Abstract
Tumor epidermal growth factor receptor (EGFR) mutation analysis is significant for making treatment decisions for metastatic pulmonary adenocarcinoma. However, less than half of patients have adequate tumor samples for mutation analysis. Patients with adenocarcinoma of the lungs who were due to receive erlotinib treatment were included in the present study. Tumor EGFR mutation status was analyzed using DNA sequencing. Plasma specimens from the patients were collected prior to erlotinib treatment. The plasma-free DNA EGFR mutation status was analyzed using the PCR clamp method. A total of 54 consecutive patients were included in the study. The plasma-free DNA EGFR mutation status of the 54 patients was analyzed. Only 30 patients had adequate tumor samples for EGFR analysis, including 15 with activating mutations (exon 19 deletions or L858R). EGFR-activating mutations were detected in the plasma-free DNA in 25 of 54 patients. The response rate was 86.7 and 33.3% in patients with and without tumor activating mutations, respectively (p=0.002). The response rate was 68 and 31% based on the patients’ plasma-free DNA EGFR mutation status, respectively (p=0.013). No significant difference in progression-free survival (PFS) was observed between patients with and without EGFR-activating mutations, according to data from tumor tissue or plasma-free DNA analysis, although the median PFS time was longer for those patients with EGFR-activating mutations in plasma samples. Plasma EGFR mutation analysis is useful for adenocarcinoma patients who have no or inadequate tumor samples available for EGFR examination. Patients with plasma EGFR-activating mutations had an improved response rate and a statistically insignificant longer PFS.
Keywords: epidermal growth factor receptor, erlotinib, targeted therapy, adenocarcinoma
Introduction
Lung cancer is the leading cause of cancer mortality in the world and non-small-cell lung cancer (NSCLC) accounts for the majority of lung cancer cases (1). Epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKI), including erlotinib, are a new class of anti-cancer agents that have been used for almost a decade (2). Erlotinib is an orally-available quinazoline that is a selective inhibitor of the tyrosine kinase of EGFR. Erlotinib is the first EGFR-TKI to demonstrate an increase in survival in a phase III trial of NSCLC patients who had failed one or two previous chemotherapy regimens (3). Several studies have demonstrated that the tumor tissue EGFR mutation status may be used to predict tumor response to EGFR-TKI treatment and also possibly predict patient survival. In addition to the usefulness of EGFR-TKI as second-line treatment (3), recent clinical trials have also revealed a high efficacy of EGFR-TKIs as a first-line treatment for EGFR-mutated NSCLC, compared with platinum-based doublets, in terms of prolongation of progression-free survival (PFS) and reduced toxicity (4–7). Thus, the use of EGFR-TKI in treating patients with tumor EGFR-activating mutations is gaining in significance and is becoming a first step in decision-making when treating patients with metastatic NSCLC. However, the best way to find patients with tumor EGFR-activating mutations remains undetermined (8). In addition, only approximately 36% of lung cancer patients had adequate tissue samples available for EGFR mutation analysis (4).
Plasma-free DNA EGFR mutation analysis is a novel tool that provides an easy, convenient and safe way of testing patients, and is particularly useful for those patients who do not have or have only limited pathological specimens (9–11). In the present study, we prospectively collected and examined the plasma-free DNA mutation status of our lung cancer patients with adenocarcinoma who were due to receive erlotinib treatment, to determine whether or not this method is effective in the detection of the EGFR mutation status and in the prediction of treatment response and PFS.
Patients and methods
Patients
The study protocol was approved by the institutional review board of our hospital (VGHIRB No. 98-11-05). Patients with adenocarcinoma of the lung who were due to receive erlotinib treatment and who had measureable lesion(s) were entered into the present study after informed consent had been obtained. Blood samples were collected prior to delivery of the first dose of erlotinib.
Tumor tissue EGFR sequencing
EGFR mutation analysis was performed using nucleotide sequence analysis. The VarientSEQrTM Resequencing Primer Set was selected for mutational analysis of the tyrosine kinase domain, exons 18–21 of the EGFR gene. Genomic DNA was extracted from paraffin blocks, exons 18–21 were amplified, and uncloned polymerase chain reaction (PCR) fragments were sequenced and analyzed in sense and antisense directions for the presence of heterozygous mutations. Normal control DNA, provided by Applied Biosystems (Carlsbad, CA, USA), was used for the wild-type control. The sequence variations were confirmed by multiple, independent PCR amplifications and repeated sequencing reactions. EGFR-activating mutations were defined as those with exon 19 deletions or exon 21 L858R.
Plasma sample collection, DNA extraction and sequencing
Plasma samples were collected in EDTA tubes and centrifuged at 1500 rpm for 5 min. DNA was extracted from 1 ml of plasma using the High Pure Viral Nucleic Acid kit (Roche Diagnostics, Almere, The Netherlands). The PCR reaction is based on the peptide nucleic acid-locked nucleic acid (PNA-LNA) PCR clamp-based test (12). Briefly, PCR reactions were performed in a total volume of 20 μl consisting of 10 μl 2X Master mix, 1 μm clamp primer and 0.4 μm of each primer. Thermocycling was performed in PCR tubes. Following a 10-min activation step at 95˚C, reactions were subjected to 45 cycles of 30 sec at 95˚C, 30 sec at 60˚C and 30 sec at 72˚C. The amplification product of the PCR clamp was purified using Shrimp alkaline phosphatase (SAP) and Exonuclease I (EXOI) treatments, and was then subjected to DNA sequencing using an automatic DNA sequencer. This type of PNA-LNA PCR clamp-based test is capable of detecting mutations, including exon 18 G719C or G719S, exon 19 deletions between E746 and S752, exon 20 T790M, and exon 21 L858R or L861Q mutations. As with tumor tissue EGFR sequencing, only exon 19 deletions and exon 21 L858R were considered to be EGFR-activating mutations.
Efficacy evaluation
Baseline assessments were performed within 3 weeks prior to erlotinib treatment. A chest computed tomography scan (including liver and adrenal glands) was performed within 3 weeks prior to starting erlotinib treatment, 1 month and 3 months following commencement of erlotinib treatment, and then every 3 months thereafter, or when confirmation of treatment response or disease progression was required. Treatment response evaluation was performed according to the Response Evaluation Criteria in Solid Tumors (RECIST) group criteria (13). PFS was calculated from the date erlotinib treatment commenced to the earliest sign of disease progression, as determined by the RECIST criteria (13), or mortality from any cause. If disease progression had not occurred at the time of the last follow-up visit, PFS was considered to have been censored at that time.
Statistical analysis
Survival curves were drawn using the Kaplan-Meier product method. Comparisons were made with the log-rank test. Hazard ratios in the overall population and in the patient subsets were calculated using the Cox proportional hazards model. The Chi-square test was used to compare the response rates according to the molecular profiles. P-values were 2-sided and p<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS software (SPSS Inc., Chicago, IL, USA).
Results
Patients and EGFR mutation analysis
A total of 54 patients who received erlotinib treatment were enrolled in the study, including 21 males and 33 females, with a mean age of 64 years (range, 30–88). ECOG performance status was 0 or 1 in 40 patients and ≥2 in 14 patients. Thirty-nine patients were non-smokers and 15 were smokers. Erlotinib treatment was the first-line treatment in 4 patients and the second-line or later treatment in 50 patients. Forty patients had pathological specimens available for tumor EGFR mutation analysis; however, 10 of the 40 patients had inadequate specimens for analysis. Thus, only 30 patients had adequate pathological specimens for tumor EGFR sequencing. The tumor EGFR mutation analysis of these 30 patients revealed that 15 (50%) had activating mutations (exon 19 deletions in 5 patients, L858R mutations in 9 and exon 19 deletions + L858R mutations in 1 patient). Among the remaining 15 patients without activating mutations, 4 had atypical mutations (exon 18 G719C + exon 20 S768I in 1, exon 19 P733L in 1, exon 19 E749X in 1 and exon 21 R836P in 1 patient), and 11 patients had wild-type mutations.
All 54 patients underwent plasma-free DNA EGFR mutation analysis prior to erlotinib treatment. Activating mutations were found in 25 patients (46.3%) (exon 19 deletions in 18 patients, L858R in 4 and exon 19 deletions + L858R in 3) and wild-type in 29 patients (53.7%). No atypical mutations or T790M mutations were detected in the plasma samples. The results of the 30 patients who had tumor tissue and plasma-free DNA EGFR mutation analysis data are shown in Table I.
Table I.
Sequencing results of 30 patients who had tumor tissue and plasma-free DNA EGFR mutation analysis.
| Tissue | Plasma | |||
|---|---|---|---|---|
| Exon 19 deletion | L858R | Exon 19 deletion + L858R | Wild-type | |
| Exon 19 deletion, n=5 | 4 | 0 | 1 | 0 |
| L858R, n=9 | 0 | 2 | 4 | 3 |
| Exon 19 deletion + L858R, n=1 | 0 | 0 | 1 | 0 |
| Atypical mutation, n=4 | 2 | 0 | 2 | 0 |
| Wild-type, n=11 | 3 | 0 | 8 | 0 |
EGFR, epidermal growth factor receptor.
Treatment response
Of the 54 patients, 26 (48.1%) patients had a partial response to erlotinib treatment, 13 (24.1%) had stable disease with erlotinib treatment and 15 (27.8%) had progressive disease with erlotinib treatment. The objective response rate was 86.7% (13 of 15) in patients with tumor tissue EGFR-activating mutations and 33.3% (5 of 15) in those without (p=0.003). The objective response rate was 68% (17 of 25) in patients with plasma-free DNA EGFR-activating mutations and 31% (9 of 29) in those without (p=0.013). Among the 30 patients who had tumor tissue and plasma-free DNA EGFR data, the response rate was 100% (10 of 10) in those with tumor tissue and plasma EGFR-activating mutations, 60% (3 of 5) in those with only tumor tissue EGFR-activating mutations, 50% (3 of 6) in those with only plasma EGFR-activating mutations, and 22.2% (2 of 9) in those without an EGFR-activating mutation from either sample (p=0.007).
Progression-free survival
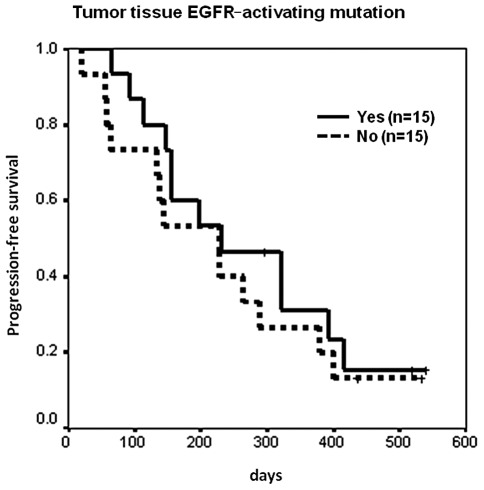
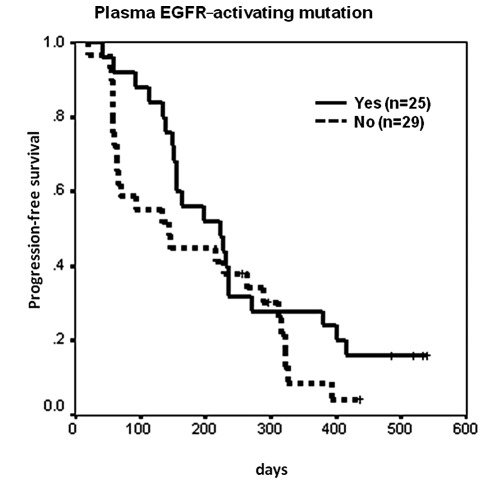
Median PFS was 9 months in 26 patients who had a partial response to erlotinib treatment [censor 5, 95% confidence interval (CI) 6.3–11.7 months], 5.4 months in 13 patients who had stable disease with erlotinib treatment (censor 2, 95% CI 2.1–8.7 months), and 1.9 months in 15 patients who had progressive disease with erlotinib treatment (censor 0, 95% CI 1.9–2 months) (p<0.0001). For those patients who had tumor tissue EGFR sequencing data, median PFS was 7.7 months in 15 patients with EGFR-activating mutations (censor 3, 95% CI 1.1–14.2 months), and 7.6 months in 15 patients without EGFR-activating mutations (censor 2, 95% CI 3.8–11.4 months) (p=0.4826, Fig. 1). The hazard ratio was 0.76 (95% CI 0.34–1.66, p=0.484). Median PFS was 7.4 months in 25 patients with plasma EGFR-activating mutations (censor 4, 95% CI 3.9–10.9 months), and 4.8 months in 29 patients without EGFR-activating mutations (censor 3, 95% CI 1.7–7.9 months) (p=0.1428, Fig. 2). The hazard ratio was 0.65 (95% CI 0.36–1.17, p=0.1471).
Figure 1.
Kaplan-Meier analysis of progression-free survival (PFS) by tumor tissue EGFR mutation status. Median PFS was 7.7 months in 15 patients with EGFR-activating mutations (censor 3, 95% CI 1.1–14.2 months), and 7.6 months in 15 without (censor 2, 95% CI 3.8–11.4 months) (p=0.4826). EGFR, epidermal growth factor receptor.
Figure 2.
Kaplan-Meier analysis of progression-free survival (PFS) by plasma-free DNA EGFR mutation status. Median PFS was 7.4 months in 25 patients with plasma EGFR-activating mutations (censor 4, 95% CI 3.9–10.9 months), and 4.8 months in 29 without (censor 3, 95% CI 1.7–7.9 months) (p=0.1428). EGFR, epidermal growth factor receptor.
Of the 30 patients who had tumor tissue and plasma EGFR sequencing data available, median PFS was 7.7 months in 21 patients who had EGFR-activating mutations detected in at least one tumor or plasma specimen (censor 4, 95% CI 1.7–13.6 months), and 4.8 months in 9 patients who had no activating mutations detected in either type of specimen (censor 1, 95% CI 3.8–5.8 months) (p=0.2115). The hazard ratio was 0.59 (95% CI 0.25–1.37, p=0.2168).
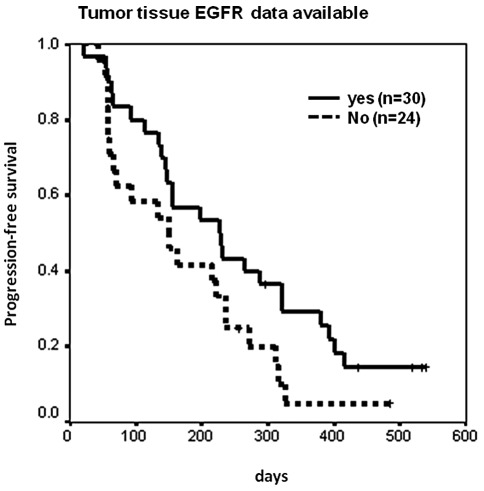
Patients without tumor tissue EGFR mutation data
The tumor tissue EGFR mutation status of 24 patients, 44.4% of the total study population, was unavailable (10 had inadequate tissue samples and 14 had no tissue samples). The overall response rate to erlotinib treatment of those 24 patients was 33.3% (8 of 24), and 60% (18 of 30) for those patients who had adequate tumor tissue samples for EGFR mutation analysis (p=0.046 and 0.061 for 1-sided and 2-sided χ2 tests, respectively). PFS was also shorter for the 24 patients whose tumor tissue EGFR mutation data were unavailable compared to the 30 patients who had adequate tissue samples for tumor EGFR analysis (median 5 vs. 7.6 months, p=0.0704, Fig. 3). With regard to the 24 patients who had no tissue samples or inadequate samples for EGFR mutation analysis, the rate of response to erlotinib treatment was 45.5% (5 of 11) for those with plasma EGFR-activating mutations, and 23.1% (3 of 13) for those without (p=0.39). Median PFS was 5.4 months for 11 patients with plasma EGFR-activating mutations, and 2.3 months for 13 patients without mutations (p=0.6868).
Figure 3.
Kaplan-Meier analysis of progression-free survival (PFS) of patients with or without tumor tissue EGFR sequencing data. PFS was shorter for 24 patients whose tumor tissue EGFR mutation data was unavailable compared to 30 patients who had adequate tissue samples for tumor EGFR analysis (median 5 months vs. 7.6 months, p=0.0704). EGFR, epidermal growth factor receptor.
Discussion
The use of molecular predictive markers to help identify those patients who may benefit from a specific treatment remains one of the most exciting areas of research in medical oncology. The development of a rapid and sensitive test for the early assessment of treatment response is also mandatory to avoid futile treatments with ineffective agents. A number of studies have documented that tumor tissue EGFR-activating mutations are useful markers for predicting responses to EGFR-TKI treatment (4–7). Determination of the tumor EGFR mutation status is significant for non-squamous NSCLC, since first-line treatment with erlotinib or gefitinib is recommended for patients with EGFR-activating mutations, and cytotoxic chemotherapy is recommended for patients without an EGFR-activating mutation status or with an unknown status. The use of plasma or serum samples for EGFR mutation analysis has been actively studied recently since more than half of the patients did not have or had inadequate pathological samples for tumor EGFR mutation analysis (9,10).
Several methods are used to detect EGFR mutations in lung cancer specimens (14). The sensitivity of direct sequencing is relatively poor and requires more mutant DNA to detect EGFR mutations in specimens (10–25% mutant DNA). The PNA-LNA PCR clamp is more sensitive and is able to detect ≥1% mutant DNA in the specimens, as is the Scorpion Amplified Refractory Mutation System (ARMS) method, which currently has kits commercially available for testing (4,10,12,14). However, direct sequencing is able to detect new mutations, while the PNA-LNA PCR clamp and the Scorpion ARMS are capable of detecting known mutations only (10,12,14).
There was no statistical difference in PFS between the patients with and without a tumor EGFR-activating mutation in the present study. This is possibly due to the low number of patients enrolled and the relatively low sensitivity of the direct tumor DNA sequencing (false negative of activating mutations). Although there was also no statistical difference in the PFS of the patients with and without plasma-free DNA EGFR-activating mutations, the numerical difference in PFS between the 2 groups of patients was larger than that detected by the direct tumor DNA sequencing. This insignificant difference in the PFS detected by the plasma-free DNA EGFR examination would be significant if patient numbers were increased.
In the present study, patients with no tumor tissue EGFR mutation data were examined, and, although there was no statistically significant difference in the response rate and PFS between the patients with (n=11) and without (n=13) plasma-free DNA EGFR-activating mutations, the response rate was doubled and the PFS was more than doubled in the patients with plasma-free DNA EGFR-activating mutations. There would possibly be a statistical significance if the patient number was increased. Thus, plasma-free DNA EGFR mutation analysis using the PNA-LNA PCR clamp method is useful for those patients with inadequate tumor specimens or without tumor tissue available for tumor EGFR examinations. This test may be of benefit to patients, in terms of predicting the response to EGFR-TKI treatment. We concluded that in patients with plasma-free DNA EGFR-activating mutations, the response to erlotinib treatment was better and the PFS was longer than in those without plasma-free DNA EGFR-activating mutations.
Acknowledgements
The study was supported in part by grants from the National Science Council of the Republic of China, grant number NSC99-2314-B-075-035-MY3, and Taipei Veterans General Hospital, grant number VGH-100-C-015.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Azzoli CG, Baker S, Jr, Temin S, Pao W, Aliff T, Brahmer J, Johnson DH. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd FA, Pereira JR, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. New Eng J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Zhou C, Wu YL, Chen G, et al. Efficacy results from the randomized phase III OPTIMAL (CTONG 0802) study comparing first-line erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM), in Chinese advanced non-small-cell lung cancer (NSCLC) patients (PTS) with EGFR activating mutations. Ann Oncol. 2010;21c(Suppl 8):LBA13. [Google Scholar]
- 7.Rosell R, Gervais R, Vergnenegre A, et al. Erlotinib versus chemotherapy (CT) in advanced non-small cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) mutations: interim results of the European Erlotinib Versus Chemotherapy (EURTAC) phase III randomized trial. Proc Am Soc Clin Oncl A. 2011;7503 [Google Scholar]
- 8.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–4955. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 9.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 10.Mack PC, Holland WS, Burich RA, et al. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. J Thorac Oncol. 2009;4:1466–1472. doi: 10.1097/JTO.0b013e3181bbf239. [DOI] [PubMed] [Google Scholar]
- 11.Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Circulating cell-free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin Chemistry. 2006;52:1833–1842. doi: 10.1373/clinchem.2005.062893. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, Nagai Y, Miyazawa H, et al. Reliability of the peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based test for epidermal growth factor receptor mutations integrated into the clinical practice for non-small cell lung cancers. Cancer Sci. 2007;98:246–252. doi: 10.1111/j.1349-7006.2006.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organisation for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–4955. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]