Abstract
Mutations at codons 12 and 13 of the KRAS gene have been identified as level I predictive biomarkers against the treatment of advanced colorectal cancer with anti-epidermal growth factor receptor (EGFR) monoclonal antibodies. It is thought that the genetic analysis of KRAS mutations associated with metastatic colorectal cancer can be routinely conducted using DNA obtained on one occasion from one organ, from the primary or a metastatic site, whichever is preferentially available. However, the issue of tumor heterogeneity resulting from acquired/intratumoral mutations remains. Recently, the possibility of acquired/intratumoral mutations in the KRAS gene has been reported by two research groups and has ranged from 7.4 to 15.4%. Specimens were collected from advanced colorectal cancer patients with resected primary, and at least one metastatic, site. Direct sequence analysis was performed for KRAS, BRAF and PIK3CA, and immunohistochemistry for glutathione S-transferase II (GSTP) and EGFR. In the current study, we identified an acquired mutation rate of approximately 11.1% in the KRAS gene (1/9). This figure is not negligible. Our observation indicates, particularly in the case of metastatic recurrence after a long interval, that there may be considerable tumor heterogeneity resulting from acquired or intratumoral mutations of the KRAS gene.
Keywords: KRAS, BRAF, PIK3CA, colorectal cancer, acquired/intratumor mutation
Introduction
In the last decade, two anti-epidermal growth factor receptor monoclonal antibodies (EGFR mAbs), cetuximab and panitumumab, were approved for the treatment of EGFR-positive colorectal cancer (CRC) (1,2). EGFR signals are transduced by KRAS and follow two signaling pathways, the RAS-RAF-MEK-ERK and RAS-PI3 kinase-AKT/PKB pathways. Mutations at codons 12 and 13 of the KRAS gene have been identified as a level I predictive biomarker against the treatment of advanced CRC with anti-EGFR mAbs according to the College of American Pathologists (CAP) level of evidence classification; that is, these mutations have been definitively proven as biomarkers based on evidence from multiple, statistically robust, published trials, and they are generally used in patient management (3). BRAF is a serine-threonine kinase located downstream of KRAS, which is a component of the RAS-RAF-MEK-ERK signaling pathway (4). A valine to glutamate substitution mutation at codon 600 (V600E) of the BRAF gene is a hot spot and is observed in 5–22% of CRCs (4). BRAF has a level IIA CAP predictive value, which means that extensive biological and clinical studies have repeatedly shown it to have predictive value for therapy; however, this remains to be validated in statistically robust studies (3). Phosphatidylinositol 3 kinase (PI3K) is composed of a regulatory and a catalytic subunit (5). The latter is encoded by the PIK3CA gene. Mutations in PIK3CA are observed in 15% of CRCs (6); approximately 70% of PIK3CA mutations are located at exon 9 [a glutamic acid to lysine substitution at codons 542 (E542K) and 545 (E545K)] and 20% at exon 20 [a histidine to arginine substitution at codon 1047 (H1047R)] (7). PIK3CA has a level IIB CAP predictive value, indicating that it has shown promise in multiple studies; however, sufficient data for its inclusion in categories I or IIA are lacking (3). Although EGFR is a direct target of EGFR mAbs, the EGFR expression level does not have any predictive value in a clinical setting (3). Glutathione S-transferase II (GSTP) is involved in detoxification and may be used as a cancer marker (8). Overexpression of GSTP has been reported to be closely correlated with KRAS mutations; the GSTP expression level is higher in CRCs with KRAS mutations compared to wild-type KRAS (9). Expression of mutant KRAS activates GSTP at a transcriptional level. If this observation is reproducible in a clinical setting, the presence of a KRAS mutation may be distinguishable by GSTP immunohistochemistry (IHC).
One report, analyzing 233 genes, indicated that there may be differences in as few as 3% of genes between primary and metastatic sites (10). Moreover, mutations in the KRAS, BRAF and PIK3CA genes occur around the adenoma stage (10). In these situations, it is thought that the routine performance of one genetic test for KRAS mutations associated with metastatic CRC using DNA obtained from one organ, either from the primary or a metastatic site, whichever is preferentially available, is sufficient. However, the possibility of considerable tumor heterogeneity remains an issue. Recently, the possibility of acquired or intratumoral mutations of the KRAS gene was reported (11,12). Although the number of cases surveyed was small, the frequency of acquired mutations identified was not negligible. In our study, we identified 9 cases in which synchronous or metachronous metastasis was resectable, together with the primary CRC, and determined the status of target genes, including KRAS, BRAF, PIK3CA, EGFR and GSTP at each of these sites to determine the incidence of acquired mutations that may affect treatment with EGFR mAbs.
Materials and methods
Patient selection
Samples were collected from the primary site, and from at least one site of distant synchronous or metachronous metastasis, from 9 patients with colorectal adenocarcinoma whose tumors were resected at Akita University Hospital (Japan). This study was approved by the institutional ethics committee for clinical studies at Akita University, Graduate School of Medicine, on July 20th, 2010, and each of the patients gave their informed consent to the procedure.
Direct sequencing
Direct sequencing of codons 12 and 13 of KRAS, codon 600 of BRAF, and exons 9 and 20 of PIK3CA was outsourced to SRL Inc. (Tokyo, Japan) or Falco Biosystems Ltd. (Kyoto, Japan). Briefly, the tumor cell-rich area of a hematoxylin and eosin-stained section was identified by microscopy. Tissue was then removed from the same area of a deparaffinized, unstained section. DNA from sections of that tissue sample was then isolated using the QIAamp FFPE Tissue kit (QIAGEN K.K.; Tokyo, Japan) and exon 1 of the KRAS gene, exon 15 of the BRAF gene, and exons 9 and 20 of the PIK3CA gene were amplified by polymerase chain reaction (PCR). The PCR products were visualized using agarose gel electrophoresis with ethidium bromide staining. PCR DNA fragments were directly sequenced using an ABI 3130 Genetic Analyzer (Applied Biosystems; Foster City, CA, USA) according to the manufacturer’s instructions.
Immunohistochemistry
Almost all of the procedures were performed using a BenchMark XT IHC/ISH Staining Module (Roche Diagnostics K.K; Tokyo, Japan). Deparaffinized 4-μm specimens were used for IHC along with anti-human EGFR mouse monoclonal antibody (clone 2-18C9, Dako Japan; Tokyo, Japan), anti-human KRAS mouse monoclonal antibody (clone ab55391, Abcam Japan; Tokyo, Japan), and polyclonal rabbit anti-human GSTP (311-H, Medical and Biological Laboratories Co., Ltd.; Nagoya, Japan). Immunopositivity for EGFR was judged as positive if there were >0.1% positive cells. Immunoreactivities for KRAS and GSTP were graded as negative (0 to <10% positive cells), + (≥10 to <30% positive cells), ++ (>30 to <70% positive cells) and +++ (>70% positive cells). The percentage of immunopositive cells was calculated by counting at least 400 cancer cells in contiguous fields with the greatest immunopositivity.
Results
Patient characteristics
A total of 9 patients (3 females and 6 males) were included in this observational study. The median age was 67 years (range, 56–75). The patients were diagnosed as having CRC adenocarcinomas (2 rectal and 7 colon cancers). Three synchronous and 5 metachronous liver metastases, 2 synchronous and 5 metachronous lung metastases, and 1 synchronous ovarian metastasis were included. Resection of the primary region and at least one metastasis site was conducted either simultaneously or independently (Table I).
Table I.
Clinical profiling of 9 mCRC patients and their biomarker status.
| Characteristic | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 |
|---|---|---|---|---|---|---|---|---|---|
| Age | 72 | 66 | 65 | 56 | 67 | 75 | 71 | 72 | 57 |
| Gender | M | M | M | F | M | F | M | M | F |
| Primary | R | A | R | A | A | A | A | T | S |
| Hist | Wel-mod | Mod | Mod>wel | Mod | Mod | Wel | Mod | Wel | Wel>mod |
| Meta | Liver | Liver | Liver | Liver | Liver | Liver | Lung | Liver | Ovary |
| Lung | Lung | LN | |||||||
| Occurence | S | S | M | S | S | M | M | M | S |
| M | S | M | |||||||
| Interval (D) | - | - | 483 | - | - | 382 | 1652 | 2321 | - |
| 1435 | - | 2321 | |||||||
| KRAS | G13D | Wild | Wild | Wild | G12D | Wild | Wild | Wild | Wild |
| G13D | Wild | Wild | Wild | G12D | Wild | Wild | G12V | Wild | |
| Wild | G12D | G12V | |||||||
| BRAF | Wild | Wild | Wild | Wild | Wild | Wild | L597R | Wild | Wild |
| Wild | Wild | Wild | Wild | Wild | Wild | L597R | Wild | Wild | |
| Wild | Wild | ND | |||||||
| PIK3CA | Wild | Wild | Wild | Q546E | Wild | Wild | Wild | Wild | Wild |
| Wild | Wild | Wild | Q546E | Wild | Wild | Wild | Wild | Wild | |
| Wild | Wild | ND | |||||||
| EGFR | (−) | (+) | (+) | (+) | (−) | (−) | (−) | (−) | (+) |
| (−) | (+) | (+) | (+) | (−) | (−) | (+) | (+) | (+) | |
| (+) | (−) | ND | |||||||
| GSTP | (+++) | (+++) | (+++) | (+++) | (++) | (−) | (+) | (−) | (−) |
| (++) | (++) | (++) | (+++) | (+++) | (−) | ND | (−) | (−) | |
| ND | ND | ND |
S, synchronous metastasis; M, metachronous metastasis; Wel, well-differentiated; Mod, moderately differentiated; ND, non-defined. Interval indicates days between primary and metastatic lesion resection. CRC, colorectal cancer; LN, lymph node; GSTP, glutathione S-transferase II; EGFR, epidermal growth factor receptor; R, rectal; A, ascending; T, transverse; S, sigmoid.
Sequence analyses of the KRAS, BRAF and PIK3CA genes
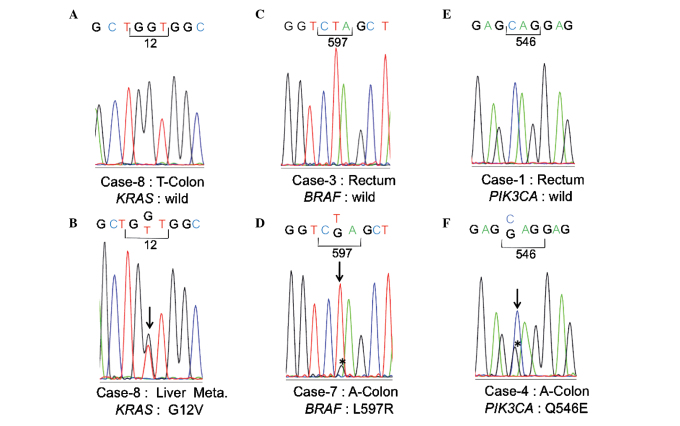
Regarding KRAS mutations, a glycine to aspartic acid mutation at codon 12 (G12D) was observed in the primary region of case 5, and a glycine to aspartic acid mutation at codon 13 (G13D) was observed in the primary region of case 1. In the remaining cases, no mutations were observed in the primary regions (Table I, Fig. 1A). The KRAS mutation frequency in the primary region was thus estimated to be 22.2% (2/9). At the metastatic sites, a G12D mutation was observed in both the lung and liver metastatic sites of case 5, and a G13D mutation was observed in the liver metastatic site of case 1. In case 8, a KRAS mutation involving a glycine to valine substitution at codon 12 (G12V) was observed in the liver metastatic site (Fig. 1B). In the remaining cases, no mutations were observed in the metastatic regions. The mutation frequency of KRAS at each metastatic site was thus estimated to be 27.3% (3/11).
Figure 1.
Sequence analyses of KRAS, BRAF and PIK3CA. The representative sequence analysis is shown for each case. Heterozygous mutations are shown by perpendicular lines.
No BRAF mutations were observed at exon 15 in the primary regions of all cases other than for case 7 (Fig. 1C)
In case 7, a leucine to arginine mutation was observed at codon 597 (L597R) (Fig. 1D). This L597R mutation was also observed at the site of lung metastasis in case 7. No other mutations were observed at any of the remaining metastatic sites. According to the genomic information found in the Catalogue of Somatic Mutations of Cancer (COSMIC), released by the Sanger Institute, L597R was confirmed as a somatic variant (http://www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=sample&id=749760).
As for PIK3CA, no mutations were observed at exons 9 and 20 in the primary region of all cases with the exception of case 4 (Fig. 1E). In case 4, a glutamine to glutamic acid mutation was observed at codon 546 (Q546E) at exon 9 (Fig. 1F). This Q546E mutation was also observed at the site of liver metastasis in case 4. No mutations were observed at the remaining metastatic sites. Q546E was confirmed as a somatic variant by COSMIC (http://www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=mut_summary&id=6147).
Immunohistochemical analyses of EGFR, KRAS and GSTP
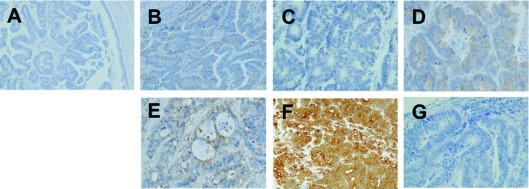
Immunopositivity for EGFR was observed at the primary site in 4 out of the 9 cases (cases 2, 3, 4 and 9) (Table I). A corresponding immunopositivity was observed at the metastatic sites in these 4 cases. However, a different immunopositivity was observed for cases 7 and 8, where the immunoreacitivity for EGFR was negative at the primary site but positive at the metastatic site (Fig. 2A and B). No immunopositivity was observed at the primary or metastatic sites in the remaining cases.
Figure 2.
Immunohistochemical analyses of EGFR, KRAS and GSTP. (A) Negative immunoreactivity for EGFR at the primary lesion and (B) positive immunoreactivity at the site of lung metastasis in case 7, (C) negative immunoreactivity for KRAS (G12D) at the primary lesion and (D) positive immunoreactivity for KRAS (G12D) at the site of lung metastasis in case 5, (E) immunoreactivity for GSTP is denoted by (++) at the primary lesion, (F) (+++) at the site of liver metastasis, and (G) (−) at the site of lung metastasis in case 5; all sites had the same KRAS G12D mutation. EGFR, epidermal growth factor receptor; GSTP, glutathione S-transferase II; G12D, glycine to aspartic acid mutation at codon 12.
Immunoreactivity for KRAS is apparently not dependent on the mutational status of KRAS (Table I, Fig. 2C and D)
Moreover, a correlation between the immunoreactivities or mutational status was not observed between KRAS and GSTP in this study (Fig. 2E-G). Therefore, we were unable to diagnose the mutational status of KRAS by GSTP IHC in a clinical setting.
Case presentation
In this observational study, a difference in the KRAS gene status between the primary and metastatic sites was observed in 1 (case 8) out of 9 cases (11.1%). This was independently confirmed by a separate analysis. Furthermore, the same KRAS mutation was detected in the resected mediastinal lymph node in case 8. Differences in immunopositivity for EGFR were observed in 2 (cases 7 and 8) out of 9 cases (22.2%). Case 7 was a 71-year-old male with ascending colon cancer. Following the primary resection, a 5-fluorouracil (5-FU) regimen (RPMI regimen) was administered for 6 months as adjuvant chemotherapy but 1,652 days following resection of the primary site, metastasis was evident in one lung and the site was resected (13). Modified leucovorin, fluorouracil and oxaliplatin (mFOLFOX6) was then administered as adjuvant chemotherapy for 180 days following resection until completion (April, 2011). Case 8 was a 72-year-old male with transverse colon cancer. Following primary resection, adjuvant chemotherapy was similarly administered for 6 months. However, 2,321 days after the primary resection, metastasis was detected at one site in the liver and in one mediastinal lymph node. The two sites were resected and mFOLFOX6 initiated as adjuvant chemotherapy (14). However, on day 159, mFOLFOX6 was terminated due to lung and abdominal lymph node metastases. A folinic acid-fluorouracil-irinotecan (FOLFIRI) regimen was then initiated and continued for 232 days up to the time of writing (15).
Discussion
In this study, we identified a possibility that acquired or intratumoral mutations may occur in the EGFR signaling pathway during CRC progression. Regarding KRAS, mutations in codons 12 and 13 were observed in 2 out of 9 cases at the primary site, and an acquired mutation was found in 1 case at a distal metastatic site. In previous reports, the mutation frequency of KRAS at codons 12 and 13 has ranged from 27 to 53% in CRC, which is similar to our finding (30%). The mutation frequencies of BRAF (V600E) and PIK3CA (exons 9 and 12) have been reported as 5–22% and 15%, respectively, in CRC. In our study, no oncogenic mutations of BRAF or PIK3CA were observed at either the primary or the metastatic sites. Differences in EGFR immunoreactivity were observed between the primary and metastatic sites in two instances, cases 7 and 8. In these two cases, the duration between the date of resection of the primary site and the date of metastatic recurrence was much longer (1,652 and 2,321 days, respectively) than that for the other cases (7 synchronous and 7 metachronous metastatic sites). In the remaining cases, the duration between the date of resection of the primary site and the date of onset of metastatic recurrence ranged from 217 to 952 days (median, 395). Since protein is easily degraded, the IHC analysis of EGFR may be affected by long-term storage. Therefore, the failure to detect immunoreactivity at the primary sites in cases 7 and 8 may be due to protein degradation during long-term storage. However, the direct sequencing of KRAS was successfully performed using DNA obtained from the archived specimens of the primary sites for these cases (Fig. 1). DNA is more stable than protein over longer periods; therefore, the quality of DNA in this study was sufficient for direct sequencing. In case 8, the possibility of an acquired or intratumoral mutation was suspected. The overall incidence of acquired or intratumoral mutations of KRAS was approximately 10% in this study, which is nearly identical to that of previous reports. Bouchahda et al reported acquired KRAS mutations (G12D and G13D) in 2 out of 13 cases (15.4%) (11). Richman et al reported intratumoral KRAS mutations at codons 12 and 13 in 5 out of 68 cases (7.4%) and an intratumoral BRAF mutation (V600E) in 2 cases (2.9%). Thus, in total, mutations in the EGFR pathway were identified in 7 out of 68 cases (10.3%) in their study (12). Although only 9 cases were analyzed in our study, each case had at least one resectable metastatic site and the total number of sites (combining primary and metastatic sites) was 18. Thus, it may be better to report an acquired mutation rate of approximately 11.1% (2/18). In conclusion, when metastatic recurrence occurs after a long interval, it is likely that acquired KRAS mutations may be identified.
Acknowledgements
This study was supported by the Ministry of Health, Labour, and Welfare.
References
- 1.Vincenzi B, Zoccoli A, Pantano F, Venditti O, Galluzzo S. Cetuximab: from bench to bedside. Curr Cancer Drug Targets. 2010;10:80–95. doi: 10.2174/156800910790980241. [DOI] [PubMed] [Google Scholar]
- 2.Giusti RM, Cohen MH, Keegan P, Pazdur R. FDA review of a panitumumab (Vectibix) clinical trial for firat-line treatment of metastatic colorectal cancer. Oncologist. 2009;14:284–290. doi: 10.1634/theoncologist.2008-0254. [DOI] [PubMed] [Google Scholar]
- 3.Ross JS, Torres-Mora J, Wagle N, Jennings TA, Jones DM. Biomarker-based prediction of response to therapy for colorectal cancer. Am J Clin Pathol. 2010;134:478–490. doi: 10.1309/AJCP2Y8KTDPOAORH. [DOI] [PubMed] [Google Scholar]
- 4.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 5.Garcia Z, Kumar A, Marques M, Cortes I, Carrera AC. Phosphoinositide 3-kinase controls early and late events in mammalian cell division. EMBO J. 2006;25:655–661. doi: 10.1038/sj.emboj.7600967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Aliya S, Reddanna P, Thyagaraju K. Does glutathione S-transferase Pi (GST-Pi) a marker protein for cancer? Mol Cell Biochem. 2003;253:319–327. doi: 10.1023/a:1026036521852. [DOI] [PubMed] [Google Scholar]
- 9.Miyanishi K, Takayama T, Ohi M, et al. Glutathione S-transferase-pi overexpression is closely associated with K-ras mutation during human colon carcinogenesis. Gastroenterology. 2001;121:865–874. doi: 10.1053/gast.2001.27982. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchahda M, Karaboué A, Saffroy R, et al. Acquired KRAS mutations during progression of colorectal cancer metastases: possible implications for therapy and prognosis. Cancer Chemother Pharmacol. 2010;66:605–609. doi: 10.1007/s00280-010-1298-9. [DOI] [PubMed] [Google Scholar]
- 12.Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, Quirke P. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol. 2011;34:61–66. doi: 10.3233/ACP-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller DG, Catalano PJ, Macdonald JS, O’Rourke MA, Frontiera MS, Jackson DV, Mayer RJ. Phase III study of fluorouracil, leucovorin, and levamisole in high-risk stage II and III colon cancer: final report of Intergroup 0089. J Clin Oncol. 2005;23:8671–8678. doi: 10.1200/JCO.2004.00.5686. [DOI] [PubMed] [Google Scholar]
- 14.André T, Boni C, Navarro M, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 15.Tournigand C, André T, Achille E, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR Study. J Clin Oncol. 2004;22:229–237. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]