Abstract
Clinically high-grade prostate cancers (PC) with high Gleason scores of 8–10 exhibit rapid growth and are more likely to spread beyond the prostate. These cancer types demonstrate a poor response to androgen deprivation therapy and eventually acquire a castration-resistant phenotype. To identify novel molecular cancer drug targets, we previously analyzed the gene expression profiles of high-grade PC using a cDNA microarray combined with laser microbeam microdissection and found a number of genes that are transactivated in high-grade PC. Among these genes, we report the identification of a novel molecular target, small nuclear ribonucleoprotein polypeptide E (SNRPE). Semi-quantitative RT-PCR confirmed that SNRPE is overexpressed in high-grade PC cells compared with normal prostatic epithelial cells. Knockdown of SNRPE expression by short interfering RNA (siRNA) resulted in the marked suppression of PC cell proliferation. By contrast, SNRPE overexpression promoted PC cell proliferation, indicating its oncogenic effects. Furthermore, we demonstrated that SNRPE regulates androgen receptor (AR) mRNA expression in PC cells. Knockdown of SNRPE expression by siRNA resulted in the marked suppression of AR and its downstream target genes at the mRNA level. We suggest that the regulation of AR expression by SNRPE is essential for cell proliferation and progression of high-grade PC and that it may be a novel molecular target for cancer drugs.
Keywords: small nuclear ribonucleoprotein polypeptide E, prostate cancer
Introduction
Prostate cancer (PC) is the most common malignancy in males and the second leading cause of cancer-related mortality in the United States and Europe (1). The incidence of PC has significantly increased in most developed countries, probably due to the prevalence of the Western lifestyle in these countries and the age distributions of their populations (1,2). Surgical and radiation therapies are effective for treating localized disease, but almost 30% of treated PC patients still suffer disease relapses (3–5). Clinically high-grade PC cases with Gleason scores of 8–10 exhibit rapid growth and are more likely to spread beyond the prostate. Most patients with advanced disease respond poorly to androgen deprivation therapy and often acquire a castration-resistant phenotype. Castration-resistant PC progresses aggressively, resulting in mortality. Thus, the development of new therapies based on the molecular mechanisms of PC progression is crucial.
In a previous study, we analyzed the gene expression profiles of high-grade PC using a cDNA microarray combined with laser microbeam microdissection to enrich populations of cancer cells (6). In this study, among the genes that are transactivated in high-grade PC cells, we focused on small nuclear ribonucleoprotein polypeptide E (SNRPE) as a novel molecular target of treatment for PC, to validate SNRPE overexpression in clinically high-grade PC with a Gleason score of 8–10.
Materials and methods
Cell lines
The human PC cell line 22Rv1 was obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA). Cell lines were cultured as monolayers in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) for 22Rv1 with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Sigma-Aldrich). The cells were maintained in incubators containing humidified air with 5% CO2 at 37°C.
Semi-quantitative RT-PCR
Tissue samples were obtained after receiving informed consent from high-grade PC patients undergoing prostatic needle biopsy before androgen ablation therapy. Purification of PC cells and normal prostatic epithelial cells from frozen PC tissues was described previously (6). Total RNA was extracted using the RNeasy Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, treated with DNase I (Roche, Basel, Switzerland), and reverse transcribed to single-stranded cDNA using oligo(dT)12–18 primer and Superscript reverse transcriptase II (Invitrogen, Carlsbad, CA, USA). We prepared the appropriate dilutions of each single-stranded cDNA and normalized the cDNA content to that of β-actin (ACTB). PCR reactions were then performed using the single-stranded cDNA as the PCR template. The primer sequences used were: ACTB (forward: 5′-TTGGCTTG ACTCAGGATTTA-3′, reverse: 5′-ATGCTATCACCTCCCCT GTG-3′); SNRPE (forward: 5′-ACCATGGCGTACCGT GGC-3′, reverse: 5′-CTAGTTGGAGACACTTTGTAGC AGA-3′); AR (forward: 5′-CGGAAGCTGAAGAAACT TGG-3′, reverse: 5′-CCTGGAGTTGACATTGGTGA-3′); KLK3 (PSA) (forward: 5′-CCAGACACTCACAGCAAGGA-3′, reverse: 5′-ATCCCATGCCAAAGGAAGAC-3′); NKX3.1 (forward: 5′-TGGTTTGTGAATCCATCTTGC-3′, reverse: 5′-AACAGGCTGTCTGGGTGAAA-3′), and TMPRSS2 (forward: 5′-CGAGGAGAAAGGGAAGACCT-3′, reverse: 5′-CCAGTCCGTGAATACCATCA-3′). The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by numerous cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, and elongation at 72°C for 30 sec on a TGradient Thermocycler (Biometra, Goettingen, Germany) (25 cycles for ACTB, 28 cycles for SNRPE, 28 cycles for AR, 28 cycles for PSA, 28 cycles for NKX3.1 and 28 cycles for TMPRSS2).
Construction of short hairpin RNA expression vectors and the cell viability assay
Plasmids designed to express short hairpin RNA (shRNA) were prepared by cloning double-stranded oligonucleotides into the pBAsi-h6 Neo vector (Takara Bio Inc., Shiga, Japan). The target sequences for SNRPE were as follows: sense strand sequence for siSNRPE: 5′-GCTCTATGAGCAAGTGAAT-3′, and negative control EGFP siRNA (siEGFP): 5′-GAAGCAGCACGACTTCTTC-3′, as previously described (6). 22Rv1 cells (2×106) that highly expressed SNRPE were seeded in 10-cm dishes, transfected with pBA-siSNRPE or pBA-siEGFP using FuGENE6 (Roche Diagnostics) according to the manufacturer’s instructions, and then cultured in the appropriate medium containing 800 μg/ml Geneticin (Sigma-Aldrich) for 14 days. The cells were fixed with 100% methanol and stained with 0.1% crystal violet-H2O for the colony formation assay. In the cell viability assay, cell viability was measured using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) 10 days after transfection. Absorbance was measured at 450 nm as a reference with an iMark Microplate Absorbance Reader (Bio-Rad, Hercules, CA, USA). Preliminarily, the knockdown effects of these shRNA expression vectors on endogenous SNRPE expression were validated 7 days following their transfection by RT-PCR using the primers employed for semi-quantitative RT-PCR.
Generation of SNRPE-overexpressing cells and the cell proliferation assay
Full-length human SNRPE cDNA (accession no. NM003094) was amplified using primers containing Myc-tag sequences in their NH2 termini and cloned into the pIRESneo3 vector (Clontech). Human 22Rv1 cells were seeded into a 10-cm dish (50% confluent) and transfected with 6 μg pIRESneo3 empty vector or pIRESneo3-Myc-SNRPE expression vector using FuGENE6 reagent (Roche), according to the manufacturer’s instructions. The cells were selected with appropriate medium containing 400 μg/ml Geneticin (Sigma-Aldrich) for 14 days, and then discrete colonies were collected. Stable clones were maintained in selective medium and assayed for exogenous SNRPE expression by RT-PCR. The proliferation of 22Rv1 cells that stably expressed SNRPE (22Rv1 SNRPE clone mixture) and those transfected with pIRESneo3 empty vector (22Rv1 mock clone mixture) was examined using the Cell Counting Kit-8 (Dojindo). The 22Rv1 SNRPE and 22Rv1 mock cells were seeded at a concentration of 5×103 cells per well in 48-well plates. The assay was performed every 48 h for 7 days, according to the manufacturer’s instructions.
cDNA microarray analysis and data acquisition
The 22Rv1 cells were transfected with pBA-siSNRPE or pBA-siEGFP, as described above. Total RNA was extracted using the RNeasy Mini Kit 7 days after the transfection, according to the manufacturer’s instructions. The GeneChip® array data were compared using the Kurabo custom analysis service (Kurabo Industries Ltd., Osaka, Japan; Kurabo Industries Ltd. is an authorized service provider for Affymetrix Japan K.K., Tokyo, Japan). Briefly, total RNA was reverse transcribed to cDNA with T7-Oligo(dT) primer (Affymetrix, Inc.). The cDNA synthesis product was used in an in vitro transcription (IVT) reaction involving T7 RNA polymerase. An unlabeled ribonucleotide mix was used in the first cycle of IVT amplification. Unlabeled cRNA was then reverse transcribed in the first-strand cDNA synthesis step of the second cycle using random primers. Subsequently, the T7-Oligo(dT) promoter primer was used for the second-strand cDNA synthesis to generate a double-stranded cDNA template containing T7 promoter sequences. The resultant double-stranded cDNA was then amplified and labeled using a biotinylated nucleotide analog/ribonucleotide mix in the second IVT reaction. The labeled cRNA products were then fragmented, loaded onto the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix, Inc., CA, USA), and hybridized according to the manufacturer’s instructions. Streptavidin-phycoerythrin (Molecular Probes) was used as a fluorescent conjugate to detect the hybridized target sequences. Raw intensity data from the GeneChip array were analyzed using GeneChip Operating Software (Affymetrix, Inc.).
Results
SNRPE overexpression in high-grade PC cells
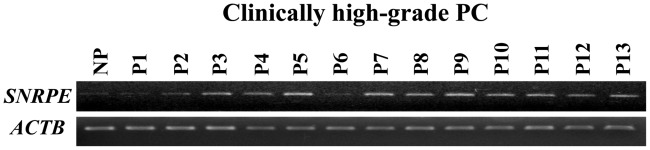
In a previous study, we analyzed the expression profiles of high-grade PC cells purified from clinically high-grade PC tissues with high PSA levels and high Gleason scores (6). Among the genes that were transactivated in the high-grade PC cells compared with normal prostatic epithelial cells, we focused on SNRPE in this study. Semi-quantitative RT-PCR (Fig. 1) confirmed that SNRPE expression was elevated in 11 out of 13 clinically high-grade PC cells.
Figure 1.
Small nuclear ribonucleoprotein polypeptide E (SNRPE) overexpression in high-grade PC cells. Semi-quantitative RT-PCR confirmed the elevated expression of SNRPE in 11 out of 13 clinically high-grade PC cells compared with normal prostatic (NP) epithelial cells. RNA content was normalized to that of β-actin (ACTB).
Knockdown of SNRPE expression by shRNA decreased the PC cell number
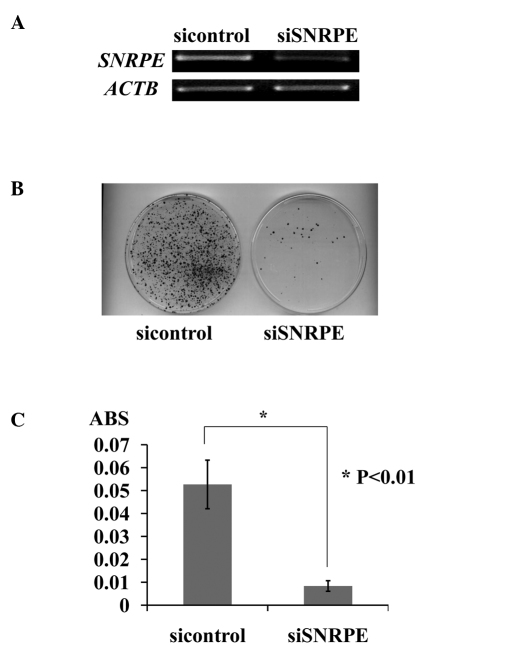
To examine the biological roles of SNRPE overexpression in PC cells, we constructed vectors designed to express shRNA specific to SNRPE and EGFP, and transfected them into 22Rv1 cells, which express endogenous SNRPE (Fig. 2A). Of the two shRNA expression vectors, siSNRPE had a significant knockdown effect on the endogenous SNRPE transcript, and its transfection resulted in a decreased number of PC colonies and a decreased number of viable PC cells as measured by the cell viability assay. By contrast, the transfection of the negative control (siEGFP) had little or no effect on SNRPE expression and did not affect the viability of 22Rv1 cells (Fig. 2B and C).
Figure 2.
Knockdown of small nuclear ribonucleoprotein polypeptide E (SNRPE) expression by siRNA decreased the prostate cancer cell number. (A) Effect of siRNA knockdown of SNRPE expression in 22Rv1 cells. Semi-quantitative RT-PCR was performed using cells that had been transfected with shRNA-expressing vectors specific to SNRPE and EGFP. RNA content was normalized to that of β-actin (ACTB). (B) Colony formation assay of 22Rv1 cells transfected with the indicated shRNA-expressing vectors specific to SNRPE and EGFP. The cells were visualized with 0.1% crystal violet staining after 14 days of incubation with Geneticin. (C) Cell viability assays of the 22Rv1 cells transfected with the indicated shRNA-expressing vectors specific to SNRPE and EGFP. Each mean value is plotted together with error bars, indicating standard deviation. ABS on the Y-axis indicates absorbance at 450 nm. These experiments were carried out in triplicate (*P<0.01, Student’s t-test).
SNRPE overexpression promoted PC cell proliferation
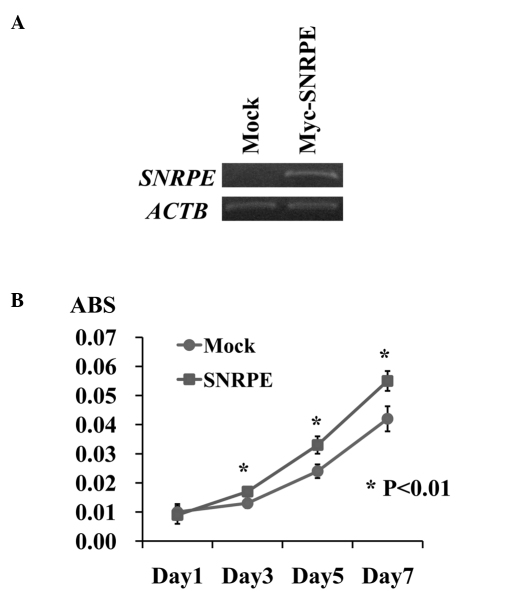
To investigate the potential oncogenic function of SNRPE, we established stable transformants from 22Rv1 cells, which constitutively expressed exogenous SNRPE. We also prepared control 22Rv1 cells, which were transfected with the empty vector (mock), and compared their proliferation. RT-PCR detected a high level of exogenous SNRPE expression in the stable clone. A cell proliferation assay revealed that the 22Rv1 SNRPE-overexpressing clones grew more rapidly than the 22Rv1 mock clones (*P<0.01, Student’s t-test), indicating that SNRPE overexpression promoted PC cell proliferation (Fig. 3).
Figure 3.
Small nuclear ribonucleoprotein polypeptide E (SNRPE) overexpression promoted cancer cell proliferation. (A) RT-PCR detected a high level of exogenous SNRPE expression in the stable clones. (B) Cell proliferation assay revealed that the 22Rv1 SNRPE-overexpressing clone grew more rapidly than the 22Rv1 mock clone (*P<0.01, Student’s t-test), indicating that SNRPE overexpression promoted prostate cancer cell proliferation.
SNRPE regulates AR signaling pathways in PC cells
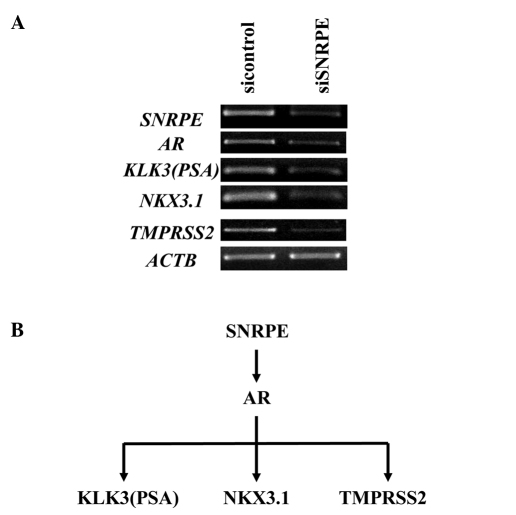
To identify the RNA targets regulated by SNRPE to explain its effect on cell proliferation, we performed a gene expression analysis using a cDNA microarray. The gene expression patterns of the 22Rv1 cells transfected with the two shRNA expression vectors (siSNRPE and siEGFP) were compared. The gene expression patterns of the 22Rv1 SNRPE-overexpressing clone and 22Rv1 mock clone were also compared. As a result, we found that the expression of the AR gene was downregulated by ≥80% in the SNRPE knockdown cells and upregulated by ≥20% in the SNRPE-overexpressing clone. Semi-quantitative RT-PCR confirmed the significant downregulation of AR and its downstream target genes (PSA, NKX3.1 and TMPRSS2) in the SNRPE knockdown clone (Fig. 4).
Figure 4.
Small nuclear ribonucleoprotein polypeptide E (SNRPE) regulates androgen receptor (AR) signaling pathways in prostate cancer cells. (A) Semi-quantitative RT-PCR confirmed that the expression levels of AR and its downstream target genes (PSA, NKX3.1 and TMPRSS2) were significantly downregulated by SNRPE knockdown. (B) SNRPE regulates AR signaling pathways in prostate cancer cells.
Discussion
We identified SNRPE as a novel molecular target of cancer drug therapy for high-grade PC. SNRPE, which is also known as spliceosomal protein E (SmE), was originally identified as an essential component of the spliceosomal complex, which is involved in pre-mRNA processing. SNRPE (SmE) belongs to a large family of polypeptides containing Sm and Sm-like (Lsm) proteins, which are conserved in eukaryotes and archaebacteria (7,8). Seven Sm proteins (SmB/B’, SmD1, SmD2, SmD3, SmE, SmF and SmG) form a core ring structure that encompasses RNA. All Sm proteins contain the Sm domain, which consists of two blocks of conserved amino acids (Sm motif 1 and Sm motif 2) that are responsible for the assembly of snRNAs (U1, U2, U4, U5 and U6) in an ordered manner to form the Sm core of the spliceosomal snRNPs (8,9) and are thereby involved in pre-mRNA processing (10). Generally, mRNA processing factors are considered to function only during the control of global gene expression, during which they are involved in essential pre-mRNA splicing. However, several studies have revealed that Sm and Lsm proteins perform other physiological activities that are distinct from pre-mRNA splicing (11–13).
In this study, we confirmed that high-grade PC cells overexpress SNRPE. The knockdown of SNRPE expression by siRNA resulted in the marked suppression of PC cell proliferation. By contrast, exogenous SNRPE expression in transfected cells promoted PC cell proliferation. These findings demonstrate the involvement of SNRPE in the proliferation and viability of PC cells.
Notably, using cDNA microarray analysis we have shown that SNRPE regulates AR mRNA expression in PC cells. The knockdown of SNRPE expression by siRNA resulted in the marked suppression of AR and its downstream target genes at the mRNA level. AR plays a key role in the modulation of prostate cell proliferation and is involved in the development and progression of PC. Several studies using in vitro prostate cancer cell lines and mouse models have suggested that the progression to castration-resistant PC is associated with increased levels of AR expression, indicating that AR downregulation using siRNA or other methods may inhibit tumor growth even in castration-resistant PC (14–17). A study by Tamura et al (18) provided new insights into the molecular characteristics that distinguish castration-resistant PC from castration-naïve PC. These authors revealed a significant association between high-levels of AR, SNRPE expression and castration-resistant PC. High-grade PC has a similar molecular profile to castration-resistant PC, which may reflect dedifferentiation and explain the clinical association between grade and prognosis (19). Our findings regarding SNRPE overexpression in high-grade PC have shown that SNRPE is crucial in PC progression through the regulation of AR expression.
In conclusion, we suggest that the regulation of AR expression by SNRPE is essential for the cell proliferation and progression of high-grade PC, although the detailed mechanisms responsible for these effects remain unknown and should be elucidated by further investigations. We also demonstrated its positive involvement in cell proliferation and the regulation of AR expression. Our findings could provide new insights into the molecular mechanisms of PC progression and indications that will aid the development of new therapeutic drugs for PC.
Acknowledgements
This study was supported in part by a grant for Research for Promoting Technological Seeds A (discovery type) no. 14-063 (to K. Tamura) from the Japan Science and Technology Agency.
References
- 1.Grönberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–64. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 2.Hsing AW, Devesa SS. Trends and patterns of prostate cancer: what do they suggest? Epidemiol Rev. 2001;23:3–13. doi: 10.1093/oxfordjournals.epirev.a000792. [DOI] [PubMed] [Google Scholar]
- 3.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 5.Han M, Partin AW, Piantadosi S, et al. Era specific biochemical recurrence-free survival following radical prostatectomy for clinically localized prostate cancer. J Urol. 2001;166:416–419. [PubMed] [Google Scholar]
- 6.Satake H, Tamura K, Furihata M, et al. The ubiquitin-like molecule interferon-stimulated gene 15 is overexpressed in human prostate cancer. Oncol Rep. 2010;23:11–16. [PubMed] [Google Scholar]
- 7.Mayes AE, Verdone L, Legrain P, et al. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, et al. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermann H, Fabrizio P, Raker VA, et al. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrecht M, Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 12.Barbee SA, Evans TC. The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev Biol. 2006;291:132–143. doi: 10.1016/j.ydbio.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Bilinski SM, Jaglarz MK, Szymanska B, et al. Sm proteins, the constituents of the spliceosome, are components of nuage and mitochondrial cement in Xenopus oocytes. Exp Cell Res. 2004;299:171–178. doi: 10.1016/j.yexcr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 15.Gregory CW, Hamil KG, Kim D, et al. Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen regulated genes. Cancer Res. 1998;58:5718–5724. [PubMed] [Google Scholar]
- 16.Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to anti-androgen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 17.Zegarra-Moro OL, Schmidt LJ, Huang H, et al. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 2002;62:1008–1013. [PubMed] [Google Scholar]
- 18.Tamura K, Furihata M, Tsunoda T, et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 19.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]