Abstract
FOXP3+ regulatory T (Treg) cells are a broadly acting and potent anti-inflammatory population of CD4+ T cells essential for maintaining immune homeostasis and preventing debilitating autoimmunity. Based on chemokine receptor expression, we identified distinct populations of Treg cells in human blood expected to colocalize with different Th cell subsets. Although each population was functionally suppressive, they displayed unique patterns of pro- and anti-inflammatory cytokine production, differentially expressed lineage-specifying transcription factors, and responded differently to antigens associated with Th1 and Th17 responses. These results highlight a previously unappreciated degree of phenotypic and functional diversity in human Treg cells that allows subsets with unique specificities and immunomodulatory functions to be targeted to defined immune environments during different types of inflammatory responses.
Introduction
The quality of the immune response against a given pathogen depends on the function of antigen-specific CD4+ effector T cells. Because the mechanisms used to eliminate or control different types of pathogens can vary widely, it is not surprising that these effector T cells are functionally heterogeneous and can be divided into distinct subsets defined by the cytokines they produce and the transcription factors essential for their differentiation.1 IFN-γ–producing Th1 cells require the lineage-specifying transcription factor T-bet for their differentiation and help to eliminate intracellular pathogens, whereas IL-4–producing Th2 cells express the transcription factor GATA-3 and help to expel large, extracellular parasites. Likewise, the transcription factors RORγt and RORα are necessary for the development of IL-17–producing Th17 cells that mediate responses to extracellular bacteria and fungi, whereas recently described IL-22–producing Th22 cells are targeted to the skin and may contribute to skin homeostasis and inflammation.2–4
In addition to the cytokines they produce, effector T cells can be distinguished by their differential expression of chemokine receptors that direct them to distinct inflammatory environments. For example, Th1 cells express CXCR3,5,6 whereas Th17 cells express the chemokine receptors CCR6 and CCR4,7–9 which together promote their migration to inflamed tissues during Th17-mediated autoimmunity.10 Moreover, IFN-γ induces the expression of the CXCR3 ligands CXCL9, CXCL10, and CXCL11, and expression of the CCR6 ligand CCL20 is induced by IL-17.11 Therefore, these chemokine receptors are thought to function in positive feedback loops to amplify and segregate Th1 and Th17 cell migration during specific inflammatory responses. More recently, IL-22–producing Th22 cells have been identified, and these express the cutaneous lymphocyte antigen (CLA), a functional E-selectin ligand that is involved in lymphocyte rolling on the endothelial cells of cutaneous postcapillary venules, along with the chemokine receptors CCR6, CCR4, and CCR10, which together facilitate the constitutive migration of these cells to the skin.2
Several mechanisms have evolved to restrain CD4+ T-cell responses to avoid unwanted tissue destruction, immunopathology, and autoimmunity. Among these, CD4+ regulatory T (Treg) cells are characterized by their ability to inhibit T-cell proliferation in vitro and by their constitutive expression of the IL-2 receptor component CD25. Highlighting their essential function in maintaining immune tolerance in vivo, the absence or depletion of Treg cells causes severe autoimmune and inflammatory disease.12 The development of Treg cells depends on the transcription factor FOXP3, which coordinates the expression of genes essential for Treg cell homeostasis and function, and blocks the production of proinflammatory cytokines.13,14
Although Treg cells are generally considered to be a separate lineage of CD4+ T cells, recent murine studies have indicated that they use different transcriptional programs to regulate Th1, Th2, or Th17 responses, and that these are associated with the expression or activation of specific Th-associated transcription factors.15–17 This suggests that, like conventional Th cells, Treg cells differentiate into specialized subsets during different types of immune responses, and that this is essential for the appropriate regulation of different Th cell populations. In addition, Treg cells are found throughout the body in both lymphoid and nonlymphoid tissues and, like conventional effector/memory T cells, they express diverse patterns of chemokine receptors expected to target them to these sites.18 We and others have shown that the ability of Treg cells to maintain immune homeostasis and prevent autoimmunity depends on their appropriate colocalization with effector T cells.19–23 However, the degree of phenotypic and functional concordance between different Th and Treg cell subsets has not been examined carefully and systematically.
To better understand how Treg cells modulate different types of effector T cell responses, we performed a comprehensive phenotypic and functional analysis of human Treg cells directly ex vivo. Based on expression of the chemokine receptors CCR6, CXCR3, CCR4, and CCR10, we identified 4 separable populations of FOXP3+ Treg cells in human peripheral blood that mirror different Th cell subsets phenotypically. Although all of the Treg cell populations showed suppressive activity in vitro, they differed in their production of both pro- and anti-inflammatory cytokines, had distinct expression patterns of Th-associated transcription factors, and proliferated differentially in response to recall antigens associated with either Th1 or Th17 cell responses. Therefore, Th and Treg cells appear to undergo functional specialization in parallel, resulting in the development of Treg cell subsets capable of colocalizing with and effectively regulating different types of Th cell responses in vivo.
Methods
Cell purification and sorting
Blood samples were obtained from healthy donors participating in the Benaroya Research Institute Immune-Mediated Disease Registry. Informed consent was obtained from all subjects according to institutional review board–approved protocols at Benaroya Research Institute and following the Declaration of Helsinki. CD4+CD25high Treg cells were enriched from PBMCs after staining with PE-cyanine 5 (PE-Cy5)–labeled anti-CD25 Ab (BioLegend), followed by positive selection using anti-PE and anti-Cy5 microbeads (Miltenyi Biotec). On the negative fraction, CD4+CD25− Th cells were purified by positive selection with CD4-specific microbeads (Miltenyi Biotec). Memory T-cell subsets were sorted to more than 97% purity as CD4+CD45RO+CD127+CD25− using PerCP/Cy5.5–conjugated anti-CD45RO, Alexa Fluor 488–conjugated anti-CD127, PE-Cy5–conjugated anti-CD25, and Alexa Fluor 700–conjugated anti-CD4 Abs (all from BioLegend). Memory Treg cells were sorted to more than 97% purity as CD4+CD127lo/−CD25highCD45RO+ using PerCP/Cy5.5–conjugated anti-CD45RO, Alexa Fluor 488–conjugated anti-CD127, PE-Cy5–conjugated anti-CD25, and Alexa Fluor 700–conjugated anti-CD4 Abs. Abs used for sorting of memory Th and Treg cell subsets were: biotin-conjugated anti-CCR6 Ab (BD Pharmingen), followed by streptavidin-allophycocyanin-cyanine 7 Ab (streptavidin-APC-Cy7; BioLegend), PE-conjugated anti-CCR10 Ab (R&D Systems), PE-Cy7–conjugated anti-CCR4 Ab (BD Pharmingen), and Alexa Fluor 647–conjugated anti-CXCR3 Ab (BioLegend). Cells were sorted with a FACSAria (BD Biosciences). CD14+ monocytes were isolated from PBMCs by positive selection with CD14-specific microbeads (Miltenyi Biotec). Cells were cultured in RPMI 1640 medium supplemented with 2mM glutamine, 1% (vol/vol) nonessential amino acids, 1% (vol/vol) sodium pyruvate, penicillin (50 U/mL), streptomycin (50 μg/mL; all from Invitrogen), and 5% heat-inactivated human serum.
Intracellular cytokine staining
Intracellular staining for FOXP3, Helios, RORγt, T-bet, IL-22, IL-17, IFN-γ, IL-10, and IL-4 was performed on sorted T cells stimulated for 5 hours with phorbol 12-myristate 13-acetate (PMA) and ionomycin in the presence of brefeldin A (all from Sigma-Aldrich) for the final 2.5 hours of culture. Cells were fixed and permeabilized with fixation/permeabilization solution (eBiosciences) according to the manufacturer's instructions. Cells were stained with eFluor 450–conjugated anti-FOXP3 Ab (eBiosciences), Alexa Fluor 647–conjugated anti-Helios Ab (BioLegend), PE-conjugated anti-RORγt Ab (eBiosciences), Alexa Fluor 647–conjugated anti–T-bet Ab (eBiosciences), PE-conjugated anti–IL-22 Ab (R&D Systems), PE-conjugated anti–IL-10 Ab (BioLegend), PE-conjugated anti–IL-4 Ab (BioLegend), Alexa Fluor 647- or Alexa Fluor 488–conjugated anti–IL-17 Ab (eBiosciences), and FITC- or APC-conjugated anti–IFN-γ Ab (BD Pharmingen and BioLegend, respectively), and then analyzed on an LSRII flow cytometer (BD Biosciences). FACS data were analyzed with FlowJo Version 9.4.4 (TreeStar).
Phenotype analysis of Th and Treg cell subsets
Th and Treg cell subsets were evaluated for expression of CTLA-4, CLA, ICOS, Helios, and Ki-67. Cells were stained with biotin-conjugated anti–CTLA-4 Ab (BD Pharmingen) followed by streptavidin PE-Cy5, FITC-conjugated anti-CLA Ab (BioLegend), FITC- or biotin-conjugated anti-ICOS Ab (BioLegend), FITC-conjugated anti-Helios Ab (BioLegend), and Alexa Fluor 488–conjugated anti–Ki-67 Ab (BD Pharmingen).
Suppression assays
CFSE-labeled (Sigma-Aldrich), sorted 5 × 104 CD4+ CD25− T cells were used as responders and cocultured with 5 × 104 sorted Treg cells. These cells were stimulated in round-bottomed 96-well plates with irradiated 5 × 104 autologous monocytes and anti-CD3 (OKT3). Cellular proliferation was assessed after 4 days by flow cytometry.
In vitro cell expansion
Sorted Th and Treg cell subsets were activated with anti-CD3/anti-CD28–coated microbeads (Invitrogen) at a 1:1 ratio and cultured for 9 days. IL-2 (300 U/mL) was added at day 0 (Treg cells only) and at days 2, 4, 6, and 8. After 9 days of culture, cells were analyzed for cytokine production by intracellular cytokine staining and for transcription factor expression by real-time quantitative RT-PCR.
Real-time quantitative RT-PCR
Total RNA from sorted T-cell subsets was extracted using the RNeasy kit (QIAGEN) and treated with DNase I (QIAGEN) to avoid genomic DNA contamination. cDNA were synthesized with M-MuLV reverse-transcriptase and oligo(dT) primers (Fermentas) and gene expression was examined with the ABI 7500 Fast Real-Time PCR system (Applied Biosystem) using a SYBR Green real-time PCR kit (Fermentas). The data were normalized to β-actin (ACTB) gene expression. The primers used were (5′-3′): RORC forward: GCATGTCCCGAGATGCTGTC and reverse: CTGGGAGCCCCAAGGTGTAG; TBX21 forward: CCGTGACTGCCTACCAGAAT and reverse: ATCTCCCCCAAGGAATTGAC; GATA-3 forward: GAACCGGCCCCTCATTAAG and reverse: ATTTTTCGGTTTCTGGTCTGGAT; FOXP3 forward: CCAGCCATGATCAGCCTCAC and reverse: CCGAAAGGGTGCTGTCCTTC; IL17A forward: GAAGGCAGGAATCACAATC and reverse: GCCTCCCAGATCACAGA; IFNG forward: CCAGGACCCATATGTAAAAG and reverse TGGCTCTGCATTATTTTTC; IL10 forward: CAAATGAAGGATCAGCTGGACAA and reverse: GCATCACCTCCTCCAGGTAAAAC; ACTB forward: GGACTTCGAGCAAGAGATGG and reverse: AGCACTGTGTTGGCGTACAG.
Antigen-specific proliferation assays
CFSE-labeled (Sigma-Aldrich), sorted Th and Treg cell subsets (5 × 104) were cocultured in round-bottomed 96-well plates with irradiated autologous monocytes (5 × 104), which were either unpulsed or pulsed for 3 hours with Candida albicans (Greer Laboratories) or human cytomegalovirus (HCMV; EastCoast Bio) antigens. For Treg cells, anti-CD28 (0.5 μg/mL) and IL-2 (20 U/mL) were added to the cultures. Cellular proliferation was assessed after 5-6 days by flow cytometry. The percentage of CFSElo cells (defined as cells that had undergone 3 or more divisions) is represented after subtraction of the background proliferation with autologous monocytes. In some experiments, after 6 days of culture, cells were restimulated with PMA/ionomycin for 4 hours and their cytokine production was analyzed by flow cytometry.
Statistics
Statistical tests were performed using Prism Version 5 software (GraphPad). Significance was determined by paired 2-tailed Student t test or 1-way ANOVA analysis with Tukey correction, as noted in the figure legends.
Results
Identification of phenotypically distinct subsets of human Treg cells
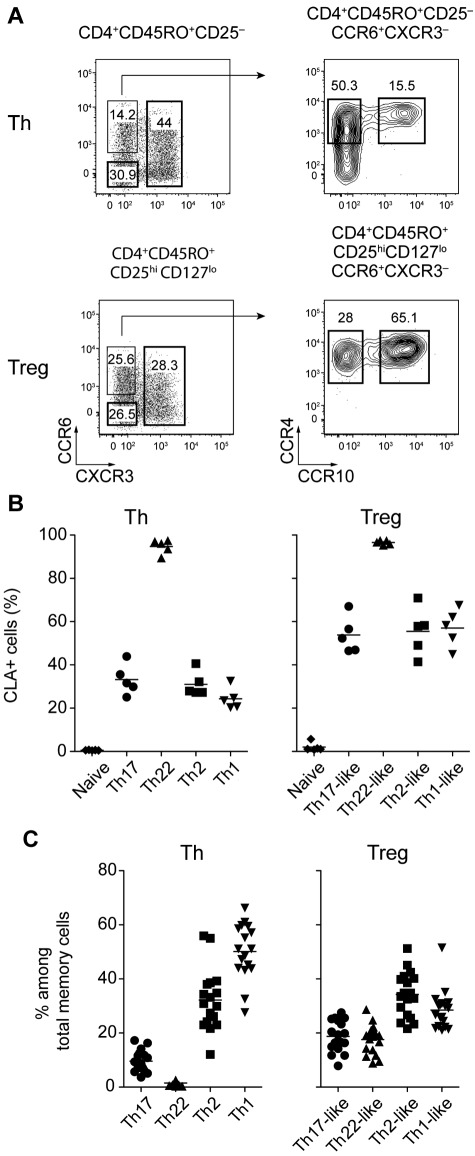
Human effector and memory CD4+ T-cell subsets express different chemokine receptor profiles enabling them to migrate to specific tissues such as the skin and to respond to chemokines produced during different types of inflammatory responses. Based on differential expression of the chemokine receptors CCR6, CXCR3, CCR4 and CCR10, 4 subsets of CD4+CD45RO+CD25−CD127+ Th cells can be defined in the peripheral blood of healthy donors: CXCR3+ Th1 cells that produce IFN-γ, CXCR3−CCR6+CCR4+CCR10− IL-17–producing Th17 cells, CXCR3−CCR6+CCR4+CCR10+ Th22 cells that secrete IL-22 but not IL-17, and CCR6−CXCR3− cells that express CCR4 (data not shown), and are enriched in IL-4–producing Th2 cells (Figure 1A top panels and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Figure 1.
Chemokine receptor expression defines human Treg cell subsets. (A) Representative flow cytometric analysis of CCR6, CXCR3, CCR4, and CCR10 expression by gated CD4+CD45RO+CD25− Th cells (top panels) and CD4+CD45RO+CD25hiCD127lo Treg cells (bottom panels) from peripheral blood. (B) Expression of CLA by the indicated Th and Treg cell subsets. Each symbol represents 1 donor; horizontal bars indicate the mean. Data are from 5 donors. (C) Frequency of cells expressing the indicated chemokine receptor combinations among Th and Treg cells. Each symbol represents 1 donor; horizontal bars indicate the mean. Data are from 15 donors.
By applying a similar gating strategy to ex vivo CD4+CD45RO+CD25hiCD127lo memory Treg cells, we identified 4 distinct Treg cell subsets phenotypically analogous to the Th populations described above (Figure 1A bottom panels). Based on their phenotypic resemblance to the different effector T-cell subsets, we refer to these populations as Th1-, Th17-, Th22-, and Th2-like Treg cells. Underscoring the phenotypic link between the Th and Treg cell subsets, both Th22 cells and Th22-like Treg cells expressed CLA homogeneously, whereas CLA expression in all of the other Th and Treg cell populations was heterogenous (Figure 1B). However, although similar phenotypically, the frequency of each subset differed among the Th and Treg cells. For example, the Th17- and Th22-phenotype cells were found at a substantially higher frequency in the Treg cell compartment, whereas the proportion of CXCR3+ cells was decreased in the Treg cells (Figure 1C). In addition, although a small fraction (approximately 10%) of CD4+CD45RA+CD45RO−CD25hiCD127lo naive Treg cells expressed low levels of CXCR3, expression of CCR6, CCR4, and CCR10 was restricted to CD45RO+ Treg cells (data not shown). Therefore, as in Th cells, the acquisition of these homing receptor phenotypes likely occurs after activation of Treg cells in the periphery.
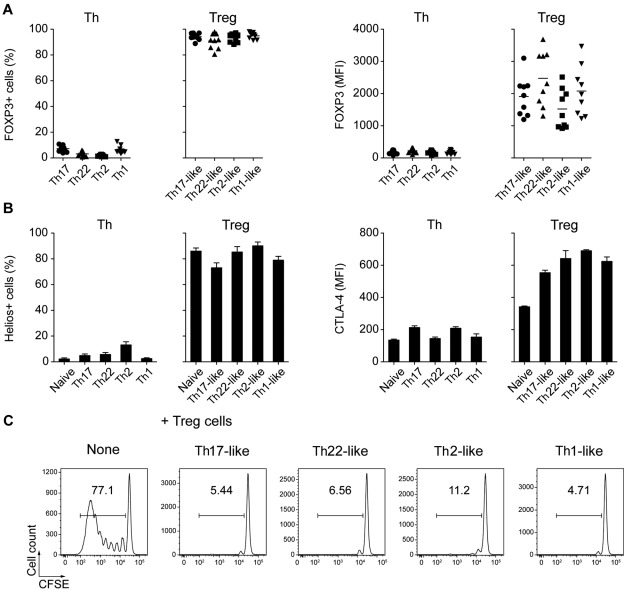
Consistent with their identification as Treg cells, greater than 90% of the cells in each of the CD25hiCD127lo cell populations were FOXP3+ (Figure 2A). In addition, most of these cells expressed the transcription factor Helios, which is thought to be expressed selectively on “natural” Treg cells of thymic origin,24 and high levels of CTLA-4, an inhibitory receptor crucial for the suppressive function of Treg cells in vivo25 (Figure 2B). Moreover, all of the CD25hiCD127lo subsets effectively blocked the proliferation of autologous anti-CD3–stimulated CD4+CD25− T cells effectively, confirming their identity as functionally suppressive Treg cells (Figure 2C).
Figure 2.
Treg cell subsets are functionally suppressive. (A) Expression of FOXP3 by the indicated Th and Treg cell subsets. Each symbol represents 1 donor; horizontal bars indicate the mean. Data are from 9 donors. (B) Expression of Helios (left) and CTLA-4 (right) by the indicated Th and Treg cell subsets. Data are from 3 or 5 donors. (C) CFSE-labeled CD4+CD25− responder T cells were cultured with autologous monocytes plus anti-CD3 (OKT3) in the presence or absence of the indicated sorted Treg cell subset at a 1:1 suppressor/responder ratio. Numbers indicate the frequency of proliferating CFSElo T cells after 5 days of culture. Data are representative of 3 independent experiments.
Coexpression of pro- and anti-inflammatory cytokines by Treg cells
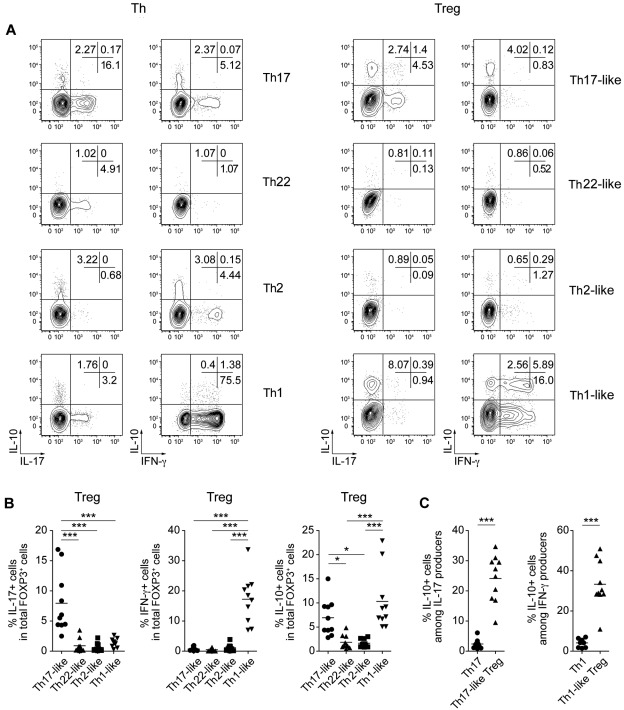
Treg cells use several functional immunomodulatory mechanisms to limit immune responses and prevent autoimmunity. Of these, production of the anti-inflammatory cytokine IL-10 was shown in several models to be required for Treg cell function.26,27 In addition, several studies have indicated that Treg cells can produce pro-inflammatory effector cytokines such as IL-17 and IFN-γ.28–33 To determine whether the Treg cell subsets we identified were functionally heterogeneous, and to compare their cytokine production profiles with the corresponding effector cell populations, we examined the production of pro- and anti-inflammatory cytokines by both Th and Treg cells directly ex vivo after stimulation with PMA/ionomycin (Figure 3A-B). As expected, Th17 cells contained a high frequency of IL-17–producing cells, whereas IFN-γ–producing cells were enriched in the CXCR3+ Th1 population. Likewise, although present at much lower frequencies, FOXP3+ Treg cells producing IL-17 and IFN-γ were found in the phenotypically equivalent Th17- and Th1-like Treg subsets. Surprisingly, the production of IL-10 was also limited to the Th17- and Th1-like Treg populations, where it was frequently coproduced with either IL-17 or IFN-γ (Figure 3C). In addition, as reported for IFN-γ and IL-17, IL-10 production was enriched dramatically in the relatively small population of Helios− cells in each of these Treg cell populations (supplemental Figure 2). In contrast to the Treg cells, conventional Th1 and Th17 cells rarely made IL-10. In addition, little IL-10 production was observed in either the Th22- or Th2-like Treg cells, and these cells also did not produce IL-22 or IL-4, the canonical effector cytokines associated with their cellular phenotypes (supplemental Figure 3A). The patterns of IFN-γ, IL-17, and IL-10 production among the Treg cell subsets were confirmed by ELISA after in vitro activation of sorted cells (supplemental Figure 3B). Moreover, quantitative RT-PCR analysis of cytokine mRNA expression by sorted Th and Treg cells showed that Th1- and Th17-like Treg cells expressed IL17A, IFNG, and IL10 mRNA ex vivo (supplemental Figure 3C), indicating that the production of these cytokines is not an artifact of strong PMA/ionomycin stimulation. These results demonstrate that Treg cells phenotypically associated with different types of Th responses are also functionally distinct, producing unique combinations of both pro- and anti-inflammatory cytokines.
Figure 3.
Th17- and Th1-like Treg cells coproduce pro- and anti-inflammatory cytokines. (A) Representative flow cytometric analysis of IL-10, IL-17, and IFN-γ production by sorted Th (left) and Treg (right) cell subsets stimulated for 5 hours with PMA/ionomycin. (B) Frequency of IL-17–, IL-10–, and IFN-γ–producing cells among gated FOXP3+ Treg cells in each of the indicated Treg cell subsets. Each symbol represents 1 donor; horizontal bars indicate the mean. Data are from 10 donors. *P < .05; ***P < .001 (ANOVA). (C) Frequency of IL-10–producing cells among IL-17+ cells in Th17 cells or Th17-like Treg cells (left) or among IFN-γ+ cells in Th1 cells or Th1-like Treg cells (right). Data are from 10 donors. ***P < .001 (2-tailed paired t test).
Transcription factor expression by Treg cell subsets
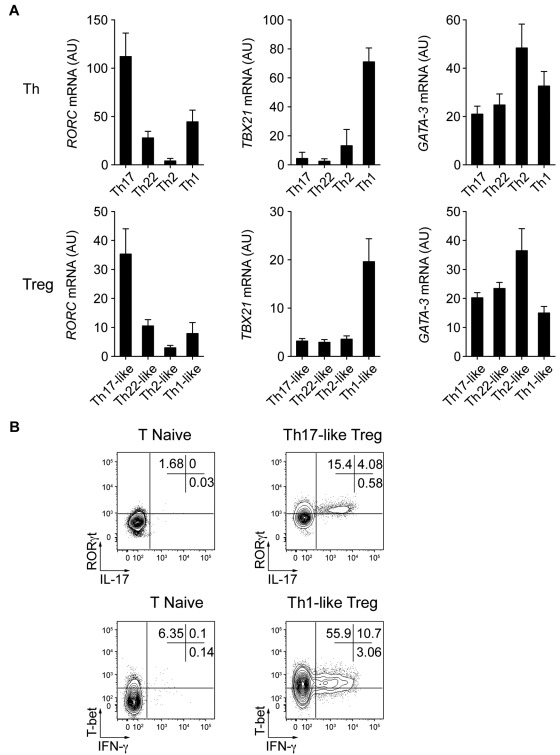
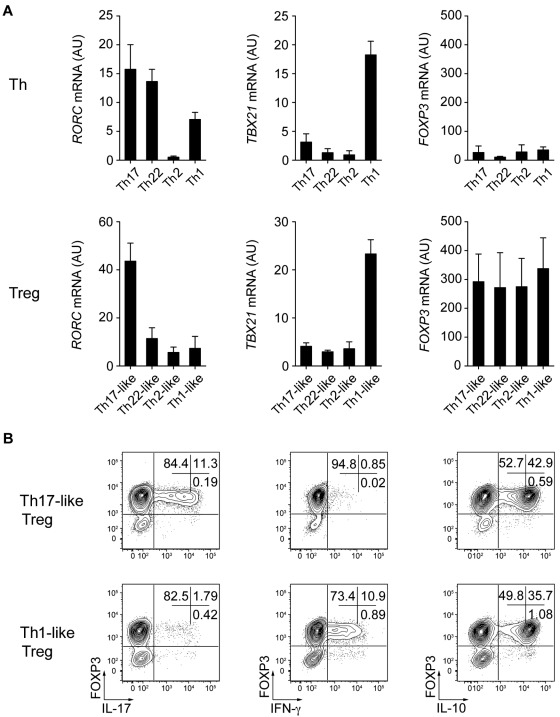
The characteristic phenotypic and functional properties of the various Th cell subsets are largely due to the selective expression of transcription factors such as T-bet and RORγt, which induce specific programs of gene expression controlling both cytokine production and migratory potential of these cells.1 Given their phenotypic resemblance to specific Th cell subsets and their ability to produce effector cytokines such as IL-17 and IFN-γ, we hypothesized that the Treg cell subsets we identified might also express these lineage-specifying transcription factors. Therefore, we used quantitative RT-PCR to measure the expression of transcription factors known to regulate Th cell differentiation and function (Figure 4A). As in Th1 and Th17 cells, expression of TBX21 (the gene encoding T-bet) and RORC (the gene encoding RORγt) were found in the Th1- and Th17-like Treg populations, respectively. The Th2-associated transcription factor GATA3 was found recently to have an essential function in Treg cells34,35 and indeed was expressed by all of the memory Treg cell subsets we describe. However, there was a trend toward higher expression of GATA3 in the Th2-like Treg cells. Moreover, although the transcription factors IRF4 and STAT3 are required for Treg cell–mediated suppression of Th2 and Th17 responses in mice,15,17 each Treg cell subset expressed equivalent amounts of IRF4 or STAT3 mRNA (data not shown).
Figure 4.
Th17- and Th1-like Treg cells express the lineage-specifying transcription factors RORγt and T-bet. (A) Quantitative RT-PCR analysis of RORγt (RORC), T-bet (TBX21), and GATA-3 (GATA3) expression by the indicated Th and Treg cell subsets. AU indicates arbitrary units. Data are means ± SEM of 7 donors. (B) Flow cytometric analysis of IFN-γ, IL-17, T-bet, and RORγt expression by sorted naive T cells (T naive), Th17-like, and Th1-like Treg cells stimulated for 5 hours with PMA/ionomycin. Data are representative of 3 independent experiments.
We used flow cytometry to confirm the expression of RORγt and T-bet by Th17- and Th1-like Treg cells and correlate it with production of IL-17 or IFN-γ (Figure 4B). IL-17–producing Th17-like Treg cells expressed detectable RORγt, and T-bet was expressed by the majority of both IFN-γ+ and IFN-γ− Th1-like Treg cells. Therefore, as has been suggested recently in murine systems, the selective expression of Th-associated lineage-specifying transcription factors likely contributes to the phenotypic and functional specialization of human Treg cells.
Human Treg cell subsets are phenotypically and functionally stable
An ongoing issue in the study of human Treg cells is the lack of a definitive Treg cell marker. Although most Treg cells are CD25hiCD127loFOXP3+, activated Th cells can also acquire this phenotype transiently. This raises the possibility that a portion of the cells we identify as Treg cells, especially those producing effector cytokines, are actually contaminating Th cells. In addition, recent studies have shown that Treg cells can down-regulate FOXP3 expression and interconvert between effector and regulatory phenotypes.36,37 However, the amount of FOXP3 expressed by Th1- and Th17-like Treg cells producing IFN-γ or IL-17 was similar to that observed in cytokine-negative Treg cells, and therefore expression of these pro-inflammatory cytokines is not due to decreased or impaired FOXP3 expression (supplemental Figure 4). In addition, after in vitro activation and expansion, FOXP3 expression was stable in all of the Treg cell subsets and was not up-regulated substantially in any of the Th populations (Figure 5A). Moreover, Th1- and Th17-like Treg cells expanded in vitro by anti-CD3/28 stimulation in the presence of IL-2 maintained their preferential expression of T-bet and RORγt, respectively, as well as their ability to produce IFN-γ, IL-17, and IL-10 (Figure 5A-B). Therefore, under neutral stimulation conditions, the phenotypic and functional characteristics of these Treg cell populations appear to be relatively stable. However, as with other Th populations, they may maintain some functional plasticity when activated in the presence of polarizing cytokines in vivo.
Figure 5.
Th17- and Th1-like Treg cells are phenotypically and functionally stable. (A) Quantitative RT-PCR analysis of RORγt (RORC), T-bet (TBX21), and FOXP3 (FOXP3) expression by the indicated Th and Treg cell subsets after 9 days of in vitro expansion. AU indicates arbitrary units. Data are means ± SEM of 4 donors. (B) Flow cytometric analysis of FOXP3, IFN-γ, IL-17, and IL-10 expression on expanded Th17- and Th1-like Treg cells stimulated for 5 hours with PMA/ionomycin. Data are representative of 4 independent experiments.
Antigen specificity of human Treg cells
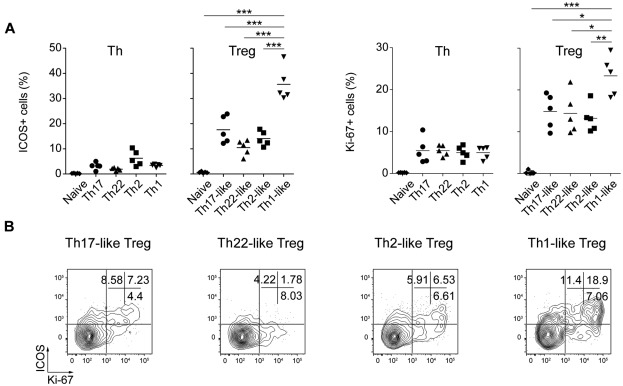
Despite their anergic phenotype in vitro, Treg cells undergo rapid homeostatic proliferation in vivo that reflects their recognition of antigens derived from the “extended self.” This not only includes auto-antigens, but also antigens derived from either benign commensal microorganisms or persistent pathogens. We evaluated the activation status of the various Th and Treg cell subsets by examining their expression of ICOS, a CD28-like costimulatory molecule that is expressed inducibly by recently activated T cells,38 and the proliferation-associated nuclear antigen Ki-67. Whereas little or no ICOS expression was detected on any of the Th cell subsets, ICOS+ cells were present in all 4 Treg cell populations and were particularly enriched among the CXCR3+ Th1-like Treg cells (Figure 6A left panels). Although expressed by fewer cells, the pattern of Ki-67 expression mirrored closely that of ICOS (Figure 6A right panels), and ICOS and Ki-67 were concordantly expressed by a large portion of the Th1-, Th2-, and Th17-like Treg cells (Figure 6B). Therefore, consistent with the notion that Treg cells recognize ubiquitously presented antigens, each of the Treg cell subsets contained a large population of activated, cycling cells.
Figure 6.
Constitutive activation of human Treg cells. (A) Expression of ICOS (left) and Ki-67 (right) by the indicated Th and Treg cell subsets. Data are from 5 donors. (B) Flow cytometric analysis of ICOS and Ki-67 expression by the indicated Treg cell subsets. Data are representative of 4 donors. *P < .0.05; **P < .01; ***P < .001 (ANOVA).
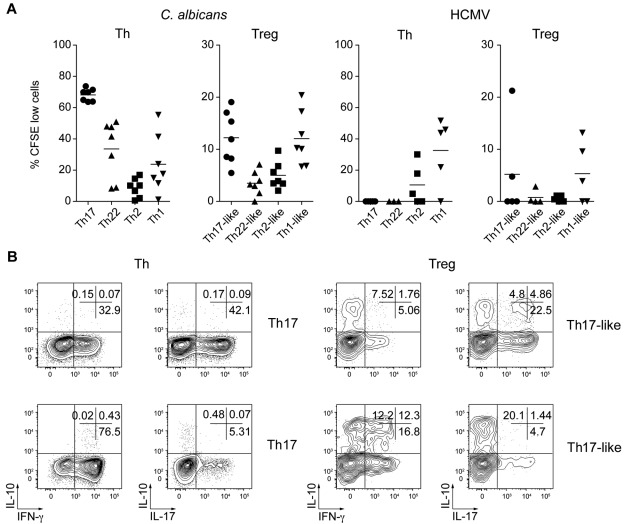
To determine whether the Treg cell subsets shared antigen specificities with their Th counterparts, we stimulated sorted CFSE-labeled cells in vitro with antigens from organisms that provoke strong Th17 (C albicans) or Th1 (HCMV) responses.7,39 All Th and Treg cell subsets displayed strong proliferation when stimulated with anti-CD3/28 Ab (supplemental Figure 5). However, results from 8 donors showed that Th cells specific for C albicans were present mainly in the Th17 population but could also be found among Th22 and Th1 cells, albeit with more donor-to-donor variability (Figure 7A left panel). C albicans–induced proliferation of Treg cells was also detected in most donors examined for the Th17- and Th1-like Treg cell subsets, but was low or undetectable in the Th22- and Th2-like Treg cells. Similarly, HCMV-specific effector T cells were found in the Th1 cell population in 4 donors and, of these, 3 also showed substantial proliferative responses in the Th1-like Treg cell population (Figure 7A right panel).
Figure 7.
C albicans– and HCMV-specific Treg cells are present in Th17- and Th1-like subsets. (A) Proportion of CFSElo cells (that had undergone 3 or more divisions) among the indicated Th and Treg cell subsets after 6 days of stimulation with autologous monocytes pulsed with C albicans or HCMV antigens. Each symbol represents 1 donor; small horizontal bars indicate the mean. Data are from 7 (C albicans) or 5 (HCMV) donors. (B) Flow cytometric analysis of cytokine production by gated CFSElo C albicans–specific Th and Treg cells after restimulation with PMA/ionomycin for 5 hours. Data are representative of 3 independent experiments using 3 different donors.
We also compared the cytokine production profile of the CFSElo C albicans–specific Th and Treg cells by flow cytometry after restimulation with PMA/ionomycin (Figure 7B). After culture with C albicans, CFSElo Th17 cells contained both IL-17+ and some IFN-γ+ cells, whereas CFSElo Th1 cells were predominantly IFN-γ+. Similarly, IL-17 and IFN-γ–producing cells were also present in the CFSElo Th17- and Th1-like Treg populations, respectively. However, consistent with their regulatory phenotype, these cells also contained significant populations of IL-10 single-producing cells and IL-10/IL-17 or IL-10/IFN-γ–coproducing cells. Therefore, Treg cells that proliferate in response to C albicans or HCMV are found in the Th17- and Th1-like Treg subsets, and these cells differ from pathogen-specific Th cells in the balance of pro- and anti-inflammatory cytokines they produce.
Discussion
As part of their functional specialization, CD4+ effector T cells acquire expression of different combinations of adhesion and chemokine receptors that enable their migration to specific tissues and inflammatory sites. Through a series of ex vivo analyses, we now extend this paradigm to FOXP3+ Treg cells and have identified and characterized functionally distinct Treg cell subsets that mimic different Th cell populations phenotypically. This diversity strongly suggests that Th and Treg cells undergo phenotypic and functional specialization in parallel during different types of inflammatory responses and likely reflects the specialized role that each Treg cell subset has in colocalizing with and suppressing specific Th cell populations in vivo.
Although the phenotypic diversity of Treg cells has been well established, the distinct functional characteristics of different Treg cell subsets are still poorly understood. By performing comprehensive functional analyses of the various Treg cell subsets, we found that although each subset was suppressive in vitro, they differed dramatically in their ability to produce both pro- and anti-inflammatory cytokines. Despite the ability of FOXP3 to inhibit IL-17 and IFN-γ expression directly,40,41 several recent studies have reported the production of these pro-inflammatory cytokines by Treg cells in both mice and humans.28–33 In humans, production of these cytokines is restricted to the Helios− fraction of Treg cells that are thought to be induced from FOXP3−–naive T cells by suboptimal stimulation in the presence of TGF-β,24 and this was taken to indicate that these cells are functionally unstable and could readily convert to pro-inflammatory effector lineages. Furthermore, both type-1 diabetes and multiple sclerosis have been associated with an increase in IFN-γ–producing Treg cells, suggesting that redirected Treg cells may contribute to autoimmune pathogenesis.30,33 In the present study, we found that the production of IL-17 and IFN-γ is restricted to the Th17- and Th1-like Treg cells that phenotypically mimic conventional Th17 and Th1 cells. Moreover, unlike previous studies examining effector cytokine production by Treg cells, we found herein that cytokine production in these Th1- and Th17-like Treg cells is qualitatively different from that observed in their effector Th counterparts. For example, whereas IL-22 was frequently coproduced with IL-17 in Th17 cells, coproduction of these cytokines was not observed in the Th17-like Treg population. In addition, unlike FOXP3− Th1 and Th17 cells, significant fractions of the IFN-γ– and IL-17–producing Treg cells coproduced IL-10, and therefore appear to retain at least some regulatory activity. Indeed, these Treg cells functionally resemble the FOXP3− IFN-γ/IL-10–coproducing cells that modulate Th1 responses during persistent parasite infection in mice42,43 and that were recently described as a small population of CD127loCD25− Th cells in humans.44 In addition, both IFN-γ and IL-17 can have an immunomodulatory functions in certain immune contexts45,46 and, rather than being pro-inflammatory or potentially pathogenic, production of these cytokines by Treg cells in conjunction with IL-10 may actually help to dampen inflammation. Therefore, the increased frequency of IFN-γ–producing Treg cells in type-1 diabetes and multiple sclerosis patients may be a response to inflammatory disease in these individuals rather than a cause of it.
Studies in murine systems have led to the notion that Treg cells use specific Th-associated transcription factors to maintain or restore immune homeostasis during Th1, Th2, and Th17 immune responses.15–17 In the present study, we show that T-bet and RORγt are expressed specifically by Th1- and Th17-like Treg cells, thereby extending this paradigm to human Treg cells and demonstrating that phenotypically distinct Treg cell populations coopt portions of the transcriptional programs of specific effector T-cell subsets. Indeed, T-bet and RORγt induce the expression of CXCR3 and CCR6, respectively,47,48 and therefore expression of these transcription factors may act to link the migratory properties of these Th and Treg cell populations. Interestingly, although FOXP3 can antagonize RORγt-mediated production of IL-17 in vitro,41 the fraction of RORγt-expressing cells that produced IL-17 was similar in the Th17 cells and the Th17-like Treg cells. In addition, the amount of IL-17 expressed per cell was comparable in the FOXP3+ and FOXP3− cells, indicating that FOXP3-mediated inhibition of IL-17 expression is impaired in Th17-like Treg cells. In humans, a second isoform of FOXP3 exists in which exon 2, including the domain essential for inhibition of RORγt, is removed by alternative splicing.41 It is possible that the IL-17–producing Treg cells we observed overexpress this FOXP3 isoform, and further studies are needed to delineate precisely how RORγt, T-bet, and other transcription factors involved in effector Th differentiation interact functionally with FOXP3 to control the balance of pro- and anti-inflammatory gene expression in Treg cells.
The mechanisms by which Treg cells acquire distinct patterns of transcription factor and chemokine receptor expression are important to define. Their shared phenotypes suggest that the Th and Treg cell subsets differentiate in parallel in response to inflammatory cues from the immune environment. Consistent with this, we found that IFN-γ and STAT1 were required for Treg cell expression of T-bet and CXCR3 in mice,16 and human naive Treg cells can differentiate into IL-17 –producing cells in vitro when stimulated in the presence of specific pro-inflammatory cytokines.49 In addition, Treg cells may modify their phenotypes in response to anatomical cues such as the active metabolite of vitamin D3, 1,25-(OH)2D3, which is produced in the skin in response to sun exposure and can induce T-cell expression of the cutaneous chemokine receptor CCR10.50
Although believed to be largely self-reactive or specific for commensal microorganisms, the specificity of human Treg cells is still poorly characterized. However, consistent with the phenotypic relationship between the various subsets of Th and Treg cells, we found that Treg cells specific for C albicans and HCMV can be found largely in the Th1- and Th17-like Treg populations. Moreover, unlike the C albicans–specific memory Th cells, the production of IL-10, either alone or in conjunction with either IFN-γ or IL-17, was observed commonly in C albicans–reactive Th1- and Th17-like Treg cells. Therefore, the responses that we observed in Treg cells appear to represent an immunomodulatory response to infection. Because HCMV generally establishes lifelong latent infection, and C albicans is a common component of the oral and intestinal flora, these Treg cells may help to maintain balanced immune responses that result in pathogen control without full eradication. A broader analysis of the repertoire and specificity of Treg cells in each of the populations we have characterized will be important for determining how each contributes to the regulation of immune responses to both foreign and self-antigens.
Our identification and characterization of phenotypically and functionally distinct human Treg cell subsets provide new insights into how Treg cells modulate different types of effector T-cell responses, and suggests that specific populations of specialized Treg cells could be stably expanded and used to treat inflammatory and autoimmune diseases caused by dysregulated Th1, Th2, Th17, and Th22 responses. Further exploration of the diversity of human Treg cells is essential for determining how these cells balance the need to mount robust and effective immune responses during pathogen infection with the requirement to maintain self-tolerance and prevent collateral tissue damage and immunopathology. This will also shed light on how aberrant Treg cell function contributes to the development and progression of immune-mediated diseases and provide new avenues for the therapeutic manipulation of Treg cell activity.
Supplementary Material
Acknowledgments
The authors thank K. Arumuganathan for his help with cell sorting and flow cytometry; the Benaroya Research Institute Translational Research Program Clinical Core for obtaining donor samples; S. F. Ziegler, J. A. Hamerman, and M. A. Koch for comments on the manuscript; and M. Warren for administrative assistance.
This work was supported in part by the National Institutes of Health (grants AR055695, DK072295, and AI067750 to D.J.C.). T.D. was the recipient of a postdoctoral fellowship from the Arthritis Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.D. and R.D. performed the experiments and analyzed the results; T.D., A.L., F.S., and D.J.C. designed the research; and T.D. and D.J.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel J. Campbell, Benaroya Research Institute, 1201 9th Ave, Seattle, WA 98101; e-mail: campbell@benaroyaresearch.org.
References
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10(8):857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 3.Nograles KE, Zaba LC, Shemer A, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123(6):1244–1252.e2. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10(8):864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 5.Qin S, Rottman JB, Myers P, et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101(4):746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187(6):875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acosta-Rodriguez EV, Rivino L, Geginat J, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 8.Annunziato F, Cosmi L, Santarlasci V, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204(8):1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180(1):214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- 10.Reboldi A, Coisne C, Baumjohann D, et al. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol. 2009;10(5):514–523. doi: 10.1038/ni.1716. [DOI] [PubMed] [Google Scholar]
- 11.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3(+) regulatory T cells. Nat Rev Immunol. 2011;11(2):119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Marson A, Kretschmer K, Frampton GM, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhry A, Rudra D, Treuting P, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y, Chaudhry A, Kas A, et al. Regulatory T-cell suppressor program coopts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim HW, Broxmeyer HE, Kim CH. Regulation of trafficking receptor expression in human forkhead box P3+ regulatory T cells. J Immunol. 2006;177(2):840–851. doi: 10.4049/jimmunol.177.2.840. [DOI] [PubMed] [Google Scholar]
- 19.Dudda JC, Perdue N, Bachtanian E, Campbell DJ. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J Exp Med. 2008;205(7):1559–1565. doi: 10.1084/jem.20072594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erhardt A, Wegscheid C, Claass B, et al. CXCR3 deficiency exacerbates liver disease and abrogates tolerance in a mouse model of immune-mediated hepatitis. J Immunol. 2011;186(9):5284–5293. doi: 10.4049/jimmunol.1003750. [DOI] [PubMed] [Google Scholar]
- 21.Sather BD, Treuting P, Perdue N, et al. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J Exp Med. 2007;204(6):1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villares R, Cadenas V, Lozano M, et al. CCR6 regulates EAE pathogenesis by controlling regulatory CD4+ T-cell recruitment to target tissues. Eur J Immunol. 2009;39(6):1671–1681. doi: 10.1002/eji.200839123. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki T, Yang XO, Chung Y, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181(12):8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thornton AM, Korty PE, Tran DQ, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wing K, Onishi Y, Prieto-Martin P, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 26.Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190(7):995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubtsov YP, Rasmussen JP, Chi EY, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Ayyoub M, Deknuydt F, Raimbaud I, et al. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the T(H)17 lineage-specific transcription factor RORgamma t. Proc Natl Acad Sci U S A. 2009;106(21):8635–8640. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beriou G, Costantino CM, Ashley CW, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113(18):4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClymont SA, Putnam AL, Lee MR, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186(7):3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voo KS, Wang YH, Santori FR, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106(12):4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldenhove G, Bouladoux N, Wohlfert EA, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31(5):772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17(6):673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35(3):337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlfert EA, Grainger JR, Bouladoux N, et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121(11):4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey-Bucktrout SL, Bluestone JA. Regulatory T cells: stability revisited. Trends Immunol. 2011;32(7):301–306. doi: 10.1016/j.it.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 39.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies pre-T helper (Th)1, pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200(6):725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono M, Yaguchi H, Ohkura N, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446(7136):685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 41.Zhou L, Lopes JE, Chong MM, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453(7192):236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jankovic D, Kugler DG, Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. 2010;3(3):239–246. doi: 10.1038/mi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(-)Foxp3(-) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204(2):285–297. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Häringer B, Lozza L, Steckel B, Geginat J. Identification and characterization of IL-10/IFN-gamma-producing effector-like T cells with regulatory function in human blood. J Exp Med. 2009;206(5):1009–1017. doi: 10.1084/jem.20082238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Connor W, Jr, Kamanaka M, Booth CJ, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10(6):603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willenborg DO, Fordham S, Bernard CC, Cowden WB, Ramshaw IA. IFN-gamma plays a critical down-regulatory role in the induction and effector phase of myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. J Immunol. 1996;157(8):3223–3227. [PubMed] [Google Scholar]
- 47.Lord GM, Rao RM, Choe H, et al. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106(10):3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9(6):641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valmori D, Raffin C, Raimbaud I, Ayyoub M. Human RORgammat+ TH17 cells preferentially differentiate from naive FOXP3+Treg in the presence of lineage-specific polarizing factors. Proc Natl Acad Sci U S A. 2010;107(45):19402–19407. doi: 10.1073/pnas.1008247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigmundsdottir H, Pan J, Debes GF, et al. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8(3):285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.