Abstract
Alzheimer's disease (AD) is linked to the aberrant assembly of the amyloid β-protein (Aβ). The 21AEDVGSNKGA30 segment, Aβ(21-30), forms a turn that acts as a monomer folding nucleus. Amino acid substitutions within this nucleus cause familial forms of AD. To determine the biophysical characteristics of the folding nucleus, we studied the biologically relevant acetyl-Aβ(21-30)-amide peptide using experimental techniques (limited proteolysis, thermal denaturation, urea denaturation followed by pulse proteolysis, electron microscopy) and computational methods (molecular dynamics). Our results reveal a highly stable foldon and suggest new strategies for therapeutic drug development.
Keywords: Amyloid β-protein structural stability, molecular dynamics, Alzheimer's disease, folding nucleus, foldon
Alzheimer's disease (AD) is a fatal neurodegenerative disease postulated to be caused by the aberrant assembly of the amyloid β-protein (Aβ)1. Structure-activity relationship (SAR) studies on Aβ40 and Aβ42 have shown the 21AEDVGSNKGA30 segment, Aβ(21-30), forms a turn that acts as a monomer folding nucleus2. This nucleus was found to be protease resistant, and amino acid substitutions within it that cause familial forms of AD and cerebral amyloid angiopathy (CAA) were shown to alter its stability3. Studies of the isolated Aβ(21-30) decapeptide folding nucleus have shown that this peptide segment behaves as a “foldon,”4 demonstrating protease resistance and conformational characteristics similar to those found in the full-length holoproteins 2;3;5–9. However, these prior studies employed the peptide with free N-terminal amino and C-terminal carboxyl groups. In its native state, i.e., within the Aβ holoprotein, Aβ(21-30) exists in its peptide amide form. The charge neutralization of the peptide termini caused by peptide bonds may affect the conformational dynamics within this region. To resolve this question, we studied the blocked (N-terminal acetyl, C-terminal amide) form of Aβ(21-30) using experimental techniques (limited proteolysis, thermal denaturation, urea denaturation followed by pulse proteolysis, electron microscopy) and computational methods (molecular dynamics). Our results reveal that the blocked peptide is substantially more stable than is the unblocked alloform.
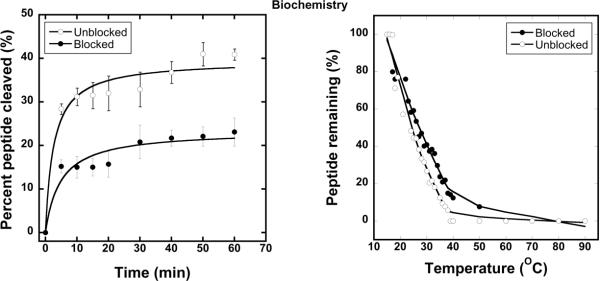
Limited proteolysis is a sensitive probe of folded peptide and protein conformation. We performed limited proteolysis using modified (trypsin-resistant) trypsin (1:100 (w/w) E:S ratio) at room temperature (RT)3. Unblocked Aβ(21-30) was cleaved rapidly, and within 60 min displayed ≈40% cleavage (Fig. 1A). In contrast, blocked Aβ(21-30) was cleaved ≈20% by this time. The cleavage difference was significant (*p<0.001).
Figure 1.
(A) Initial rates of cleavage of Aβ(21–30) by modified trypsin. (B) Thermal denaturation profile of blocked and unblocked Aβ(21–30) probed with thermolysin.
Experiments also were performed with proteinase K, a non-specific protease (Fig. S2). Results from these experiments were similar to those from the trypsin digestion. Initial rates of cleavage, and final cleavage levels, of the unblocked Aβ(21-30) were greater than those of the blocked peptide. These data suggest that turn stability is increased in the blocked peptide, which is interesting considering that Coulombic interactions between the N-terminal amine cation and the C-terminal carboxyl anion might be predicted to stabilize the turn. Such interactions have been shown to be critical in mediating the anti-parallel orientation of Aβ(16–22)10, another important segment of the holopeptide. Instead, our data argue that non-Coulombic interactions, perhaps hydrophobic interactions or H bonding, are more important in turn stabilization. These interactions would be expected to occur in the holoprotein.
We next examined turn stability using pulse proteolysis, a simple method for determining the stability of peptides and proteins that involves digestion of proteins in the unfolded state in equilibrium mixtures in which both folded and unfolded states are populated11; 12. The equilibrium is perturbed systematically by controlled protein denaturation. For Aβ(21-30), thermolysin proteolysis was performed at different temperatures. Results show significant (p<0.001, t-test) differences in thermal denaturation curves (Fig. 1B) between blocked and unblocked Aβ(21-30). The “melting temperature,” T, at which the peptide is 50% folded, was ≈24°C for unblocked Aβ(21-30) and ≈28°C for blocked Aβ(21-30). The increased thermal stability of the blocked peptide is consistent with the results of the limited proteolysis studies.
An orthogonal technique for measuring the conformational stability of proteins is denaturation using chaotropic agents such as guanidinium salts or urea13–15. We monitored the unfolding of the Aβ(21-30) alloforms in urea. Human neutrophil elastase (HNE) was used to reveal the unfolded state because this enzyme retains its activity under denaturing conditions (even in 8M urea).
Pulsing the peptide solutions with HNE for 5 min during the urea-induced unfolding process revealed significant differences between the two peptides (Fig. S1). Blocked Aβ(21-30) displayed ≈8% cleavage in 1M urea, a level that remained essentially constant, within experimental error, over the entire urea concentration range (up to 8 M). Unblocked Aβ(21-30), in contrast, displayed a monotonic increase in cleavage that was proportional to urea concentration (Fig. S1). Approximately 50% cleavage was observed using 8M urea, a highly significant (*p<0.001) difference compared with the blocked peptide.
To determine whether the primary structure differences responsible for the observed differences in stability also affected peptide assembly morphology, transmission electron microscopy (TEM) was used to visualize structures formed by the peptides immediately after their solubilization and after 7 d of incubation. Significant differences in morphologies were seen between the blocked and unblocked peptides.
Unblocked Aβ(21-30) at day 0 formed globular or slightly extended (low aspect ratio) structures of 10–30 nm in diameter that often were associated into small accretions (Fig. S4). The blocked peptide formed larger, more globular structures (40–80 nm diameter), as well as short, cylindrical structures (35–40 nm diameter). These assemblies also tended to accrete.
On day 7, the accretions formed by unblocked Aβ(21-30) were larger than those at day 0, with their composite units displaying diameters of 25–120 nm. Isolated globular assemblies of diameter 20–60 nm also were observed. The blocked peptide formed quasi-spherical (≈60 nm diameter), globular (≈30–65 nm diameter), and thread-like structures (≈20–40 nm diameter). Interestingly, the structures of assemblies formed by the blocked peptide were smaller than those of the unblocked peptide and remained similar in size to the assemblies formed at day 0. One explanation for this observation is increased stability of the folded state of the peptide. Kinetically, this would facilitate rapid self-association (the larger structures observed at day 0) but inhibit formation of more extended structures over time (day 7). This explanation is consistent with the proteolysis data and, as we discuss below, with results of computational studies.
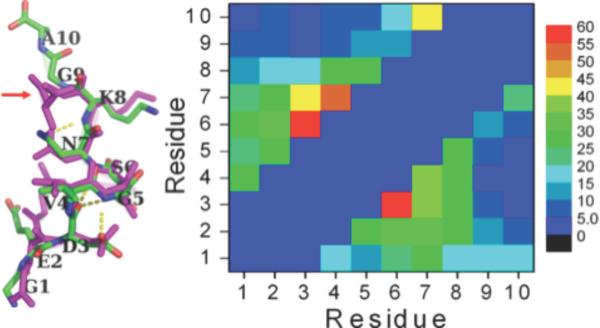
To obtain atomic resolution structural information, we performed all-atom molecular dynamics simulations. Both peptides displayed little regular β-strand or α-helix structure, but each had a significant propensity for turn14 formation (Fig. S3). Overall, residues in the blocked peptide displayed higher tendencies to exist within turns relative to those in the unblocked peptide, with residues 28–30 displaying the largest differences. FIG 2
Figure 2.
Left: the most populated structures of the blocked (magenta; 35%) and unblocked (green, 23%) peptides. Right: the intra-molecular contact maps for the blocked (lower right) and unblocked (upper left) peptides.
We next clustered the collected conformations of each peptide. In Fig. 2 (left), we superimpose the most populated structures of the two peptides (≈35% for the blocked peptide (magenta) and ≈23% for the unblocked peptide (green)). The peptide backbones of residues 1–8 are almost identical conformationally. However, a substantial difference is observed at the C-terminus of the blocked peptide, where the C-terminal Ala bends over to contact residue 7, Asn (Fig. 2, left, arrow). In Fig. 2 (right), we show the intramolecular contact maps of the two peptides. The contours are similar, which suggests that the conformations of the two peptides are similar. However, in blocked Aβ(21-30), additional intramolecular contacts can be observed between residues 3 and 6, 4 and 7, and 7 and 10.
Our simulations provide an explanation of why N-terminal acetylation and C-terminal amidation increase Aβ (21-30) stability. The blocked peptide displays more intramolecular contacts and its turn structure population (≈35%) is larger than that of the unblocked peptide (≈23%). However, their tertiary structures remain similar, as evidenced by the similarity between their contact map, and small RMSD (0.55Å) between their most populated turn structures.
Taken together, the in vitro and in silico data are consistent in their demonstration that acetylation and amidation of Aβ(21-30) significantly increases the stability of the dominant turn conformer. The 21-30 peptide segment has been shown to be the folding nucleus of full length Aβ. Initial studies of the unblocked 21-30 peptide did reveal a metastable turn, a surprising observation for such a short, unmodified (e.g., non-disulfide containing) peptide3. In addition, free peptides containing single amino acid substitutions causing FAD or CAA significantly reduce turn stability, facilitating Aβ assembly3; 6. A key feature of this assembly process is intermolecular interactions between Lys28 and Asp239. Our studies here suggest that the turn formed within full-length Aβ is more stable than previously predicted. This suggestion assumes similar behavior of the isolated decapeptide and the decapeptide segment of the holoproteins. We believe this is a reasonable assumption based on prior work on Aβ40, Aβ42, and the decapeptide, and on the fact that the decapeptide appears to be a foldon, an independent folding unit of a holoprotein2–9. If so, then the development of small molecule stabilizers of this turn element may be of therapeutic value, as has been shown in the transthyretin amyloid system16.
Supplementary Material
Acknowledgments
Funding Sources This work was supported by NIH Grants AG027818, NS038328, AG041295, AG027465, and by the Jim Easton Consortium for Drug Discovery and Biomarkers at UCLA.
ABBREVIATIONS
- Aβ40
amyloid β-protein 40
- Aβ42
amyloid β-protein 42
- Aβ(21-30)
amyloid β-protein 21-30
- SAR
structure activity relationship.
Footnotes
ASSOCIATED CONTENT Detailed experimental procedures, materials and methods and Figures S1– S4. This material is available free of charge via the internet at http://pubs.acs.org.
Author Contributions D.B.T. conceived the work. M.Y. performed and analyzed the computational work. R.R. performed and analyzed the experimental work. M.M.C. worked on peptide design and chemistry. D.B.T., M. Y., and R.R. wrote the manuscript. All authors have approved the final version of the manuscript.
REFERENCES
- 1.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Proc Natl Acad Sci U S A. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Proc Natl Acad Sci U S A. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borreguero JM, Urbanc B, Lazo ND, Buldyrev SV, Teplow DB, Stanley HE. Proc Natl Acad Sci U S A. 2005;102:6015–6020. doi: 10.1073/pnas.0502006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea JE. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarus B, Straub JE, Thirumalai D. J Mol Biol. 2008;379:815–829. doi: 10.1016/j.jmb.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Mousseau N, Derreumaux P. J Chem Phys. 2006;125:084911. doi: 10.1063/1.2337628. [DOI] [PubMed] [Google Scholar]
- 9.Cruz L, Urbanc B, Borreguero JM, Lazo ND, Teplow DB, Stanley HE. Proc Natl Acad Sci U S A. 2005;102:18258–18263. doi: 10.1073/pnas.0509276102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma B, Nussinov R. Proc Natl Acad Sci U S A. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park C, Marqusee S. Curr Protoc Protein Sci. 2006;Chapter 20(Unit 20):11. doi: 10.1002/0471140864.ps2011s46. [DOI] [PubMed] [Google Scholar]
- 12.Marqusee CPS. Nature Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 13.Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsell DA, Sauer RT. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 15.Tanford C. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 16.Peterson SA, Klabunde T, Lashuel HA, Purkey H, Sacchettini JC, Kelly JW. Proc Natl Acad Sci U S A. 1998;95:12956–12960. doi: 10.1073/pnas.95.22.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.