologous human enzyme, make TbRET2 a prominent system for structure-based inhibitor design studies to target the T. brucei pathogens.9 Richness of available data on the minimal terminal uridylyl transferase, TbTUT4, which has a root-mean-square-deviation (for the common domains with TbRET2) as low as 1.60 Å, despite having only 30% sequence identity, is also an asset.20 In this work, we aim to provide insight into the dynamics of TbRET2 and its specificity determinants for UTP through multi-copy all-atom explicitly solvated molecular dynamics (MD) simulations of the enzyme both with and without its native substrate. The MD-generated trajectories of TbRET2 are analyzed for variations in ligand-binding hot spots, electrostatics, binding pocket conformations and volumes.
Materials and Methods
System Preparation for TbRET2
Four systems were simulated for TbRET2: the apo form, and UTP-, CTP-, and ATP- bound forms (Table S1). The initial structural coordinates of apo and UTP-bound forms of TbRET2 were obtained from Protein Data Bank 2B4V and 2B51, respectively.9 The missing residues in the apo crystal structure (residues 308–312, and 472–473) were modeled by superimposition with the UTP-bound TbRET2 crystal structure. CTP-bound TbRET2 structure was modeled simply by modifying the ligand in the UTP-bound TbRET2 crystal structure. ATP-bound TbRET2 structure was modeled using ATP-bound TbTUT4 crystal structure (PdbID:2Q0D) as a template. The selenomethionines used for crystal structure refinement in the apo TbRET2 were replaced with methionines. If there were multiple conformations for a residue, the A conformers were adopted. The crystallographic waters were retained and the catalytic manganese ion in PdbID:2B51 was replaced by a magnesium ion. A second magnesium was added while modeling the ATP-bound TbRET2 system (by replacing Wat828 based on superimposition of TbRET2 with TbTUT4, a homologous enzyme) because 2 catalytic metal cations were observed in ATP-bound TbTUT4 crystal structure (PdbID:2Q0D),11 while only a single catalytic metal cation was observed in UTP-bound TbTUT4 crystal structure (PdbID:2IKF).20 The residues were checked whether the side chains needed flipping using the Molprobity web server.21 The histidine protonation states were determined using the Whatif Web Interface22 and manually verified. UTP and CTP parameters were generated with the Antechamber module of Amber1023 using the Generalized Amber Force Field (GAFF)24,25 with RESP HF 6–31G* charges, and manually compared with ATP/GTP parameters of Carlson et al. 26 Then, the torsional and angle parameters for the triphosphate tail of UTP and CTP were modified to adopt the triphosphate parameters by Carlson et al. ATP/GTP parameters were obtained from Carlson et al.26 The Leap module of Amber10 was used to add the missing atoms including hydrogens. Each system was then solvated in a TIP3P27 water box forming a 10 Å buffer between the protein and the periodic boundary. Chloride ions were added to each system for neutralization. Amber FF99SB force field28 was used to construct the topology files for each of the sytems. The final systems consisted of 63950, 65384, 65385 and 65387 atoms for the apo, UTP-bound, CTP-bound and ATP-bound TbRET2 systems, respectively.
Molecular Dynamics Simulations
A total of 26,000 steps of energy minimization were carried out to remove artificial contacts and relax the systems. Only the hydrogen atoms were relaxed in the first 2,000 steps of minimization having all other atoms fixed. In the second 2,000 steps, all water atoms and ions were relaxed in addition to the hydrogen atoms. And in the following 2,000 steps, all atoms were relaxed while the backbone atoms are held fixed. In the last 20,000 steps, the entire system was relaxed.
The initial one-nanosecond-long molecular dynamics simulation at 310 K was carried out as constrained molecular dynamics to prevent artificial structural effects due to introducing kinetic energy to the system. For this purpose, positional restraints were used for the heavy atoms of the protein backbone and the ligand, and were decreased from 4.0 to 1.0 kcal/(mol * Å2) in four consecutive steps that were 250-picosecond-long molecular dynamics simulation each.
Following the constrained dynamics, unconstrained molecular dynamics were carried out for 50 nanoseconds with a time step of 1 femtoseconds. Temperature was maintained by Langevin dynamics at 310 K with a collision frequency of 5 ps−1, and the pressure was maintained at 1 atm by the Nose Hoover-Langevin piston method29,30 using period and decay times of 100 and 50 femtoseconds, respectively. The long-range electrostatics was treated by the Particle Mesh Ewald method.31 The interatomic distances within water molecules were constrained using the SHAKE algorithm.32,33 A multiple-time step algorithm was employed, in which bonded interactions were computed at every time step, short-range non-bonded interactions were computed at every 2 time steps, and full electrostatics was computed at every 4 time steps. All minimizations and molecular dynamics simulations were performed using the highly parallelized NAMD2.734 on the Teragrid XSEDE Ranger cluster. The simulations scaled as 7.1 ns per day using 128 processors.
The analysis was performed on 10,000 snapshots that were extracted from the two copies of 50-ns trajectories of apo and UTP-bound TbRET2 with regular 10 ps intervals. 5,000 snapshots again with regular 10 ps intervals were analyzed for CTP-bound and ATP-bound TbRET2 systems. Conformational clustering was performed using the GROMOS algorithm35,36,37 with GROMACS4.0.5 analysis software.38 The snapshots in the MD trajectory were superimposed with respect to all Cα atoms to remove overall translation and rotation, then clustered at various RMSD cutoff values based on atomic coordinates of the active site. Eventually, RMSD cutoffs of 1.75 Å and 1.55 Å were chosen for the apo and UTP-bound TbRET2 ensembles, respectively. The active site was defined here as all atoms of residues that have at least one atom in 10 Å vicinity of UTP in PdbID:2B51 which correspond to residues 21, 22, 27, 31, 52–72, 74, 79, 111, 114–118, 120, 235–253, 256, 267, 268, 270–275, 281, 283, 286–294, 297, 314, 317, 320, 324, 361, 390–395, 402–407. In each cluster, the structure that has the smallest average RMSD to all of the other structures in the cluster was selected as the cluster centroid, or cluster representative structure. H-bond analysis was performed with HBonanza program39 using a hydrogen bond distance cutoff of 3.0 Å and a hydrogen bond angle cutoff of 30°.
Computational Fragment Mapping
To identify druggable sites of the TbRET2 enzyme, we performed computational fragment mapping using the FTMap program (http://ftmap.bu.edu).40 This method is similar to the X-ray crystallography-based technique called Multiple Solvent Crystal Structures (MSCS) that is based on resolving the crystal structures of a particular protein in aqueous solutions with different organic solvent molecules.41 The FTMap program utilizes 16 solvent probe molecules to identify and rank the most favorable binding sites energetically. To achieve this, FTMap initially performs rigid body docking of probe molecules with a fast Fourier transform method; then minimizes and rescores the docked poses, and clusters them for each probe using a simple greedy algorithm; finally locates the consensus sites of binding sites identified by all probes. Using the FTMap program, we located consensus binding sites on the TbRET2 crystal structure PdbID:2B51 as well as the 3 most-populated cluster representatives obtained from the apo and UTP-bound TbRET2 MD trajectories. The populations of the most-populated three clusters add up to 71.16% and 69.95% of the entire apo and UTP-bound TbRET2 trajectories, respectively.
Ensemble-Averaged Electrostatics
The ensemble-averaged electrostatic potential of TbRET2 was calculated using 10,000 frames of the UTP-bound TbRET2 trajectory (extracted at regular 10 ps intervals) as well as using only the UTP-bound crystal structure. For this purpose, a new VMD42 plugin called DelEnsembleElec43 interfacing with DelPhi44,45, a numerical Poisson-Boltzmann (PB) equation solving suite, was utilized. The electrostatic potential was calculated on a 111×111×111 static grid with an interior dielectric constant of 1, an exterior dielectric constant of 80 and at 0 salt concentration. The grid scale was set to 1.0 Å and the probe radius defining the dielectric boundary was 1.4 Å. The convergence was achieved once the change of potential decreased below 0.0001 kT/e.
Binding Pocket Volumes
The volume of each frame in the MD trajectories was measured using POVME,46 a pocket volume measuring algorithm. A single inclusion sphere that encompasses the TbRET2 binding cleft was used to measure the pocket volume. The same inclusion sphere was used for all systems after superimposing the first frames of the trajectories. POVME algorithm computes the pocket volume by subtracting the volume occupied by the protein atoms in each frame from the inclusion sphere volume.
MM-GB/PBSA Binding Energy Calculations
Binding affinities of TbRET2 were calculated for each of UTP (2 copies), CTP and ATP using 500 frames (with regular 200 ps intervals) of each MD trajectory. The MM-PBSA (python version) program in AmberTools1.547 was used for this purpose using a single trajectory approach. Although there were two Mg+2 ions in ATP-bound TbRET2 MD simulations, only one Mg+2 ion was used in the calculations to be comparable to the other NTP-bound TbRET2 binding energy calculations. In addition, the binding energy values using two Mg+2 were calculated for comparison. The Mg+2 ion was considered to be part of the receptor and only the NTP molecule was considered to be the ligand. No water molecules were included in computations. The gas phase protein-ligand interaction energy that consists of electrostatic and van der Waals components was computed using the force field parameters in the topology files. The electrostatic solvation energy in the MM-PBSA calculations was computed at a solute dielectric constant of 1 and an exterior dielectric constant of 80 using the program’s internal PB solver, the force field radii, a grid spacing of 0.5 Å and a probe radius of 1.4 Å. The electrostatic solvation energy in the MM-GBSA calculations was calculated using a solute dielectric constant of 1 and an exterior dielectric constant of 80, mbondi2 radii and the GBOBC2 model.48 The nonpolar solvation energy was computed using the LCPO method.49 No entropic contribution was calculated due to its high computational cost and convergence issues; furthermore, similar entropic contributions are predicted for similarly-sized NTP ligands. The default values of the program were used for all other parameters.
Results and Discussion
Molecular Dynamics Simulations
We investigated the dynamical properties of TbRET2 in its apo and NTP-bound forms using all-atom molecular dynamics simulations in explicit solvent at 1 atm and 310 K for each system. Two copies of 50 ns MD simulations for each of apo and UTP-bound TbRET2 systems were performed as well as one copy of 50 ns MD simulation for each of ATP-bound and CTP-bound TbRET2 systems (Table S1 in the Supporting Material). The time evolution of the root-mean-square-deviation (RMSD) of Cα atoms of the apo and ligand-bound forms of TbRET2 presented in Figure S1 shows that the simulations are stable.
Ensemble-averaged Electrostatics
The ensemble-averaged electrostatic potential was computed for the UTP-bound TbRET2 trajectory as well as the crystal structure (Figure S2). The ensemble-averaged electrostatic potential demonstrates a more uniform and pronounced positive electrostatic patches at the periphery of the binding cleft compared to that of the crystal structure, which may serve to attract the RNA.
UTP-TbRET2 Interaction Analysis
UTP retains the binding pose observed in the crystal structure PdbID:2B51 throughout the MD simulations reflected by the time evolution of the root-mean-square-deviation (RMSD) of the UTP heavy atoms (Figure S3). The interactions of UTP with TbRET2 in the MD simulations are analyzed here by dividing them into three subsets; the interactions of UTP in the nucleotide base, the ribose and the triphosphate regions. The atom-naming map of UTP is provided in Figure S4.
In the triphosphate region, Mg+2 cation retains an octahedral coordination. One oxygen atom from each of Asp97 carboxylate, Asp99 carboxylate of TbRET2, and α-phosphate, β-phosphate, γ-phosphate of UTP, and a water molecule form the coordination sphere of the Mg+2 cation. The water molecule H-bonded to the metal cation (Wat525 in PdbID:2B51) does not exchange with the bulk water in either of the two 50-ns MD simulations (average distance between O atom water and the metal cation: 2.07 ± 0.07 Å). The γ-phosphate has persistent H-bonding interactions with Lys300, Lys304 and Ser318 side chains, the last one being the most persistent interaction of all (Table 3). The H-bonding interaction between Ser96 side chain and γ-phosphate observed in the crystal structure is persistent for only about 37% of the simulation time overall (Table 3) although the lifetime of the interaction can be as long as 10 ns. The α-phosphate and β-phosphate of UTP are not involved in any direct interactions with the protein, and their only interactions are with the water molecules and the metal cation.
TABLE 3.
Percentages of H-bonds between TbRET2 and UTP in MD simulations.
| H-bond donor | H-bond acceptor | Percentage |
|---|---|---|
| UTP.O2′ | Asn277.OD1 | 99.94% |
| Ser318.OG | UTP.O3G | 99.71% |
| Asn277.ND2 | UTP.O2 | 83.98% |
| Lys304.NZ | UTP.O3G | 80.63% |
| Lys300.NZ | UTP.O1G | 84.21% |
| Ser96.OG | UTP.O1G | 36.78% |
| UTP.O3′ | Ser278.OG | 27.92% |
| Lys304.NZ | UTP.O1G | 18.96% |
| Lys300.NZ | UTP.O3B | 15.47% |
| Gly84.N | UTP.O3′ | 5.67% |
| Ser278.OG | UTP.O2′ | 4.05% |
In the ribose region of UTP, residues Asn277 and Ser278 are the only two residues, which directly interact with the nucleotide substrate. Ser278 accepts a H-bond from the 3′ OH group, and simultaneously donates a hydrogen bond to the 2′ OH group of UTP’s ribose but these interactions persist for only 28% and 4% of the simulation time, respectively (Table 3). Unlike Ser278, Asn277 forms a persistent hydrogen bond between its side chain carbonyl group and the 2′ OH group of the UTP ribose (Table 3). Ser278 side chain also participates in persistent hydrogen bond interactions with two glycine backbone atoms (Gly274 and Gly84), which together constrain it to the optimum position to interact with the ribose of UTP. Based on their interactions with the 2′ OH group of UTP, Asn277 and Ser278 appear to be the key residues for TbRET2 in discriminating between UTP and dUTP.
In the nucleotide base region, the essential stacking interaction with Tyr319 phenol ring is persistently observed over the entire simulation. The only direct interaction observed between the protein and the nucleotide base region of UTP is the one between Asn277 side chain amino group and the O2 of the UTP nucleotide base (average distance: 2.88 ± 0.14 Å). Also, we observed a water molecule that interacts with N3 of the UTP nucleotide base, as well as with the Asp421 and Glu424 side chains. This water molecule was suggested by Deng et al50 to have a role in TbRET2’s ability to discriminate UTP from CTP. In the MD simulations, this water molecule frequently exchanged with the bulk water, however, its position was conserved. Additionally, the O4 atom of UTP nucleotide base forms a water-mediated hydrogen bonding interaction with the Arg435 guanidinium group.
Ligand-binding Hot Spots in TbRET2
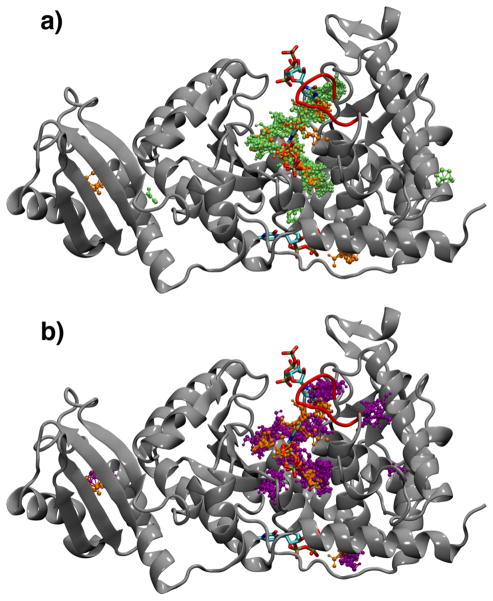
The computational fragment mapping technique identifies and ranks the most favorable binding sites of a protein using small molecule probes. FTMap and its predecessor CSMap51 have been shown to successfully identify consensus sites that correlate strongly with ligand binding hot spots identified by experimental methods.40,52,53 We used the FTMap program to locate consensus binding sites on the TbRET2 crystal structure PdbID:2B51 as well as the 3 most-populated cluster representatives obtained from the apo and UTP-bound TbRET2 MD trajectories (Figure 1).
FIGURE 1.
Consensus sites identified by computational fragment mapping. (a) Consensus sites identified by mapping 3 UTP-bound MD structures are shown altogether in lime with ball-and-stick representation. (b) Consensus sites identified by mapping 3 apo MD structures are shown altogether in magenta in ball-and-stick representation. Consensus sites identified by mapping X-ray crystal structure PdbID:2B51 are shown in orange with ball-and-stick representation. Mapping results for both the apo and UTP-bound ensembles are superimposed onto the X-ray crystal structure PdbID:2B51 which has 3 UTP molecules bound (one UTP bound to the primary UTP binding site, the other two bound to secondary sites) colored by atom type. The most flexible loop extending between residues 308 and 315 is colored red.
X-ray crystallization studies showed that at low UTP concentrations, a single UTP binds to TbRET2 at the bottom of the deep binding cleft (site A).9 At high UTP concentrations or at high concentrations of UMP in addition to low concentration of UTP, two additional sites, site B (which is at the periphery of the deep binding cleft) and site C (which is distinct and far outside the deep binding cleft), are ligated with either UTP or UMP.9
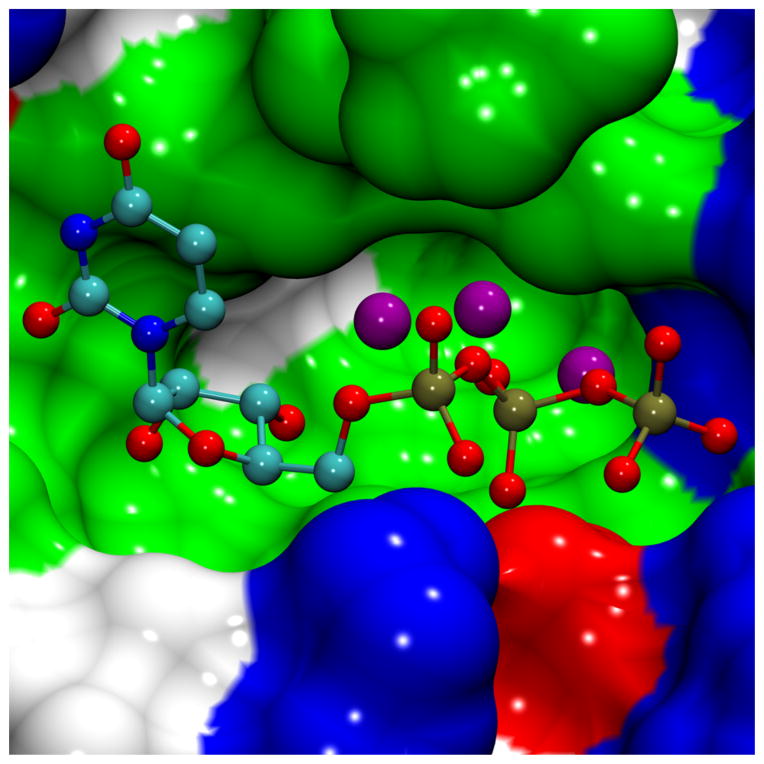
Computational fragment mapping successfully locates the UTP binding site A, the uridine part of the UTP binding site B, and the RNA primer binding site in all three cases: using the crystal structure, the apo TbRET2 MD structures and the UTP-bound TbRET2 MD structures. Site A found by FTMap using the apo TbRET2 MD structures extends further into the region that became available due to conformational motion of Tyr319. Additionally, a consensus site is located (and ranked high and even as the best binding site in some cases) in all cases in the deep polar pocket right next to the β-phosphate of UTP (Figure 2).
FIGURE 2.
Polar water pocket in the vicinity of the UTP binding site. The three water molecules that reside in the polar water pocket are illustrated with purple spheres at the location of the water oxygen atom. UTP atoms are depicted in ball-and-stick representation and colored according to atom names. The MSMS surface representation of neighboring residues is shown colored by residue type: blue-basic, red-acidic, green-polar, white-nonpolar.
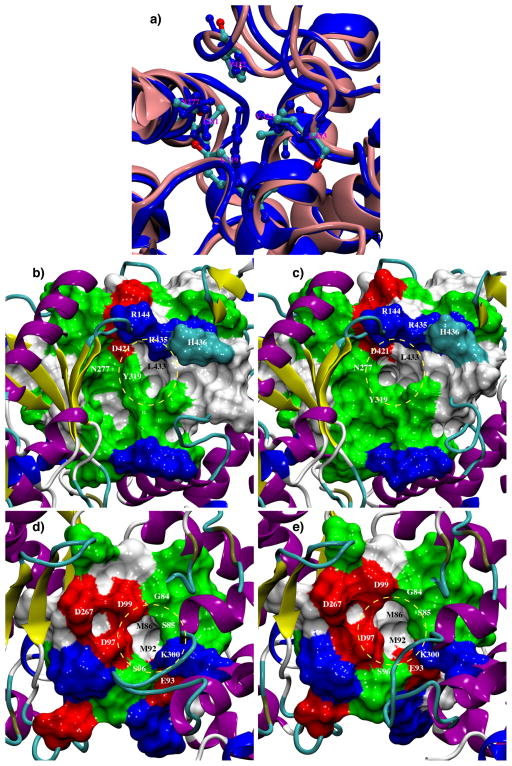
A consensus site in the pocket formed by Asp97, Asp99 and Asp267 (Figure 3e) is located only in the apo TbRET2 MD structures. In addition, a consensus binding site in the vicinity of Site C is found only in the UTP-bound TbRET2 trajectories.
FIGURE 3.
Two new binding pockets are revealed during the apo TbRET2 MD simulations. (a) Structural alignment of the crystal structure (backbone depicted in blue cartoon, residues depicted in blue ball-and-stick representation) and the representative frame of the most-populated cluster from the apo simulation (backbone depicted in pink cartoon, residues depicted in ball-and-stick representation and colored according to atom names) at the new pocket 1 region. Residues Tyr319, Leu433, Val323, Ile281, Asn277 and Pro422 are depicted in this figure. (b) MSMS surface representation of residues in the most-populated representative frame that line the new pocket 1. Residues are colored by residue type in (b), (c), (d) and (e): blue-basic, red-acidic, green-polar, white-nonpolar. (c) Same representation as in (b) in the apo TbRET2 crystal structure. (d) MSMS surface representation of residues in the most-populated representative frame that line the new pocket 2. (e) Same representation as in (d) in the apo TbRET2 crystal structure.
Role of Water Molecules in the Binding Site
All crystallographic water molecules were retained in our simulations and the dynamics of deeply buried water molecules in the active site are of interest for inhibitor design purposes. The water numbering in PdbID: 2B51 will be adopted here.
The three crystallographic waters, Wat519, Wat521 and Wat528, lie in a deep polar pocket right next to the β-phosphate of UTP in PdbID: 2B51 structure (Figure 2). During the UTP-bound TbRET2 MD simulations, the presence of UTP hinders the exit of water molecules from this pocket, preventing their exchange with the bulk solvent — although the water molecules do exchange position with each other. One of these water molecules interacts with both the UTP’s ribose 2′ OH group and the β-phosphate. There is only one extra water molecule that enters and remains in this pocket in the final 18 ns of one copy of the 50-ns MD simulations. This polar pocket is identified by the FTMap program to be the most favorable binding site in TbRET2 (using the crystal structure as well as the most-populated 1st and 3rd cluster representative structures of UTP-bound TbRET2 simulations), and thus we propose it could be utilized for inhibitor design.
Effect of UTP Binding on the Dynamics of TbRET2
All atoms of the active site, which are defined here to be all residues that have at least one atom within a 10 Å vicinity of UTP in the initial crystal structure, are used to cluster the snapshots of each system obtained from the MD simulations. The number of clusters at the same RMSD cutoff values for apo and UTP-bound TbRET2 MD simulations differ significantly reflecting the reduced flexibility in TbRET2 active site upon UTP binding (Table 1).
TABLE 1.
Number of clusters found in 10,000-frame-MD-trajectory of apo and UTP-bound TbRET2 systems at different RMSD cutoff values (in Å).
| 1.25 | 1.35 | 1.45 | 1.55 | 1.65 | 1.75 | |
| Apo | 741 | 373 | 187 | 95 | 50 | 28 |
| UTP-bound | 375 | 158 | 60 | 27 | 14 | 7 |
The organization and stabilization provided by UTP binding is also reflected in the time evolution of calculated binding cleft volume during the MD simulations. The volume of the TbRET2 binding cleft is stable in the UTP-bound TbRET2 MD simulations unlike the apo or other NTP-bound TbRET2 MD simulations (Figure S5). The standard deviation of the binding cleft volume calculated for the UTP-bound TbRET2 simulations are the lowest among all (Table S2).
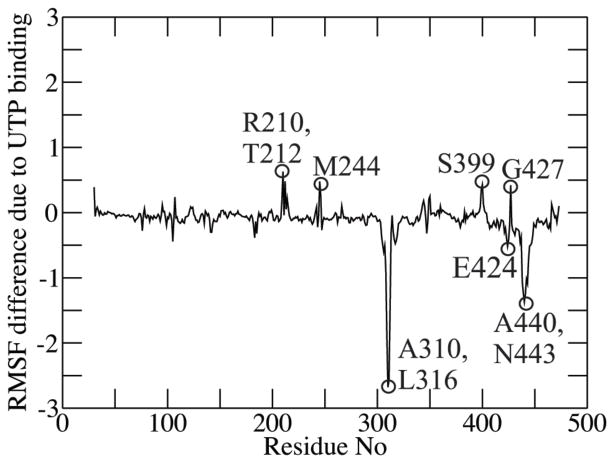
To understand the effect of UTP binding on TbRET2 dynamics, the difference between Cα root-mean-square-fluctuation (RMSF) of the UTP-bound TbRET2 MD trajectory and that of the apo MD trajectory is calculated for each residue. Remarkably, the peripheral loop that extends from residue 308 to residue 315 shows the largest difference upon UTP binding and is much more flexible in the apo TbRET2 simulations (Figure 4). The terminal turn of neighboring α20 (residues 440 to 443) also demonstrates a similar trend. This observation is in line with the fact that the residues 308 to 312 could not be resolved in the apo TbRET2 crystal structure (PdbID:2B4V), unlike the case in the UTP-bound crystal structure (PdbID:2B51). In the UTP-bound TbRET2 crystal structures (PdbID:2B51 and PdbID:2B56), an additional UTP or UMP molecule binds exactly at this loop (called “site B” in Deng et al9), interacting with Ser312, Gly313, Ala314, Met315 as well as residues Arg144, Arg435 and His436, which reside on neighboring loops. Interaction of these residues with a UTP or UMP molecule may stabilize this flexible loop, facilitating the x-ray crystallization of the UTP-bound TbRET2 structure. Based on its location, being bound to a UTP/UMP molecule in the crystal structures, and its remarkable flexibility in the simulations, this peripheral loop is a good candidate to play a role in RNA primer binding.
FIGURE 4.
Cα root-mean-square-fluctuation difference per residue between UTP-bound and apo TbRET2 MD simulations (in Å).
The RMSF difference analysis also shows that UTP binding increases the flexibility of residues 210, 212 and 244 that lie on peripheral loops of the TbRET2 middle domain. It has been shown recently that TbRET2 integrates into the RNA editing core complex (RECC) of the editosome via interaction of the TbRET2 middle domain with the structural protein MP81.54 These residues that demonstrate the largest RMSF difference in the middle domain may participate in interdomain signaling between TbRET2 and the RECC in the presence of UTP. Additionally, UTP binding destabilizes residues 399 and 427 (Figure 4), which are at the periphery of the UTP binding site of TbRET2.
Remarkable Conformational Change Observed in Apo TbRET2 Active Site
In one of the two copies of the 50-ns MD simulations of apo TbRET2, a significant conformational change occurs at around 20 ns and persists for approximately 15 ns before the active site returns back to its initial conformation (in total, this new conformation accounts for 38% of the trajectory). This conformation is also sampled for short periods of time in the other MD copy, representing a total of 28% of the trajectory. In this new conformation, the side chain of Tyr319, which provides the main hydrophobic contact with the nucleotide base of UTP, swings into the active site changing the χ1 dihedral angle of Tyr319 from −180° up to −120° in the apo system. This motion opens up a small pocket adjacent to the UTP binding site, which is surrounded by Asn277, Asp421 and Leu433 (Figure 3a, b and c). This pocket opening is not seen in the presence of UTP because the interactions between the nucleotide base and the protein confine Tyr319 side chain into one conformation and a persistent hydrogen bond is observed between the side chains of Tyr319 and Asp421.
In addition, the side chain of catalytic Asp99 moves by changing χ1 from −80° to −170°, sampling this conformation for about 96% of the simulation time. This conformational motion widens a second small pocket surrounded by Ser85, Met92, Ser96, Asp97 and Asp99 during the simulations (Figure 3, d and e). This pocket is adjacent to where the triphosphate tail of UTP binds and Asp99 and Asp97 side chains are both coordinated to the divalent metal cation in the UTP-bound state. In the apo TbRET2 crystal structure, no divalent metal cation is observed and the electron density required double conformations to be modeled for all three aspartic acid side chains, Asp97, Asp99 and Asp267. A strategy targeting a unique inactive conformation of an enzyme proved to be very successful in the discovery of the FDA-approved cancer drug Gleevec that inhibits Abelson tyrosine kinase.55,56 We propose that this alternative, predominant conformation sampled in the MD could be an inactive conformation of TbRET2 useful in a similar drug design strategy. To assist the rational design of inhibitors against the inactive form of TbRET2, the structure of TbRET2 with the reorganized active site is provided in the Supporting Material.
Both of these structural rearrangements are observed in the most-populated cluster representative structure when the apo TbRET2 MD ensemble is clustered using the GROMOS clustering algorithm at 1.75 Å RMSD cutoff. Using FTMap on this most-populated representative structure, the first new pocket around Tyr319 and the second new pocket around Asp99 are ranked to be the 3rd and 4th most favorable binding sites in this conformation of TbRET2, respectively. Based on the fragment solvent mapping results, both pockets can accommodate hydrophobic and polar solvent molecules alike.
Nucleic acid specificity of TbRET2 revealed in NTP-bound MD simulations
Although residual catalytic activity is observed with CTP, ATP and GTP in other TUTases, such as TbTUT4,11 TbRET2 strictly requires UTP for catalytic activity.57 Among different NTPs, the only substrate that could be entirely resolved in ligand-bound TbRET2 crystal structures is UTP.9 Although all four NTPs were bound to TbRET2 in the crystals, CTP, ATP and GTP did not have clearly defined electron densities except for the triphosphate tails. In addition, no crystal structure of the ternary complex of TbRET2 with UTP and RNA primer is yet available. The minimal TUTase of T. brucei, TbTUT4, is very similar to TbRET2 and a wealth of information including all NTP-bound crystal structures as well as pre-reaction ternary complex (PdbID:2Q0F) and pro-reaction complex (PdbID:2Q0G) crystal structures of TbTUT411 provide valuable insights for TbRET2. Still, due to the lack of direct evidence from crystal structures, the determinants of exclusive nucleic acid specificity of TbRET2 are not well understood.
The experimental binding constants of different NTPs to TbRET2 are not available in the literature. MM-PBSA and MM-GBSA methods can estimate binding free energies of ligands by combining molecular mechanics calculations and either Poisson Boltzmann (PB) or Generalized Born (GB) surface area (SA) implicit solvent models, respectively. Based on the MD simulations of TbRET2, the MM-PBSA method predicts that UTP binding is approximately 8 kcal/mol more favorable than CTP binding (Table S3). The MM-GBSA scheme differs from the MM-PBSA method in that it utilizes an approximation to solving the finite-difference Poisson Boltzmann to compute the polar component of the solvation free energy. Although its performance can be quite good, in this case, MM-GBSA appears to be less accurate, predicting a much larger difference of 23 kcal/mol. Unfortunately, the presence of a second metal cation in ATP-bound TbRET2 simulations prevents us from obtaining a comparable relative binding energy for ATP. When only a single metal cation (metal cation B) was included in the calculations, the resulting binding energy was highly unfavorable compared to other NTPs due to the proximity of the negatively charged residues brought together by metal cation A in the simulations. When both metal cations were included in the calculations, the resulting binding energy became highly favorable compared to others due to the strong interactions of two metal cations with the negatively charged residues in the surrounding area. Thus, neither calculation is able to provide a one-to-one comparable binding energy value for ATP and the other nucleotides.
The nucleic acid specificity of TbRET2 observed experimentally is reflected also in the dynamics of the NTP-bound TbRET2 systems. The average heavy-atom RMSD of the NTP ligands with respect to the initial conformation are measured after aligning the trajectories with respect to all Cα atoms of TbRET2 (Figure S3). The average RMSD values in both copies of the UTP-bound TbRET2 simulations are significantly lower compared to the CTP-bound and especially the ATP-bound systems (Table 2). Unlike other NTP-bound TbRET2 systems, the nucleotide base of ATP samples conformations that would interfere with the RNA primer binding. This motion, reflected in Figure S6, demonstrates the time evolution of the distance between the geometric centers of aromatic rings of NTP and Tyr319 and the angle between the two aromatic ring planes.
TABLE 2.
Average RMSD deviations of heavy-atoms of NTP in NTP-bound TbRET2 simulations when the trajectories are aligned with respect to the coordinates of all TbRET2 Cα atoms. The first frame of each MD trajectory is used as the reference frame for RMSD calculations. Standard deviations are also reported.
| RMSD (Å) | |
|---|---|
| UTP-bound (copy1) | 0.736 ± 0.156 |
| UTP-bound (copy2) | 0.697 ± 0.167 |
| CTP-bound | 1.013 ± 0.289 |
| ATP-bound | 1.935 ± 0.246 |
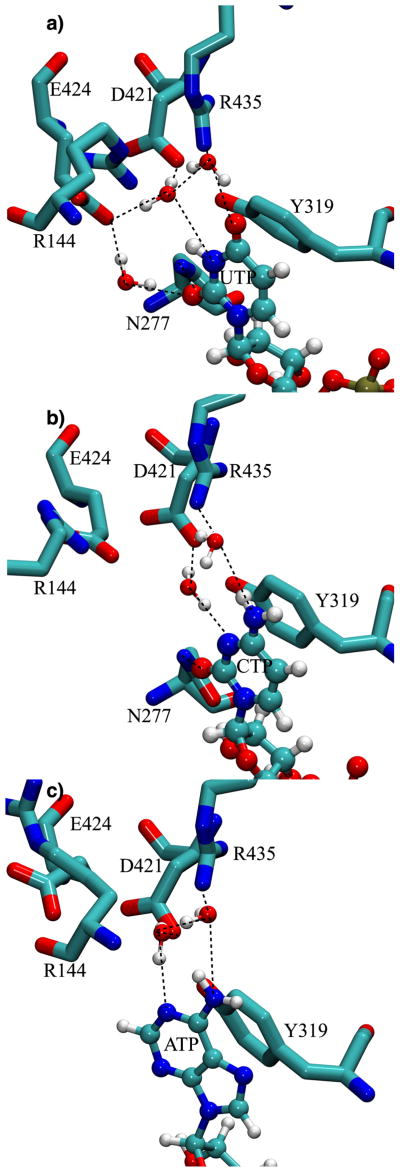
Due to the large TbRET2 binding site and the lack of direct enzyme-ligand interactions exhibited in the crystal structures, the exclusive UTP specificity of TbRET2 for catalysis remains an outstanding question. In our MD simulations, the binding poses of all three NTPs overlapped perfectly (Figure S7). However, a detailed analysis of the trajectories reveals that an exquisite water-mediated hydrogen-bonding network likely provides the nucleic acid specificity of TbRET2. In the UTP-bound TbRET2 systems, uridine interacts with Asp421, Glu424 and Arg435 side chains via water-mediated hydrogen bonds. The contribution of the water-mediated interaction of Arg435 decreases from 69.05% in UTP-bound MD trajectory to 39.00% in CTP-bound MD simulation, and finally to 27.12% in ATP-bound MD trajectory (Figure 5). Glu-424 is locked into position by two water-mediated interactions with uridine (Figure 5a). One of them is a water-mediated interaction between Glu424 side chain and the O2 atom of UTP, and is sampled 77.04% of the MD trajectory. Glu424 side chain has a second water-mediated interaction with the N3 atom of UTP as well as Asp421 side chain which is sampled 90.25% of the MD trajectory. Interestingly, the Glu424 side chain does not have similar water-mediated interactions with cytidine in CTP-bound TbRET2 system. Instead, Glu424 interacts with the Arg144 side chain after a conformational rearrangement (Figure 5b). Due to this conformational motion, the O2 atom of CTP interacts with the water molecules around in the active site rather than a specific water-mediated interaction with a TbRET2 residue (which is sampled only 0.99% of the MD trajectory). Due to the conformational motion of Glu424 as well as the difference in hydrogen-bonding properties of CTP compared to UTP, the N3 atom of CTP interacts only with the Asp421 side chain via a water-mediated interaction sampled for 96.38% of the MD trajectory. In the ATP-bound TbRET2 system, the situation is more pronounced since both the Glu424 and Arg144 side chains move away from the binding cleft towards bulk solvent, and none of the persistent water-mediated interactions observed in other NTPs between the nucleotide base and the protein exist (Figure 5c). Still, a similar water-mediated interaction is sampled between a nitrogen atom of ATP and Asp421 side chain in 67.22% of the MD trajectory. Based on the crystal structure of the homolog TbTUT4 ternary complex, Glu424 and Arg144 in TbRET2 are among the residues that would interact with the RNA primer’s terminal nucleotide and favor ATP energetically compared to other NTPs.11 Thus, the reason for TbRET2’s exclusive UTP preference for catalysis appears to be that only UTP, with its high binding affinity, is able pre-organize the binding site for productive RNA primer binding. Mutations of Arg144 and Arg435, which are conserved in all trypanosomal TUTases, each abolish enzymatic activity of TbRET2 both in vitro and in vivo,58 supporting the idea that both residues are essential for TbRET2 due to their role in intricately positioning UTP as well as the RNA primer.
FIGURE 5.
Water-network responsible for nucleotide specificity of TbRET2. Representative frames of MD trajectories are depicted for each of NTP-bound TbRET2.
Conclusions
All-atom molecular dynamics simulations of different NTP-bound TbRET2 systems revealed valuable insights on the determinants of NTP specificity of the enzyme. UTP constructs an exquisite water-mediated interaction network with the residues surrounding uridine, constraining Glu424 in an optimum position for RNA primer binding. Based on the crystal structures of the TbTUT4 ternary complex, Glu424 and Arg144 in TbRET2 would interact with the terminal nucleotide of the RNA primer. However, the hydrogen-bonding network formed by CTP or ATP in the TbRET2 binding site cannot orient Glu424 similarly and hence is likely to fail productive RNA primer binding. Additionally, “hot spots” of TbRET2 are identified for the apo and UTP-bound structural ensembles using computational fragment mapping. Several binding sites that are not observed in crystal structures are revealed during the MD simulations and may be targeted for inhibitor design.
Supplementary Material
Acknowledgments
This work was funded in part by the National Institutes of Health through the NIH Director’s New Innovator Award Program DP2-OD007237, a K22 Career Transition Award K22-AI081901, and through the NSF TeraGrid Supercomputer resources grant RAC CHE060073N to R.E.A. Molecular dynamics simulations were performed at the Texas Advanced Computing Center.
Footnotes
Supporting Information Available
Seven figures, three tables and a coordinate set of reorganized TbRET2. This information is available free of charge at http://pubs.acs.org.
References
- 1.WHO. Working to overcome the global impact of neglected tropical diseases -Summary. Wkly Epidemiol Rec. 2011;86(13):113–120. [PubMed] [Google Scholar]
- 2.WHO. Technical report 0512–3054 World Health Organ Tech Rep Series. Vol. 881. Switzerland: 1998. Control and surveillance of African trypanosomiasis; pp. I–VI.pp. 1–114. [PubMed] [Google Scholar]
- 3.Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, Ghabri S, Baudin E, Buard V, Kazadi-Kyanza S, Ilunga M, Mutangala W, Pohlig G, Schmid C, Karunakara U, Torreele E, Kande V. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. 2009;374(9683):56–64. doi: 10.1016/S0140-6736(09)61117-X. [DOI] [PubMed] [Google Scholar]
- 4.Carnes J, Stuart K. Working Together: the RNA Editing Machinery in Trypanosoma brucei. In: Göringer HU, editor. RNA Editing. Nucleic Acids and Molecular Biology. Vol. 20. Springer; Berlin Heidelberg: 2008. pp. 143–164. [Google Scholar]
- 5.Ochsenreiter T, Hajduk S. RNA Editing. Nucleic Acids and Molecular Biology. Vol. 20. Springer; Berlin Heidelberg: 2008. The Function of RNA Editing in Trypanosomes; pp. 181–197. [Google Scholar]
- 6.Aphasizhev R. RNA Editing. Molecular Biology. 2007;41(2):227–239. [Google Scholar]
- 7.Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11(6):1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 8.Aphasizhev R, Aphasizheva I. RNA editing uridylyltransferases of trypanosomatids. Methods Enzymol. 2007;424:55–73. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- 9.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBOJ. 2005;24(23):4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steitz TA. A mechanism for all polymerases. Nature. 1998;391(6664):231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 11.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci U S A. 2007;104(37):14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci U S A. 2003;100(19):10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138(4):696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13(6):860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22(1):50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 2007;27(10):3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickens M, Kwak JE. A tail tale for U. Science. 2008;319(5868):1344–1345. doi: 10.1126/science.1154946. [DOI] [PubMed] [Google Scholar]
- 18.Bai Y, Srivastava SK, Chang JH, Manley JL, Tong L. Structural basis for dimerization and activity of human PAPD1, a noncanonical poly (A) polymerase. Mol Cell. 2011;41(3):311–320. doi: 10.1016/j.molcel.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12(8):1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007;366(3):882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Homology modeling model and software evaluation: three related resources. Bioinformatics. 1998;14(6):523–528. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 23.Case DA, TAD, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. Amber10. University of California San Francisco; San Francisco, CA: 2008. [Google Scholar]
- 24.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graphics Modell. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24(9):1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 28.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins-Struct Funct Bioinf. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 30.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular-Dynamics Algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 31.Darden T, York D, Pedersen L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 32.Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13(8):952–962. [Google Scholar]
- 33.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, KalÈ L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daura X, van Gunsteren WF, Mark AE. Folding-unfolding thermodynamics of a beta-heptapeptide from equilibrium simulations. Proteins. 1999;34(3):269–280. doi: 10.1002/(sici)1097-0134(19990215)34:3<269::aid-prot1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Baron R, McCammon JA. Dynamics, hydration, and motional averaging of a loop-gated artificial protein cavity: the W191G mutant of cytochrome c peroxidase in water as revealed by molecular dynamics simulations. Biochemistry. 2007;46(37):10629–10642. doi: 10.1021/bi700866x. [DOI] [PubMed] [Google Scholar]
- 37.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput- Aided Mol Des. 2008;22(9):693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 39.Durrant JD, McCammon JA. HBonanza: a computer algorithm for molecular-dynamics-trajectory hydrogen-bond analysis. J Mol Graphics Modell. 2011;31:5–9. doi: 10.1016/j.jmgm.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25(5):621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nat Biotechnol. 1996;14(5):595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43. [accessed April 17, 2012];DelEnsembleElec. http://amarolab.ucsd.edu/delensembleelec.html.
- 44.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent ions. J Phys Chem B. 2001;105(28):6507–6514. [Google Scholar]
- 45.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 46.Durrant JD, de Oliveira CA, McCammon JA. POVME: an algorithm for measuring binding-pocket volumes. J Mol Graphics Modell. 2011;29(5):773–776. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Case DA, TAD, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. Amber11. University of California-San Francisco; San Francisco, CA: 2010. [Google Scholar]
- 48.Onufriev A, Bashford D, Case DA. Exploring protein native states and large scale conformational changes with a modified generalized born model. Proteins. 2004;55(2):383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 49.Weiser JSPS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J Comput Chem. 1999;20(2):217–230. [Google Scholar]
- 50.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol. 2004;343(3):601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 51.Kortvelyesi T, Dennis S, Silberstein M, Brown L, 3rd, Vajda S. Algorithms for computational solvent mapping of proteins. Proteins. 2003;51(3):340–351. doi: 10.1002/prot.10287. [DOI] [PubMed] [Google Scholar]
- 52.Landon MR, Lieberman RL, Hoang QQ, Ju S, Caaveiro JM, Orwig SD, Kozakov D, Brenke R, Chuang GY, Beglov D, Vajda S, Petsko GA, Ringe D. Detection of ligand binding hot spots on protein surfaces via fragment-based methods: application to DJ-1 and glucocerebrosidase. J Comput-Aided Mol Des. 2009;23(8):491–500. doi: 10.1007/s10822-009-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landon MR, Lancia DR, Jr, Yu J, Thiel SC, Vajda S. Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J Med Chem. 2007;50(6):1231–1240. doi: 10.1021/jm061134b. [DOI] [PubMed] [Google Scholar]
- 54.Ringpis GE, Aphasizheva I, Wang X, Huang L, Lathrop RH, Hatfield GW, Aphasizhev R. Mechanism of U insertion RNA editing in trypanosome mitochondria: the bimodal TUTase activity of the core complex. J Mol Biol. 2010;399(5):680–695. doi: 10.1016/j.jmb.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303(5665):1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 56.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 57.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim Biophys Acta. 2008;1779(4):270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ringpis GE, Stagno J, Aphasizhev R. Mechanism of U-insertion RNA editing in trypanosome mitochondria: characterization of RET2 functional domains by mutational analysis. J Mol Biol. 2010;399(5):696–706. doi: 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.