Abstract
Testing for mutations in the KRAS oncogene for patients with metastatic colorectal cancer (mCRC) is generally performed using DNA from formalin-fixed paraffin-embedded tumor tissue; however, access to specimens can be limited and analysis challenging. This study assessed the identification of KRAS mutations in circulating free DNA (cfDNA) using a commercially available KRAS polymerase chain reaction (PCR) kit. Matched plasma, serum and tumor samples were available from 71 patients with mCRC who had received prior therapy but whose disease progressed following therapy. Yields of cfDNA from plasma and serum samples were comparable. Analyses were successful in 70/71 plasma-extracted samples (specificity: 97%, sensitivity: 31%) and 67/71 serum- extracted samples (specificity: 100%, sensitivity: 25%). This study demonstrates that KRAS mutations can be detected in cfDNA using a commercially available KRAS PCR kit, confirming cfDNA as a potential alternative source of tumor DNA in a diagnostic setting if access to archival tumor specimens is limited.
Keywords: KRAS, mutation, cfDNA, colorectal cancer
Introduction
The human oncogene KRAS encodes a G-protein responsible for signal transduction from the epidermal growth factor receptor (EGFR) and other cell-surface proteins to various intracellular targets.1,2 Activating mutations in KRAS are observed in 30%–40% of human colorectal tumors2,3 and are a predictor of poor outcomes in patients with metastatic colorectal cancer (mCRC) who receive treatment with anti-EGFR antibodies alone or in combination with chemotherapy.4–9 Thus patients with mCRC are routinely tested for KRAS mutation before they receive anti-EGFR antibody therapy and only patients with wild-type KRAS should receive such therapy.10,11
Currently, KRAS mutations are usually assessed in DNA isolated from formalin-fixed, paraffin- embedded (FFPE) biopsy or resection samples.12 The isolation of sufficient DNA of adequate quality for biomarker analysis from such FFPE samples is, however, not always possible. The amount of tumor DNA available is often limited.13 Moreover, chemical degradation of DNA can occur in FFPE samples13,14 and delayed fixation and cold ischemia can affect the integrity of DNA samples between removal from the patient and preservation, rendering the quality of the samples unsuitable for biomarker analyses.15
An alternative method to assess KRAS mutation is the analysis of tumor-derived, cell-free or circulating free DNA (cfDNA). Tumor-derived cfDNA can be extracted from plasma or serum and has the potential to be a viable starting material for the identification of genetic markers for diagnosis and recurrence of CRC, and for the early detection of disease recurrence.16–18 The use of tumor-derived cfDNA also offers a number of potential benefits, including the fact that it is a much less invasive technique for mutation detection in comparison with acquisition of FFPE tumor samples. In addition, analysis of cfDNA provides a real-time assessment of KRAS mutation status. Moreover, cfDNA analysis may also provide better representation of the disease as a whole, as it has the potential to yield information about all subclones of a tumor and could contain DNA fragments from distant metastatic sites.17,19 However, it is important to be aware that analysis of cfDNA may not be suitable for assessment of whether patients have wild-type KRAS, since there is a risk of false negative results.
Previous studies have confirmed that cfDNA can be used for the detection of KRAS mutations in CRC.17 However, mutant DNA represents only a small fraction of total cfDNA,16 and therefore is often not assessable using commercially available polymerase chain reaction (PCR) kits. In this study we assessed whether the QIAGEN® therascreen KRAS PCR kit could be modified to enhance sensitivity, without affecting specificity, in the detection of KRAS mutations in cfDNA.
Materials and Methods
Samples
Tumor, plasma, and serum samples for this exploratory biomarker analysis were collected from a cohort of 71 patients as part of a Phase II, randomized, double-blind trial in patients with mCRC that progressed during or after first-line therapy (NCT00278889). Archival diagnostic tumor tissue was collected via biopsy or resection in routine clinical practice, formalin fixed, stored at ambient temperature in pathology laboratories and submitted as pre-study treatment specimens in the course of the Phase II trial. Thus the submitted tumor samples ranged in age from a few months to 11 years prior to the start of this study. DNA was subsequently extracted from the tumor samples approximately 3 years after study initiation. Plasma and serum samples (1 mL) were collected at the start of this study and were stored at −80 °C for approximately 5 years before cfDNA extraction. For both sample types DNA was analyzed immediately after extraction. The study was performed in accordance with the Declaration of Helsinki and was approved by the Institutional Review Boards of the participating centers, including approval for the retrospective analysis of collected samples. All patients provided written, informed consent for provision of plasma, serum, and tumor material.
Role of funding source
This study was funded by AstraZeneca who designed the study and were involved in the collection, analysis, and interpretation of data. The authors (including AstraZeneca employees and Laboratory Corporation of America® employees) were responsible for the writing of the manuscript (with medical writing support as detailed in the acknowledgments section) and the decision to publish and to which journal it should be submitted.
DNA extraction and KRAS mutation analysis
DNA was extracted from FFPE tumor samples using the QIAGEN® QIAamp® DNA FFPE tissue kit according to the manufacturer’s instructions.20 cfDNA was extracted from the plasma and serum samples using the QIAGEN® QIAamp® circulating nucleic acid kit according to the manufacturer’s instructions.21
KRAS mutations in the FFPE-tumor extracted DNA and in the cfDNA were detected using the QIAGEN® therascreen KRAS PCR kit via ARMS™ and Scorpions™ probes.22 This kit utilizes the Scorpions™ technology, which determines the number of PCR cycles necessary to detect a fluorescent signal above the background signal (‘threshold cycle’ [Ct]) as an indicator of the target molecules present in the original sample. The kit includes a control assay (used to assess the total amount of input DNA) and mutation specific or “test” assays. The ΔCt values can then be calculated as the difference between test and control assay Cts. According to the manufacturer, approximately 1% sensitivity is achievable if control Cts are >24 and <29.22 The QIAGEN® therascreen KRAS PCR kit enables the detection of seven KRAS mutations (G12A, G12R, G12D, G12C, G12S, G12V, and G13D; NCBI reference sequence: NM_004985),22 which were all assessed in the FFPE tumor DNA samples.
Serum-and plasma-extracted cfDNA samples were initially assessed in singleton; positive results were subsequently repeated in triplicate in order to verify their mutation status. Samples were classified as ‘mutation positive’ (M+) if they generated a positive result in at least one of the repeat analyses. Data were analyzed using a standard analysis criterion (‘M+ Confidence Level 1’), based on the recommendations for the QIAGEN® therascreen KRAS PCR kit for FFPE tumor samples (ie, 1% ΔCt values applied and diagnostic Ct data analyzed using ≤40 cycles of PCR),22 and three exploratory analysis criteria (M+ Confidence Levels 2, 3, and 4) (Table 1).
Table 1.
Standard and exploratory analysis criteria (M+ Confidence Levels 1–4).
Reduction in assay stringency
|
|
Increasing confidence in results
|
Abbreviations: Ct, threshold cycle; M+, mutation positive.
Statistical analyses
Five test characteristics were used to assess the performance of the KRAS PCR kit in plasma- or serum-extracted cfDNA, relative to its performance in FFPE tumor-extracted DNA, the current ‘gold standard’ for assessing KRAS mutation status in mCRC.
Sensitivity: the proportion of M+ samples identified from tumor-extracted DNA that were classified as M+ in plasma- or serum-extracted cfDNA.
Specificity: the proportion of mutation negative (M−) samples identified from tumor-extracted DNA that were classified as M− in plasma- or serum-extracted cfDNA.
Positive predictive value (PPV): the proportion of samples classified as M+ in plasma- or serum-extracted cfDNA that were identified to be M+ in tumor-extracted DNA (true positives).
Negative predictive value (NPV): the proportion of samples classified as M− in plasma- or serum-extracted cfDNA that were identified to be M− in tumor-extracted DNA (true negatives).
Tumor versus cfDNA concordance: the proportion of total true positive and true negative results out of all samples tested.
Results
FFPE tumor-extracted DNA
FFPE tumor samples for KRAS mutation analysis were available from 109 patients with mCRC. Overall, 108 (99.1%) of the FFPE samples were analyzed successfully; 48 of these (44%) were found to be KRAS M+. This mutation frequency is similar to that seen in other studies.2,3 Of the seven mutations in codons 12 and 13 of KRAS that the kit could detect (G12A, G12R, G12D, G12C, G12S, G12V, and G13D; NCBI reference sequence: NM-004985) all but the G12R mutation were detected (Fig. 1).
Figure 1.
Frequency of specific KRAS mutations in FFPE tumor samples. The PCR kit used in the analysis could detect seven mutations in codons 12 and 13 of KRAS.
Note: Samples from 109 patients were analyzed; data are shown for the 48 M+ samples.
Adaptation of the QIAGEN® therascreen KRAS PCR kit for use with cfDNA
When the QIAGEN® therascreen KRAS PCR kit was used to analyze extracted cfDNA, the mean control Ct was 31–32. According to the manufacturer’s guidance ΔCt values for the seven KRAS mutations should range from 6.5 to 9 for 1% mutant samples. This suggests that mutations present at low levels might be missed if the assay was run for the recommended 40 cycles. Therefore, the assay was run for 50 cycles of PCR to improve the likelihood of detecting mutations. Due to the limited amount of sample, plasma- and serum-extracted cfDNA were not analyzed for the G12R mutation, which was not detected in any FFPE samples.
Utility of cfDNA for KRAS mutation analysis
Plasma- and serum-extracted cfDNA yields
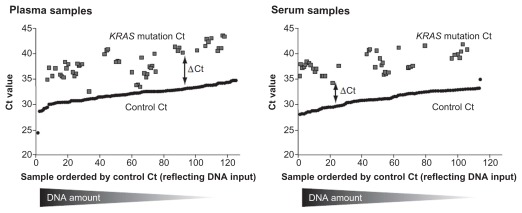
Matched plasma and serum samples were available from 71 patients with analyzed FFPE tumor samples; for the remaining 38 patients the amount of plasma and serum available was insufficient for cfDNA extraction. Control Ct values for plasma- and serum-extracted cfDNA samples, which are indicative of the total DNA yields, were similar (mean: 31.93 and 31.35; range 24 to 35 and 28 to 35, respectively; Figure 2). Mutations of KRAS continued to be detected when control Ct values were high, suggesting that mutations can still be detected even when the overall DNA yield is low (Fig. 2). Average ΔCt values were lower in plasma samples than serum samples, suggesting that the mutation load in plasma was higher (Fig. 2).
Figure 2.
Mutation threshold cycle (Ct) values in plasma- and serum-extracted cfDNA samples.
Plasma-extracted cfDNA versus tumor-extracted DNA
KRAS mutation analysis was successful in 70/71 (98.6%) of the plasma-extracted cfDNA samples. At M+ Confidence Level 1, eight of the samples were positive, all of which were also M+ in the tumor (Table 2). One additional positive plasma sample (negative in the tumor) was identified when the M+ Confidence Level was increased from Level 1 to 2. Detailed analysis of this sample suggested that this was likely to be a true positive result even though no mutation was detected in the matched tumor: ΔCt values were reproducible in four replicates and PCR plots showed exponential amplification (data not shown). In addition, there was no evidence of non-specific amplification at or near to the relevant cycle number in any other samples. At M+ Confidence Level 3, two further positive samples were detected (both of which were positive in the tumor). A false positive (tumor negative) result was detected when the M+ Confidence Level 4 analysis criterion was used and no further positive results were observed where the tumor had generated a positive result.
Table 2.
Comparison of KRAS mutation prevalence between plasma-extracted cfDNA and tumor-extracted DNA samples using four different analysis criteria.
| Plasma (n = 70) | KRAS tumor type (n = 71) | |
|---|---|---|
| Positive | Not detected | |
| Confidence Level 1 | ||
| Test outcome | ||
| Positive | 8 | 0 |
| Unknown | 24 | 39 |
| Sensitivity: 0.25 | Specificity: 1 | |
| PPV: 1 | NPV: 0.62 | |
| Confidence Levels 1 and 2 | ||
| Test outcome | ||
| Positive | 8 | 1 |
| Unknown | 24 | 38 |
| Sensitivity: 0.25 | Specificity: 0.97 | |
| PPV: 0.89 | NPV: 0.61 | |
| Confidence Levels 1, 2, and 3 | ||
| Test outcome | ||
| Positive | 10 | 1 |
| Unknown | 22 | 38 |
| Sensitivity: 0.31 | Specificity: 0.97 | |
| PPV: 0.91 | NPV: 0.63 | |
| Confidence Levels 1, 2, 3, and 4 | ||
| Test outcome | ||
| Positive | 10 | 2 |
| Unknown | 22 | 37 |
| Sensitivity: 0.31 | Specificity: 0.95 | |
| PPV: 0.83 | NPV: 0.63 | |
In summary, applying less stringent analysis criteria resulted in increased assay sensitivity. However, decreased assay specificity for plasma-extracted cfDNA was observed if the analysis criteria were adjusted too far (Table 2). These data suggest that M+ Confidence Levels 1–3 would be appropriate analysis criteria for detection of KRAS mutations in plasma-extracted cfDNA samples.
Serum-extracted cfDNA versus tumor-extracted DNA
Overall, 67/71 (94.4%) of the serum-extracted cfDNA samples were analyzed successfully. Assay sensitivity for serum-extracted cfDNA increased as the stringency of the analysis criteria was reduced (Table 3). This effect was particularly marked when the M+ Confidence Level was increased from Level 1 to 2; in this instance, an extra three positive samples were detected, all of which were also M+ in the tumor (Table 3). One additional positive sample was observed when the stringency of the analysis criteria was reduced to M+ Confidence Level 4 (Table 3). However, in order to avoid false positives and in line with the plasma sample data, we judged that M+ Confidence Levels 1–3 would be appropriate analysis criteria for the detection of KRAS mutations in serum-extracted cfDNA samples.
Table 3.
Comparison of KRAS mutation prevalence between serum-extracted cfDNA and tumor-extracted DNA samples using four different analysis criteria.
| Serum (n = 67) | KRAS tumor type (n = 71) | |
|---|---|---|
| Positive | Not detected | |
| Confidence Level 1 | ||
| Test outcome | ||
| Positive | 5 | 0 |
| Unknown | 27 | 39 |
| Sensitivity: 0.16 | Specificity: 1 | |
| PPV: 1 | NPV: 0.59 | |
| Confidence Levels 1 and 2 | ||
| Test outcome | ||
| Positive | 8 | 0 |
| Unknown | 24 | 39 |
| Sensitivity: 0.25 | Specificity: 1 | |
| PPV: 1 | NPV: 0.62 | |
| Confidence Levels 1, 2, and 3 | ||
| Test outcome | ||
| Positive | 8 | 0 |
| Unknown | 24 | 39 |
| Sensitivity: 0.25 | Specificity: 1 | |
| PPV: 1 | NPV: 0.62 | |
| Confidence Levels 1, 2, 3, and 4 | ||
| Test outcome | ||
| Positive | 9 | 0 |
| Unknown | 23 | 39 |
| Sensitivity: 0.28 | Specificity: 1 | |
| PPV: 1 | NPV: 0.63 | |
Singleton analyses
Serum-and plasma-extracted cfDNA samples were initially assessed in singleton; positive results were subsequently repeated in triplicate in order to verify their mutation status. The results from the initial singleton analyses showed that, for both sample types, sensitivity increased but specificity decreased when the stringency of analysis criteria was reduced from those recommended in the kit (ie, 1% ΔCt and diagnostic Ct ≤ 40).22 This effect was most marked for plasma-extracted cfDNA samples (Table 4). If it is only possible to perform KRAS mutation analysis in singleton, the kit guidelines should be used to ensure detection of false positives is avoided.
Table 4.
Assay sensitivity and specificity in plasma- and serum-extracted cfDNA samples using different analysis criteria (singleton analysis).
| M+ Confidence Level | ||||
|---|---|---|---|---|
| 1 | 1 and 2 | 1, 2, and 3 | 1, 2, 3, and 4 | |
| Plasma | ||||
| Sensitivity | 0.25 | 0.28 | 0.34 | 0.38 |
| Specificity | 1 | 0.90 | 0.90 | 0.85 |
| Serum | ||||
| Sensitivity | 0.19 | 0.25 | 0.28 | 0.34 |
| Specificity | 1 | 0.97 | 0.97 | 0.97 |
Abbreviation: M+, mutation positive.
Preferred medium for KRAS mutation detection
Plasma- versus serum-extracted cfDNA
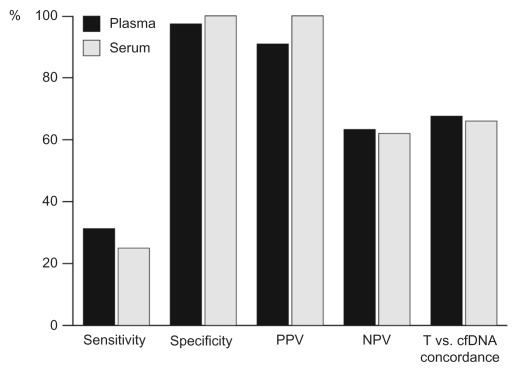
To determine which cfDNA sample type was the preferred medium for KRAS mutation detection, assay data for plasma- and serum-extracted cfDNA were compared at M+ Confidence Level 3. At this level, plasma-extracted cfDNA samples had marginally higher sensitivity (31% vs. 25%) but similar specificity (97% vs. 100%) compared with serum-extracted cfDNA samples (Fig. 3). No meaningful differences were observed between sample types in NPV or the proportion of concordance (the rate at which cfDNA and tumor sample results were in agreement) (Fig. 3). Overall, more KRAS mutations were detected in plasma-extracted samples (10 mutations) than in those from serum (8 mutations). Seven mutations were detected in both plasma- and serum-extracted cfDNA samples.
Figure 3.
Comparison of parameters determining KRAS mutation detection utility between cfDNA plasma and serum samples (M+ Confidence Level 3).
Abbreviations: NPV, negative predictive value; PPV, positive predictive value; T, tumour.
Discussion
This study demonstrates that a commercially available QIAGEN® therascreen KRAS PCR kit can be used for the identification of KRAS mutation positive CRC from plasma and serum samples. It also shows that modified analysis criteria can be used to increase assay sensitivity without reducing specificity. Analysis success rates for plasma-and serum- extracted cfDNA samples were high (>94%) and mutations in KRAS were detected in both sample types. The data also suggest that extracting cfDNA from plasma is associated with a marginal improvement in assay sensitivity compared with cfDNA from serum.
The KRAS PCR kit had high specificity when used with cfDNA (>97% at M+ Confidence Levels 1–3), which is consistent with other studies in CRC that have compared KRAS testing results from cfDNA with those from tumor samples.23,24 By adjusting the recommended analysis criteria of the PCR assay, we were able to increase the sensitivity for detection of KRAS mutations in the plasma- and serum-extracted cfDNA samples. Nevertheless, we found that sensitivity in both sample types was lower than expected.25 One reason for this might be the limited sample volume (1 mL), which had to suffice for testing in six different assays plus confirmatory analysis. Larger volumes of plasma/serum, which could feasibly be obtained in a diagnostic setting, might be needed to further increase assay sensitivity. Importantly, the plasma and serum samples analyzed in this study were derived from patients whose tumors had been resected at a time point ranging from a few months to 11 years prior to the study, and who had previously received first-line anti-cancer therapy. Despite this, KRAS mutations were still detected in the cfDNA of some patients. It is therefore possible to speculate that in a diagnostic setting prior to tumor resection the ability to detect KRAS mutations in cfDNA might be further enhanced.
A marked increase in assay sensitivity was observed among serum-extracted but not plasma-extracted cfDNA samples when analysis criteria were reduced from M+ Confidence Level 1 to Level 2. This might be explained by a lower mutation load in serum samples. Serum-derived cfDNA can be contaminated with wild-type DNA from white blood cells, due to lysis of these cells during the sample collection.
Repeat analysis of one plasma-extracted cfDNA sample demonstrated a reproducible KRAS M+ result when no mutation was detected in the FFPE tumor sample. Due to insufficient amounts of cfDNA it was not possible to verify the additional positive result with alternative methods; the tumor DNA was sequenced and no evidence of mutation was found. This additional positive plasma result might reflect tumor heterogeneity. Another possibility is that the detected mutation occurred later in disease progression, after the tumor sample was taken as a biopsy sample for diagnosis. Furthermore, it should be taken into account that patients received first-line therapy for CRC according to their local standard of care before serum and plasma samples were collected, though the impact of chemotherapy or radiation on the KRAS mutation status in CRC is still unclear.2 In addition, testing of cfDNA is theoretically representative of all tumor sites and may capture mutations occurring at sites other than the primary tumor cfDNA. Indeed, discordance in the KRAS mutation status of primary and metastatic tumors has previously been shown to occur in CRC patients, including an example of a wild-type primary tumor with a KRAS mutation in the metastasis.26
In summary, the higher specificity observed in plasma-extracted compared with serum-extracted cfDNA samples suggested that future mutation analyses might be preferentially conducted in plasma samples. In addition, based on sensitivity and specificity data of the plasma- and serum-derived cfDNA samples, we believe that future mutation analysis based on M+ Confidence Level 3 criteria might be appropriate. However, for singleton analysis (if insufficient cfDNA is available for re-analysis) assay kit guidelines and criteria for identifying M+ samples should be adhered to in order to avoid the detection of false positive results.
It is also important to note that, although high PPV gives confidence in predicting positive results, low NPV indicates that many mutations are missed in cfDNA samples. Therefore, analysis of the KRAS mutation status in serum- or plasma-extracted cfDNA cannot be used for the selection of KRAS mutation negative mCRC and a more sensitive assay is needed to accurately identify patients with wild-type KRAS mCRC.
In conclusion, this exploratory study with matched tumor, serum, and plasma samples from patients with mCRC demonstrated that a commercially available assay kit intended for use on FFPE CRC tissue can be used to detect KRAS mutations in plasma- or serum-extracted cfDNA. For the cfDNA samples, assay sensitivity was increased by modification of existing assay kit guidelines. Further verification of the revised analysis criteria is required, but the initial results presented here indicate that cfDNA may be an important source of tumor DNA in a diagnostic setting, especially if access to archival tumor specimens is limited. In addition, the use of cfDNA may avoid some of the inherent technical challenges associated with DNA analysis of FFPE tumor samples.
Footnotes
Author Contributions
Conceived and designed the experiments: SM, ED, JS. Analysed the data: JW, ME, EK, LKM. SM, ED, JW were responsible for preparation of the manuscript. All authors agreed conclusions, reviewed and approved the final manuscript.
Competing Interests
SRM, JW, and ED are employees of AstraZeneca. JS is a former employee of AstraZeneca and is now retired. MTE, EK, and LK-M are employees of Laboratory Corporation of America® Holdings. Results from this study were previously presented in an abstract and poster at the 6th Joint Meeting of the British Division of the International Academy of Pathology and the Pathological Society of Great Britain & Ireland, heldin Ghent, Belgium, 10–13 May, 2011.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
This study was sponsored by AstraZeneca. Medical writing support was provided by Rick Flemming, PhD, CMPP, on behalf of Complete Medical Communications and was funded by AstraZeneca.
References
- 1.Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:71–119. doi: 10.1016/s0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- 2.Plesec TP, Hunt JL. KRAS mutation testing in colorectal cancer. Adv Anat Pathol. 2009;16:196–203. doi: 10.1097/PAP.0b013e3181a9d4ed. [DOI] [PubMed] [Google Scholar]
- 3.van Krieken JH, Jung A, Kirchner T, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–31. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 4.De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508–15. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 5.Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–9. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 7.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 8.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 9.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 10.Van Custem E, Nordlinger B, Cervantes A. Advanced colorectal cancer: ESMO Clinical Practice Guidelines for treatment. Ann Oncol. 2010;21(Suppl 5):v93–7. doi: 10.1093/annonc/mdq222. [DOI] [PubMed] [Google Scholar]
- 11.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology Provisional Clinical Opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 12.Richman SD, Chambers P, Seymour MT, et al. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol (Amst) 2011;34:61–6. doi: 10.3233/ACP-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Carlton VE, Karlin-Neumann G, et al. High quality copy number and genotype data from FFPE samples using Molecular Inversion Probe (MIP) microarrays. BMC Med Genomics. 2009;2:8. doi: 10.1186/1755-8794-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Womack C, Mager R, Greywoode G, Revill M, Kerr D, Gray N. The effect of clinical tissue sample cold ischaemia time on immunohistochemical analysis: towards a panel to check tissue sample quality. Biopreserv Biobank. 2009;7:51. [Google Scholar]
- 16.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecomte T, Ceze N, Dorval E, Laurent-Puig P. Circulating free tumor DNA and colorectal cancer. Gastroenterol Clin Biol. 2010;34:662–81. doi: 10.1016/j.gcb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 19.Fleischhacker M, Schmidt B. Cell-free DNA resuscitated for tumor testing. Nat Med. 2008;14:914–5. doi: 10.1038/nm0908-914. [DOI] [PubMed] [Google Scholar]
- 20.QIAGEN. QIAmp® DNA FFPE Tissue Handbook. [Google Scholar]
- 21.QIAGEN. QIAmp® Circulating Nucleic Acid Handbook. Second Edition [Google Scholar]
- 22.QIAGEN. Therascreen® KRAS PCR Kit Handbook (Version 1) pp. 1–40. [Google Scholar]
- 23.Kopreski MS, Benko FA, Borys DJ, Khan A, McGarrity TJ, Gocke CD. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92:918–23. doi: 10.1093/jnci/92.11.918. [DOI] [PubMed] [Google Scholar]
- 24.Mora J, Urgell E, Farré A, Comas L, Montserrat E, González-Sastre F. Agreement between K-ras sequence variations detected in plasma and tissue DNA in pancreatic and colorectal cancer. Clin Chem. 2006;52:1448–9. doi: 10.1373/clinchem.2006.067140. [DOI] [PubMed] [Google Scholar]
- 25.Lecomte T, Berger A, Zinzindohoué F, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542–8. doi: 10.1002/ijc.10526. [DOI] [PubMed] [Google Scholar]
- 26.Knijn N, Mekenkamp LJ, Klomp M, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–6. doi: 10.1038/bjc.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]