Abstract
De novo genomic aberrations are considered an important cause of autism spectrum disorders. We describe a de novo 380-kb gain in band p22.3 of chromosome 7 in a patient with Asperger syndrome. This duplicated region contains 9 genes including the LNFG gene that is an important regulator of NOTCH signaling. We suggest that this copy number variation has been a contributive factor to the occurrence of Asperger syndrome in this patient.
Key Words: Autism spectrum disorder, Chromosome 7p22.3, Duplication, Gain, Lunatic fringe
Introduction
Asperger syndrome (OMIM 608638) is one of the autism spectrum disorders (ASDs). Patients with Asperger syndrome have qualitative impairment in social interactions and show restricted, repetitive and stereotyped patterns of behavior, interests, and activities. It differs from autism because of the higher cognitive abilities and the absence of qualitative impairments in communication [American Psychiatric Association, 2000]. ASDs are highly heritable. Based on twin and family studies the heritability is estimated to be as high as 80% [Freitag, 2007; Ronald and Hoekstra, 2011]. However, as with other complex disorders, in many patients the cause of the ASD remains unknown. One explanation for this missing heritability is the occurrence of de novo aberrations [Manolio et al., 2009; Vissers et al., 2010; O'Roak et al., 2011]. Indeed, rare de novo and inherited copy number variations (CNVs) as well as de novo point mutations are an important cause of autism [Sebat et al., 2007; Pinto et al., 2010; Levy et al., 2011; O'Roak et al., 2011; Sanders et al., 2011]. Here, we present a de novo CNV in 7p22.3 in a patient with Asperger syndrome.
Methods
Single-Nucleotide Polymorphism Array
Genome-wide single-nucleotide polymorphism (SNP) array analysis was performed on DNA isolated from peripheral blood using a 250K SNP array (Affymetrix Inc., Santa Clara, CA, USA) according to the protocol provided by the manufacturer. Data analysis and interpretation was done as previously described [de Leeuw et al., 2011].
Results
Patient Description
The patient, a 14-year-old boy, had been referred to our department because of Asperger syndrome and chronic fatigue. He was the first child of healthy, non-consanguineous parents, born after an uneventful pregnancy and delivery at 40 weeks of gestation with a weight of 3,220 g (30th centile). He developed normally. He could sit at the age of 5.5 months, walk at the age of 1 year, and said his first words appropriately for age. Before the age of 6 years, behavioral problems became evident. He showed obsessive behavioral patterns, such as repeated on and off switching of light and hiding of keys. Moreover, he often rocked back and forth and had difficulty adjusting to unexpected situations. At the age of 6 years, his parents took him to a child and adolescent psychiatrist, who subsequently diagnosed him with Asperger syndrome. The patient followed normal education, though with some extra coaching. His intelligence quotient (IQ) was estimated to be 102 (WISC-III), although there was a discrepancy between his verbal IQ (109) and performance IQ (94). At the age of 12 years, the patient sought medical help due to chronic fatigue. He also complained of myalgia and pain in his legs. Extensive clinical and laboratory investigations, including metabolic disease screening, were normal. The fatigue symptoms were so severe that the patient was referred for an inpatient rehabilitation program. He was discharged after 9 weeks of rehabilitation and transferred to a special secondary school for children with physical disabilities. At the age of 14 years, the patient had returned to a normal school curriculum, though he was still functioning at a lower academic level than he used to do. His further medical history revealed no abnormalities. He had a normal sleeping pattern, normal hearing and vision, and he used to take part in normal sports activities, without any restrictions.
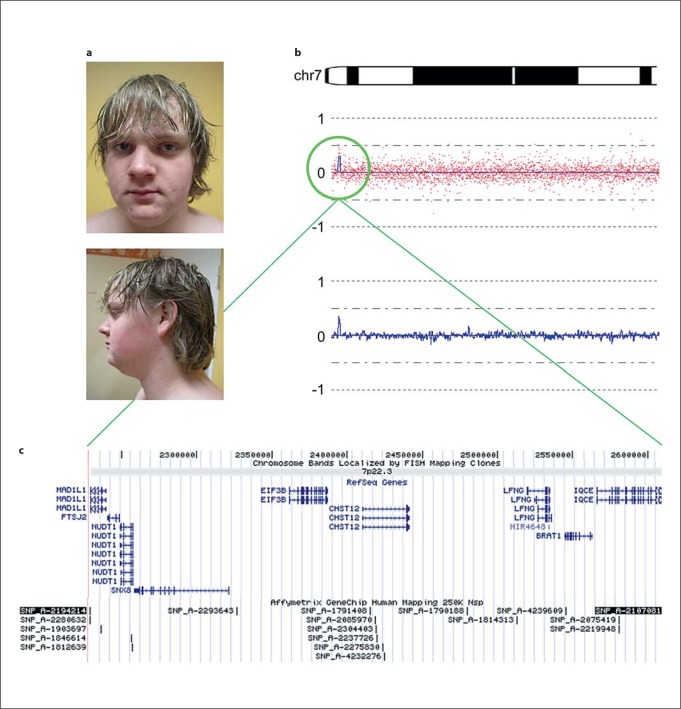
Physical examination performed at the age of 14 years showed a length of 186 cm (98th centile), a weight of 95 kg (+2.5 SD), and a head circumference of 61 cm (98th centile). There were no facial dysmorphisms, except for a broad nasal base (see fig. 1a). The muscle strength was normal, despite the poor physical condition.
Fig. 1.
a Facial features of the patient; b idiogram of the distal short arm of chromosome 7p22.3–p15.2 with an interstitial 380-kb gain in 7p22.3 detected by 250K SNP array shown in the lower part of this figure; c screen shot of the UCSC Genome Browser (Human Genome, February 2009 (GRCh37/Hg19) Assembly at http://genome.ucsc.edu/) showing the duplicated region in 7p22.3 encompassing a total of 9 coding genes.
The patient had a younger brother who developed normally. His father was 184 cm (50th centile) with a head circumference of 58 cm (50th centile). His mother was 173 cm (70th centile) with a head circumference of 57 cm (85th centile). A son of a paternal aunt was diagnosed with pervasive developmental disorder not otherwise specified, and a female second cousin on the paternal side (part of a large family) was also diagnosed with Asperger syndrome.
SNP array analysis revealed an interstitial gain in band p22.3 of chromosome 7 with a minimal size of 380 kb (18 SNPs, 2.26–2.64 Mb from the p-telomere, SNP_A-2194214 to SNP_A-2107081) and a maximal size of 450 kb (2.23–2.68 Mb from the p-telomere, SNP_A-1914583 to SNP_A-1785979) (NCBI Genome Build 37/Hg19) (fig. 1b). The duplicated region contains at least 9 known RefSeq genes, namely MAD1L1, FTSJ2, NUDT1, SNX8, EIF3B, CHST12, LFNG, BRAT1, and IQCE (fig. 1c). The absence of the same gain in the SNP array analyses of DNA from the parents revealed that the gain had occurred de novo in their son.
Discussion
We describe a de novo gain in 7p22.3 in a patient with Asperger syndrome. This region has never been associated with ASDs previously [Risch et al., 1999; Szatmari et al., 2007; Yang and Gill, 2007; Glessner et al., 2009; Pinto et al., 2010; Sanders et al., 2011]. In the Decipher (http://decipher.sanger.ac.uk/) and ECARUCA (www.ecaruca.net) databases only much larger overlapping aberrations have been described that are usually part of complex rearrangements in patients with primarily intellectual disability (ID) [Feenstra et al., 2006; Firth et al., 2009].
In healthy control persons 2 overlapping duplications of the 7p22 region have been described in the Study of Addiction: Genetics and Environment (SAGE, http://zork.wustl.edu/gei/) and the study by Park et al. [2010]. Although the controls of the SAGE study have been selected to be not addicted, ASDs were not excluded in these controls. The latter is similar for the study by Park et al. [2010]. In a recent study on autism by Sanders et al. [2011], 2 partially overlapping duplications have been found in healthy parents that were not present in their own autistic children. These duplications contained the genes MAD1L1, FTSJ2, NUDT1, SNX8, and EIF3B. However, the duplicated region in our patient contained 4 additional known RefSeq genes, namely CHST12, LFNG, BRAT1, and IQCE.
The most likely candidate gene for ASDs is LFNG, or lunatic fringe. In animal models, Lfng is essential for the demarcation of boundaries between groups of cells during development, for example in dorsoventral patterning of the Drosophila wing, the hindbrain and the somites [Irvine and Wieschaus, 1994; Johnston et al., 1997; Zeltser et al., 2001; Tossell et al., 2011]. In humans an autosomal recessive mutation in LFNG has been detected in a family with spondylocostal dysostosis, a vertebral malsegmentation disorder [Sparrow et al., 2006]. In the developing brain, Lfng is particularly expressed in the ventricular zone of the neuroepithelium where neurogenesis takes place [Johnston et al., 1997; Ishii et al., 2000]. LFNG is a glycosyltransferase that modulates the activity of the Notch signaling pathway [Panin et al., 1997; Moloney et al., 2000; Stanley and Okajima, 2010]. During neural development Notch plays an essential role in the regulation of cell fate choice through lateral inhibition [Cau and Blader, 2009]. It effectuates the maintenance of neuronal progenitors and the inhibition of neuronal differentiation in the ventricular zone [Yoon and Gaiano, 2005; Louvi and Artavanis-Tsakonas, 2006]. Both loss-of-function of lfng in zebrafish and Notch1 in mice leads to an early increase in the number of differentiating neurons leading to a reduction in the size of brain structures as a result of secondary progenitor pool depletion [Lutolf et al., 2002; Yoon and Gaiano, 2005; Nikolaou et al., 2009], while overexpression results in an increase of neuronal progenitor cells at the expense of differentiating cells [Gaiano et al., 2000; Breunig et al., 2007; Li et al., 2008; Nikolaou et al., 2009; Kato et al., 2010]. The latter may in turn result in an enlargement of brain structures [de Bellard et al., 2007]. Interestingly, ASDs are associated with an enlarged brain volume (and head circumference) [Kanner, 1943; Piven et al., 1995; Woodhouse et al., 1996]. One of the possible mechanisms to explain this increase in brain volume is an increase in neuron number [Vaccarino et al., 2009; Rubenstein, 2011]. Therefore, activation of the Notch signaling pathway might be an interesting candidate mechanism for the ASD in our patient. This is supported by the large head circumference that was present in the patient, but not in his parents. Besides, the Notch signaling pathway has been previously described to be differentially expressed in patients with ASDs [Seno et al., 2011]. In addition, a causal link with the Notch signaling pathway has been hypothesized in other psychiatric diseases, including schizophrenia, and cognition [Costa et al., 2005; Wang et al., 2006]. Therefore, it seems probable that an increased dosage of LFNG through this duplication may result in an increased brain volume and an enlarged head circumference. This may have predisposed the patient to an ASD.
Of the other 8 genes that were duplicated in the patient, 3 could also be possible candidates for Asperger syndrome, namely IQCE, FTSJ2, and EIF3B. A related gene of these 3 genes has previously been associated with a neurobehavioral phenotype. IQCE contains an IQ domain [Rhoads and Kenguele, 2005], while IQSEC2, another IQ domain containing protein, is a known X-linked ID gene [Shoubridge et al., 2010]. The homolog of FTSJ2, FTSJ1, is also a known gene for nonsyndromic X-linked ID [Freude et al., 2004; Ramser et al., 2004]. In addition, duplications of Xp11.22–p11.23, including FTSJ1, have been described in patients with ID, autistic behavior and seizures. However, autistic features have also been described in patients with smaller duplications that do not overlap FTSJ1 [El-Hattab et al., 2011]. EIF3B encodes for a subunit of one of the family members of the eukaryotic initiation factors that regulate mRNA translation [Maitra et al., 1982; Hinnebusch, 2006]. Deregulation of EIF4E expression, another family member of the eukaryotic initiation factors, has been described in patients with severe autism [Neves-Pereira et al., 2009], while EIF2B subunit mutations are found in patients with vanishing white matter [Leegwater et al., 2001]. Besides, EIF3 has recently been shown to interact with collybistin (ARHGEF9 in humans) and gephyrin, 2 postsynaptic proteins [Sertie et al., 2010]. Defects in synaptic function have already been implicated in the pathogenesis of autism [Sudhof, 2008; van Spronsen and Hoogenraad, 2010].
However, 2 partially overlapping duplications have already been found in healthy parents of an autistic child (and not in the child itself) as mentioned before. One of these duplications encompasses both FTSJ2 and EIF3B, which makes their relation to this phenotype less likely. Being that both IQSEC2 and FTSJ1 have been described in ID, while our patient had Asperger syndrome with a normal IQ, this attenuates a possible link between the related genes IQCE and FTSJ2 and the phenotype of our patient. Finally, most of these genes cause these diseases by a loss-of-function mechanism, while a duplication was found in this patient.
Two family members on the father's side of the family of the patient also have an ASD. As the prevalence of ASDs is known to be high (in the range of 6–7 per 1,000) [Fombonne, 2009], this might be due to a coincidence. However, ASDs are known to be complex genetic disorders [Folstein and Rosen-Sheidley, 2001]. It has been hypothesized that for ASDs multiple mutations together reach a certain threshold in order to result in the clinical phenotype. It is therefore possible that an increased background risk for the development of an ASD was already present in the patient's family. The gain in 7p22.3 may then be only a contributing factor to the development of Asperger syndrome. This is in line with the finding of a small CNV including LFNG and the neighboring BRAT1 in an anonymous healthy Asian female by Park et al. [2010]. Nevertheless, being that the gain in 7p22.3 occurred de novo in the patient, while his parents were healthy, without any signs of ASDs, a causal link with his phenotype could be postulated.
In conclusion, we describe a de novo submicroscopic gain in 7p22.3 in a patient with Asperger syndrome. We suggest that this CNV, which encompasses 9 genes including LFNG, has been a possible contributive factor to the occurrence of Asperger syndrome in this patient.
Acknowledgements
We are grateful to the patient and his parents for their participation. This work was supported by the European Commission: AnEUploidy Project grant 037627 under FP6 (A.T.V.-v.S., B.B.A.d.V., N.d.L., H.G.B.), and GENCODYS grant 241995 under FP7 (A.T.V.-v.S., B.B.A.d.V.), the Dutch Organisation for Health Research and Development (ZON-MW grant 917-86-319 to B.B.A.d.V.), and Hersenstichting Nederland (B.B.A.d.V.).
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. ed. 4, text revision. American Psychiatric Association: Washington DC; 2000. [Google Scholar]
- Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Blader P. Notch activity in the nervous system: to switch or not switch? Neural Dev. 2009;4:36. doi: 10.1186/1749-8104-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Drew C, Silva AJ. Notch to remember. Trends Neurosci. 2005;28:429–435. doi: 10.1016/j.tins.2005.05.003. [DOI] [PubMed] [Google Scholar]
- de Bellard ME, Barembaum M, Arman O, Bronner-Fraser M. Lunatic fringe causes expansion and increased neurogenesis of trunk neural tube and neural crest populations. Neuron Glia Biol. 2007;3:93–103. doi: 10.1017/S1740925X07000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw N, Hehir-Kwa JY, Simons A, Geurts van Kessel A, Smeets DF, et al. SNP array analysis in constitutional and cancer genome diagnostics – copy number variants, genotyping and quality control. Cytogenet Genome Res. 2011;135:212–221. doi: 10.1159/000331273. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Bournat J, Eng PA, Wu JB, Walker BA, et al. Microduplication of Xp11.23p11.3 with effects on cognition, behavior, and craniofacial development. Clin Genet. 2011;79:531–538. doi: 10.1111/j.1399-0004.2010.01496.x. [DOI] [PubMed] [Google Scholar]
- Feenstra I, Fang J, Koolen DA, Siezen A, Evans C, et al. European cytogeneticists association register of unbalanced chromosome aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur J Med Genet. 2006;49:279–291. doi: 10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, et al. DECIPHER: Database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- Freude K, Hoffmann K, Jensen LR, Delatycki MB, des Portes V, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:305–309. doi: 10.1086/422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Nakamura S, Osumi N. Demarcation of early mammalian cortical development by differential expression of fringe genes. Brain Res Dev Brain Res. 2000;119:307–320. doi: 10.1016/s0165-3806(99)00186-8. [DOI] [PubMed] [Google Scholar]
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- Kato TM, Kawaguchi A, Kosodo Y, Niwa H, Matsuzaki F. Lunatic fringe potentiates Notch signaling in the developing brain. Mol Cell Neurosci. 2010;45:12–25. doi: 10.1016/j.mcn.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Leegwater PA, Vermeulen G, Konst AA, Naidu S, Mulders J, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–388. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Li H, Chang YW, Mohan K, Su HW, Ricupero CL, et al. Activated Notch1 maintains the phenotype of radial glial cells and promotes their adhesion to laminin by upregulating nidogen. Glia. 2008;56:646–658. doi: 10.1002/glia.20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lutolf S, Radtke F, Aguet M, Suter U, Taylor V. Notch1 is required for neuronal and glial differentiation in the cerebellum. Development. 2002;129:373–385. doi: 10.1242/dev.129.2.373. [DOI] [PubMed] [Google Scholar]
- Maitra U, Stringer EA, Chaudhuri A. Initiation factors in protein biosynthesis. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Muller B, Massie D, Williams JH, O'Brien PC, et al. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- Nikolaou N, Watanabe-Asaka T, Gerety S, Distel M, Koster RW, Wilkinson DG. Lunatic fringe promotes the lateral inhibition of neurogenesis. Development. 2009;136:2523–2533. doi: 10.1242/dev.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piven J, Arndt S, Bailey J, Havercamp S, Andreasen NC, Palmer P. An MRI study of brain size in autism. Am J Psychiatry. 1995;152:1145–1149. doi: 10.1176/ajp.152.8.1145. [DOI] [PubMed] [Google Scholar]
- Ramser J, Winnepenninckx B, Lenski C, Errijgers V, Platzer M, et al. A splice site mutation in the methyltransferase gene FTSJ1 in Xp11.23 is associated with non-syndromic mental retardation in a large Belgian family (MRX9) J Med Genet. 2004;41:679–683. doi: 10.1136/jmg.2004.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads A, Kenguele H. Expression of IQ-motif genes in human cells and ASPM domain structure. Ethn Dis. 2005;15:S5–91. [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: A decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2011;52:339–355. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Sertie AL, de Alencastro G, De Paula V, Passos-Bueno MR. Collybistin and gephyrin are novel components of the eukaryotic translation initiation factor 3 complex. BMC Res Notes. 2010;3:242. doi: 10.1186/1756-0500-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge C, Walikonis RS, Gecz J, Harvey RJ. Subtle functional defects in the Arf-specific guanine nucleotide exchange factor IQSEC2 cause non-syndromic X-linked intellectual disability. Small Gtpases. 2010;1:98–103. doi: 10.4161/sgtp.1.2.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet. 2006;78:28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossell K, Kiecker C, Wizenmann A, Lang E, Irving C. Notch signalling stabilises boundary formation at the midbrain-hindbrain organiser. Development. 2011;138:3745–3757. doi: 10.1242/dev.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino FM, Grigorenko EL, Smith KM, Stevens HE. Regulation of cerebral cortical size and neuron number by fibroblast growth factors: implications for autism. J Autism Dev Disord. 2009;39:511–520. doi: 10.1007/s10803-008-0653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, Hoogenraad CC. Synapse pathology in psychiatric and neurologic disease. Curr Neurol Neurosci Rep. 2010;10:207–214. doi: 10.1007/s11910-010-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wei J, Zhang X, Guo Y, Xu Q, et al. A review and re-evaluation of an association between the NOTCH4 locus and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:902–906. doi: 10.1002/ajmg.b.30383. [DOI] [PubMed] [Google Scholar]
- Woodhouse W, Bailey A, Rutter M, Bolton P, Baird G, Le Couteur A. Head circumference in autism and other pervasive developmental disorders. J Child Psychol Psychiatry. 1996;37:665–671. doi: 10.1111/j.1469-7610.1996.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int J Dev Neurosci. 2007;25:69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zeltser LM, Larsen CW, Lumsden A. A new developmental compartment in the forebrain regulated by Lunatic fringe. Nat Neurosci. 2001;4:683–684. doi: 10.1038/89455. [DOI] [PubMed] [Google Scholar]