Abstract
In order to elucidate differences between oral cancers and cancers of the pharynx and larynx, we investigated the genetic and epigenetic changes in these tumors using molecular biology methods. Methylation of the promoter region of the p16 tumor suppressor gene was examined using methylation-specific polymerase chain reaction in specimens from 47 oral, 39 pharyngeal and 35 laryngeal squamous cell carcinomas. These specimens were also characterized for allelic loss of certain areas of the genome, i.e., 3p22, 9p21 and 17p13 (TP53). The frequency of methylation of the promoter region of the p16 gene in tongue cancers (35.3%) was significantly higher than in pharyngeal (12.8%) and laryngeal cancers (11.4%) (p=0.046 and p=0.039, respectively). The frequency of methylation in tumors of female patients (47.1%) was significantly higher compared to tumors of male patients (15.4%) (p=0.0067). In contrast, the frequency of the loss of heterozygosity (LOH) at 3p21 in pharyngeal cancers (66.7%) was significantly higher than in oral cancers (20.0%) (p=0.0006). The frequencies of LOH at 17p13 in pharyngeal (71.0%) and laryngeal cancers (73.1%) were also significantly higher than in oral cancers (36.1%) (p=0.009 and p=0.009, respectively). Our results indicate that there are marked differences in the frequencies of the hypermethylation of genes and allelic loss between oral cancers and cancer of the pharynx and larynx. Although all of these tumors were diagnosed as squamous cell carcinomas, the process of carcinogenesis may be different in tumors located in various parts of the head and neck. Loss of function of tumor suppressor genes by allelic loss gives rise to tumors in the pharynx and larynx, while loss of function due to methylation of the promoter regions of those genes is related to carcinogenesis in the oral cavity.
Keywords: head and neck cancers, loss of heterozygosity, DNA methylation, location of the tumor, gender
Introduction
Head and neck squamous cell carcinomas (HNSCCs) manifest various clinical behaviors according to their area of origin, i.e., from various parts of the head and neck mucosa. However, genetic and epigenetic factors involved in the carcinogenesis of HNSCC in various tissues have yet to be thoroughly studied and evaluated. Although there have been numerous studies on the hypermethylation of the promoter regions of tumor suppressor genes, particularly p16 (1–9), differences in the frequencies of methylation among the cancer sites in the head and neck region have yet to be elucidated, partly due to the low number of samples. However, we have previously pointed out differences in the frequencies of allelic loss of certain areas of the genome between oral cancers and cancers of the pharynx and larynx (10).
To elucidate the differences between oral cancer and cancers of the pharynx and larynx, we investigated the genetic and epigenetic changes in these tumors using molecular biology methods. We analyzed the hypermethylation of the promoter region of the p16 tumor suppressor gene as an epigenetic change. We also examined allelic loss of certain areas of the genome, i.e., 3p21, 9p21 and 17p13 (TP53) as genetic changes. Comparing the results of these two approaches reveals novel findings regarding the differences between oral cancer and cancers of the pharynx and larynx.
Materials and methods
Patients and tissue specimens
Primary tumors were removed surgically or obtained as biopsies prior to treatment during the period between May 1994 and November 2000, following an approved protocol at the Miyagi Cancer Center. The clinical features of the patients are summarized in Table I.
Table I.
Clinical features of the patients and tumor location.
| Oral cavity | Pharynx | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Overall | Tongue | FOM | BM | Gingiva | Overall | Meso | Hypo | Larynx | P-value | |
| Age | ||||||||||
| 45> | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.021 (OC vs. P) |
| 45< | 41 | 28 | 6 | 4 | 3 | 39 | 14 | 25 | 33 | |
| Gender | ||||||||||
| Female | 14 | 9 | 0 | 4 | 1 | 2 | 0 | 1 | 1 | 0.0034 (OC vs. P) |
| Male | 33 | 25 | 6 | 0 | 2 | 37 | 14 | 24 | 34 | 0.0018 (OC vs. L) |
| Tumor sizea,b | ||||||||||
| T1 | 8 | 6 | 2 | 0 | 0 | 1 | 0 | 1 | 8 | NS |
| T2 | 20 | 14 | 3 | 2 | 1 | 19 | 4 | 15 | 14 | |
| T3 | 13 | 10 | 1 | 2 | 0 | 9 | 6 | 3 | 6 | |
| T4 | 3 | 1 | 0 | 0 | 2 | 9 | 4 | 5 | 6 | |
| Lymph node involvementa | N0 vs. others | |||||||||
| N0 | 28 | 20 | 4 | 2 | 2 | 10 | 2 | 8 | 29 | 0.0016 (OC vs. P) |
| N1 | 8 | 6 | 1 | 1 | 0 | 7 | 3 | 4 | 1 | <0.0001 (P vs. L) |
| N2 | 10 | 7 | 1 | 1 | 1 | 20 | 9 | 11 | 4 | |
| N3 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 1 | |
| Stagea | I and II vs. III and IV | |||||||||
| I | 8 | 7 | 1 | 0 | 0 | 1 | 0 | 1 | 7 | 0.0004 (OC vs. P) |
| II | 17 | 12 | 3 | 1 | 1 | 5 | 0 | 5 | 11 | 0.0009 (P vs. L) |
| III | 10 | 7 | 1 | 2 | 0 | 9 | 4 | 5 | 6 | |
| IV | 12 | 8 | 1 | 1 | 2 | 24 | 10 | 14 | 11 | |
| Total | 47 | 34 | 6 | 4 | 3 | 39 | 14 | 25 | 35 | |
Patients who had recurrent lymph node metastasis were omitted from this classification. Meso, mesopharyngeal; Hypo, hypopharyngeal; NS, not significant. FOM, floor of mouth; BM, buccal mucosa; OC, oral cavity; P, pharynx; L, larynx.
This study was approved by the Institutional Review Boards of Tohoku University and the Miyagi Cancer Center. Surgical specimens were obtained under informed consent, and written informed consent was obtained from all patients.
DNA preparation
Genomic DNA was extracted from the frozen tissue samples using proteinase K digestion and phenol extraction according to standard methods (11,12).
Detection of hypermethylation in the p16 gene promoter region
The detection of gene hypermethylation was performed using sodium bisulfite for DNA modification followed by methylation-specific polymerase chain reaction (MSP). DNA modification was performed using a CpGenome DNA Modification Kit (Chemicon International, Temecula, CA, USA). The sequences of the primers used were as follows: p16U forward, 5′-TTATT AGAGGGTGGGGTGGATTGT-3′ and reverse, 5′-CAACCCC AAACCACAACCATAA-3′; p16M forward, 5′-TTATTAGA GGGTGGGCGGATCGC-3′ and reverse, 5′-GACCCCGAACC GCGACCGTAA-3′. The annealing temperatures of the PCR reactions for p16U and p16M were 60 and 69°C, respectively. Purified PCR products were separated by electrophoresis on 3% agarose gels containing 1 mg/ml ethidium bromide.
Detection of allelic loss
The polymorphic markers, D3S1067, IFNA, D3S171 and TP53, were purchased from Invitrogen (Carlsbad, CA, USA) and each forward primer of those markers was labeled using [γ-32p]ATP and polynucleotide kinase (Takara Shuzo, Kyoto, Japan). PCR was performed using a thermal cycler (Perkin-Elmer Cetus, Boston, MA, USA) for 36 cycles under optimal conditions, i.e., 1 min at 94°C, 1 min at 56°C and 1 min at 72°C, except for the first denaturation, which was performed at 94°C for 5 min. Purified PCR products were separated by electrophoresis on 6% polyacrylamide gels containing 8.3 M urea (10).
Statistical analysis
Statistical analyses of the data were performed using the χ2 test.
Results
Table I shows the clinicopathological features of the patients and their tumors. The ages of the patients with oral cancers were significantly lower than those with cancers of the pharynx and larynx. As for gender, the number of female patients was significantly higher among patients with oral cancer than those with cancers of the pharynx and larynx. Advanced stage (stage III and IV) patients were significantly higher in patients with cancers of the pharynx and larynx than those with oral cancer. This is due to the fact that lymph node metastases were significantly more frequent in patients with cancers of the pharynx and larynx than in those with oral cancer.
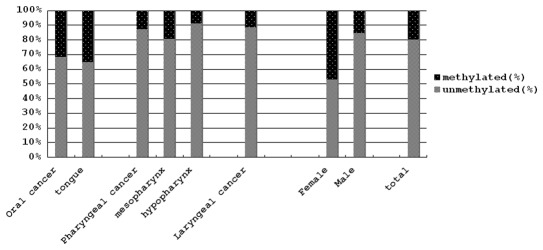
Hypermethylation of the promoter region of the p16 tumor suppressor gene was detected using the MSP method. We examined 47 oral cancers, 39 pharyngeal cancers and 35 laryngeal cancers. Representative results are shown in Fig. 1 and all results are summarized in Table II and Fig. 2. The frequencies of methylation of the promoter region of the p16 gene in oral and tongue cancers were 31.9 and 35.3%, respectively. On the contrary, those of pharyngeal and laryngeal cancers were 12.8 and 11.4%, respectively, and those of mesopharyngeal and hypopharyngeal cancers were 21.4 and 8.7%, respectively. The frequency of methylation in oral cancers was significantly higher than in hypopharyngeal cancers (p=0.047). The frequency of methylation in tongue cancers was significantly higher than in pharyngeal, hypopharyngeal and laryngeal cancers (p=0.046, 0.033 and 0.039, respectively). However, the frequency of methylation in tumors of female patients (47.1%) was significantly higher than in tumors of male patients (15.4%) (p=0.0067).
Figure 1.
Hypermethylation of the p16 gene promoter region detected by the bisulfite-MSP method. Lane 1, tongue cancer (T2N0M0); lane 2, laryngeal cancer (T4N0M0); lane 3, cancer of the floor of the mouth (T2N0M0); lane 4, hypopharyngeal cancer (T2N2aM0); lane 5, hypopharyngeal cancer (T2N2aM0). U, unmethylated; M, methylated. The methylated band was observed only in tongue cancer (lane 1).
Table II.
Hypermethylation of the p16 promoter region, tumor location and gender.
| Unmethylated | Methylated | Methylated (%) | P-value | |
|---|---|---|---|---|
| Oral cancer | 32 | 15 | 31.9 | 0.047 (vs.HP)a |
| Tongue | 22 | 12 | 35.3 | 0.046 (vs.P)b |
| Floor of mouth | 6 | 0 | ||
| Buccal mucosa | 1 | 3 | ||
| Gingiva | 3 | 0 | ||
| Pharyngeal cancer | 34 | 5 | 12.8 | |
| Mesopharynx | 11 | 3 | 21.4 | |
| Hypopharynx | 23 | 2 | 8.7 | 0.033 (vs. T)c |
| Laryngeal cancer | 31 | 4 | 11.4 | 0.039 (vs. T)d |
| Gender | ||||
| Female | 9 | 8 | 47.1 | 0.0067 |
| Male | 88 | 16 | 15.4 | |
| Total | 97 | 24 | 19.8 | |
Significant differences were observed between
oral cancer (OC) and hypopharynx (HP),
tongue (T) and pharyngeal cancer (P),
T and HP,
T and laryngeal cancer.
Figure 2.
Hypermethylation of the p16 gene promoter region in HNSCC. The frequencies of hypermethylation of the p16 gene promoter region are shown in the percentage column for each site. Columns on the left show differences between oral cancer and cancers of the pharynx and larynx. Columns on the right show differences between females and males.
Specimens of 47 oral, 39 pharyngeal and 35 laryngeal squamous cell carcinomas were characterized for allelic loss of certain areas of the genome, i.e., 3p22, 9p21 and 17p13 (TP53). The frequency of loss of heterozygosity (LOH) at 3p21 in pharyngeal cancers (66.7%) was significantly higher than in oral and tongue cancers (20.0 and 14.8%) (p=0.0006 and 0.0003, respectively) (Table IIIA). The frequencies of LOH at 3p21 in mesopharyngeal and hypopharyngeal cancers (63.6 and 68.8%) were also significantly higher than in oral cancers (p=0.018 and p=0.002, respectively). The frequencies of LOH at 3p21 in mesopharyngeal and hypopharyngeal cancers were also significantly higher than in tongue cancers (p=0.009 and p=0.001, respectively).
Table III.
LOH at 3p21, 9p21 and TP53 and tumor location.
| A, LOH at 3p21 and location of tumors. | ||||
|---|---|---|---|---|
| 3p21 | ||||
| Location | N | LOH(+) | LOH(−) | P-value |
| Oral cavity | 35 | 7 | 28 | p=0.018 (MP)a, p=0.002 (HP)b |
| Tongue | 27 | 4 | 23 | p=0.009 (MP)c, p=0.001 (HP)d |
| Floor of mouth | 4 | 1 | 3 | |
| Buccal mucosa | 2 | 1 | 1 | |
| Gingiva | 2 | 1 | 1 | |
| Pharynx | 27 | 18 | 9 | p=0.0006 (OC)e, p=0.0003 (T)f |
| Mesopharynx | 11 | 7 | 4 | |
| Hypopharynx | 16 | 11 | 5 | |
| Larynx | 28 | 12 | 16 | |
| Total | 90 | 37 | 53 | |
| B, LOH at 9p21 and location of tumors. | ||||
|---|---|---|---|---|
| 9p21 | ||||
| Location | N | LOH(+) | LOH(−) | P-value |
| Oral cavity | 26 | 9 | 17 | - |
| Tongue | 18 | 6 | 12 | - |
| Floor of mouth | 4 | 2 | 2 | - |
| Buccal mucosa | 3 | 0 | 3 | - |
| Gingiva | 1 | 1 | 0 | - |
| Pharynx | 24 | 14 | 10 | - |
| Mesopharynx | 9 | 4 | 5 | - |
| Hypopharynx | 15 | 10 | 5 | - |
| Larynx | 25 | 15 | 10 | - |
| Total | 75 | 38 | 37 | - |
| C, LOH at TP53 and location of tumors. | ||||
|---|---|---|---|---|
| TP53 | ||||
| Location | N | LOH(+) | LOH(−) | P-value |
| Oral cavity | 36 | 13 | 23 | p=0.02 (MP)a, p=0.009 (P)b |
| Tongue | 27 | 10 | 17 | p=0.03 (MP)c, p=0.02 (P)d |
| Floor of mouth | 4 | 1 | 3 | |
| Buccal mucosa | 3 | 0 | 3 | |
| Gingiva | 2 | 2 | 0 | |
| Pharynx | 31 | 22 | 9 | |
| Mesopharynx | 11 | 9 | 2 | |
| Hypopharynx | 20 | 13 | 7 | |
| Larynx | 26 | 19 | 7 | p=0.009 (OC)e, p=0.018 (T)f |
| Total | 93 | 54 | 39 | |
Significant differences were observed between
oral cavity (OC) and mesopharynx (MP),
OC and hypopharynx (HP),
tongue (T) and MP,
tongue (T) and HP,
OC and pharynx (P),
T and P. LOH, loss of heterozygosity.
No significant difference was observed. LOH, loss of heterozygosity.
Significant differences were observed between
oral cavity (OC) and mesopharynx (MP),
OC and pharynx (P),
tongue (T) and MP,
T and P,
OC and larynx (L),
T and L. LOH, loss of heterozygosity.
Although the frequencies of LOH at 9p21 in pharyngeal (58.3%) and laryngeal cancers (60.0%) were higher than in oral cancers (34.6%), significant differences were not observed. The frequencies of LOH at 9p21 in mesopharyngeal (44.4%) and hypopharyngeal cancers (66.7%) were also higher than in tongue cancers (33.3%). However, significant differences were also not observed (Table IIIB).
The frequencies of LOH at 17p13 in pharyngeal (71.0%) and laryngeal cancers (73.1%) were significantly higher than in oral cancers (36.1%) (p=0.009 and p=0.009, respectively). The frequencies of LOH at 17p13 in pharyngeal and laryngeal cancers were significantly higher than in tongue cancers (37.0%) (p=0.02 and p=0.018, respectively). The frequency of LOH at 17p13 in mesopharyngeal cancers (81.8%) was significantly higher than in oral and tongue cancers (p=0.02 and p=0.03, respectively) (Table IIIC). These results are summarized in Fig. 3.
Figure 3.
Differences in the frequencies of LOH in HNSCC. The frequencies of LOH are shown in the percentage column for each site. Significant differences were observed in comparison between the frequencies of LOH of cancers of various regions of the head and neck (Table III and relevant text). LOH, loss of heterozygosity; HNSCC, head and neck squamous cell carcinoma.
Discussion
We focused on differences in HNSCCs at various regions of origin with respect to the genetic and epigenetic aspects of the tumors. Although we have previously reported differences in the frequencies of allelic loss in several regions of head and neck cancers (10), in the present study our results indicate that there are marked differences in the frequencies of hypermethylation of genes between oral cancers and cancers of the pharynx and larynx, as well as allelic loss.
There have been numerous studies on the hypermethylation status of the promoter region of the p16 tumor suppressor gene. The frequency of methylation was found to be approximately 30% in head and neck squamous cell carcinoma specimens obtained from surgically removed tumors (1–9). Although several reports have described the hypermethylation of the promoter region of the p16 tumor suppressor gene in oral cancer and in cancers of the pharynx and larynx, there have not been any reports which point out differences in the frequencies of the methylation among those cancers. This may have been largely due to the low number of samples.
Our results indicate that the hypermethylation of the promoter region of the p16 tumor suppressor gene is related to gender, i.e., female patients tend to have more frequent hypermethylation of the p16 gene. El-Naggar et al also reported that there was no significant correlation between p16 alterations and the clinicopathological factors of the patients, with the exception of gender (3). Although their methods of characterization were different from ours and the detection of the hypermethylation itself was carried out using a method different from ours, their results are compatible with ours. Oral cancer patients included significantly more females than patients with cancers of the pharynx and larynx. We assumed that this tendency resulted in the significant differences of the frequencies of hypermethylation between oral cancer and cancers of the pharynx and larynx. This indicates that the hypermethylation of the promoter region of the p16 tumor suppressor gene may be a crucial factor in the carcinogenesis of squamous cell carcinomas of the head and neck in female patients.
In HNSCC, allelic loss on chromosome 9p occurs most frequently at 9p21 (1,13–15) and it occurs as an early event in the process of carcinogenesis in head and neck cancers (13). This region of 9p21 includes the tumor suppressor and cell cycle regulator gene, p16 (INK4a, also known as CDKN2 or MTS1) (16–19). Although homozygous deletions, monosomy and point mutations of p16 occur frequently in HNSCC cell lines, corresponding evidence of the same alterations in primary cancer specimens is less common (1,14,20,21). However, primary HNSCC shows frequent methylation of the promoter region of the p16 gene (22,23) and LOH at 9p21. It appears that these two types of gene alteration are the main mechanism underlying the inactivation of the p16 gene as a tumor suppressor. In our study, LOH at 9p21 was observed in approximately half of the specimens, and it was more frequent in cancers of the pharynx and larynx than in oral cancer. In contrast, hypermethylation of the p16 gene was more frequent in oral cancer than in cancers of the pharynx and larynx. These results indicate that the type of inactivation of the p16 gene is different in various sites of head and neck squamous cell carcinomas. It should also be noted that allelic loss at 3p and 17p is significantly more frequent in cancers of the pharynx and larynx than in oral cancers.
In conclusion, although these tumors were diagnosed as squamous cell carcinomas, the process of carcinogenesis may be different in tumors located in different parts of the head and neck. Loss of function of tumor suppressor genes by allelic loss is mainly active and give rise to tumors in the pharynx and larynx, while loss of function due to methylation of the promoter region of the genes is related to carcinogenesis in the oral cavity, which may place additional burden on allelic loss.
Acknowledgements
We are grateful to Ms. Elizabeth Hearing for her editorial work in the preparation of this manuscript.
References
- 1.Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Barterk J, Sidransky D. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3633. [PubMed] [Google Scholar]
- 2.Gonzalez MV, Pello MF, Lopez-Larrea C, Suarez C, Menendez MJ, Coto E. Deletion and methylation of the tumor suppressor gene p16/CDKN2 in primary head and neck squamous cell carcinoma. J Clin Pathol. 1997;50:509–512. doi: 10.1136/jcp.50.6.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Naggar AK, Lai S, Clayman G, Lee J-KJ, Luna MA, Goepfert H, Batsakis JG. Methylation, a major mechanism of p16/CDKN2 gene inactivation in head and neck squamous cell carcinoma. Am J Pathol. 1997;151:1767–1774. [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–895. [PubMed] [Google Scholar]
- 5.Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–4236. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 6.Weber A, Wittekind C, Tannapfel A. Genetic and epigenetic alterations of 9p21 gene products in benign and malignant tumors of the head and neck. Pathol Res Pract. 2003;199:391–397. doi: 10.1078/0344-0338-00435. [DOI] [PubMed] [Google Scholar]
- 7.Ai L, Stephenson KK, Ling W, Zuo C, Mukunyadzi P, Suen JY, Hanna E, Fan C-Y. The p16 (CDKN2a/INK4a) tumor- suppressor gene in head and neck squamous cell carcinoma: a promoter methylation and protein expression study in 100 cases. Mod Pathol. 2003;16:944–950. doi: 10.1097/01.MP.0000085760.74313.DD. [DOI] [PubMed] [Google Scholar]
- 8.Maruya S, Issa J-PJ, Weber RS, Rosenthal DI, Haviland JC, Lotan R, El-Naggar AK. Differential methylation status of tumor-associated genes in head and neck squamous cell carcinoma: incidence and potential implications. Clin Cancer Res. 2004;10:3825–3830. doi: 10.1158/1078-0432.CCR-03-0370. [DOI] [PubMed] [Google Scholar]
- 9.Puri SK, Si L, Fan C-Y, Hanna E. Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinoma. Am J Otolaryngol. 2005;26:12–17. doi: 10.1016/j.amjoto.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Shiga K, Matsuura K, Tateda M, Saijo S, Ogawa T, Miyagi T, Kobayashi T. Allelic loss correlated with tissue specificity in head and neck squamous cell carcinomas and the clinical features of patients. Tohoku J Exp Med. 2004;204:163–172. doi: 10.1620/tjem.204.163. [DOI] [PubMed] [Google Scholar]
- 11.Shiga K, Yamamoto H, Okamoto H. Isolation and characterization of the human homologue of rig and its pseudogenes: The functional gene has features characteristic of housekeeping genes. Proc Natl Acad Sci USA. 1990;87:3594–3598. doi: 10.1073/pnas.87.9.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiga K, Shiga C, Sasano H, Miyazaki S, Yamamoto T, Yamamoto M, Hayashi N, Nishihira T, Mori S. Expression of c-erbB-2 in human esophageal carcinoma cells: overexpression correlated with gene amplification or with GATA-3 transcription factor expression. Anticancer Res. 1993;13:1293–1302. [PubMed] [Google Scholar]
- 13.Van der Riet P, Nawroz H, Hruban RH, Corio R, Tokino K, Koch W, Sidransky D. Frequent loss of chromosome 9p21-22 early in head and neck cancer progression. Cancer Res. 1994;54:1156–1158. [PubMed] [Google Scholar]
- 14.Zhang S-Y, Klein-Szanto AJP, Sauter ER, Shafarenko M, Mitsunaga S, Nobori T, Carson DA, Ridge JA, Goodrow TL. Higher frequency of alterations in the p16/CDKN2 gene in squamous cell carcinoma cell lines than in primary tumors of the head and neck. Cancer Res. 1994;54:5050–5053. [PubMed] [Google Scholar]
- 15.Nawroz H, van der Riet P, Hruban RH, Koch W, Ruppert JM, Sidransky D. Allotype of head and neck squamous cell carcinoma. Cancer Res. 1994;54:1152–1155. [PubMed] [Google Scholar]
- 16.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 17.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, Depinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 18.Kamb A, Gruis NA, Weaver-Fejdhaus J, Lin Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 19.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 20.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Frequency of homozygous deletion at p16/CDK2 in primary human tumors. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 21.Lydiatt WM, Murty VV, Davidson BJ, Xu L, Dyomina K, Sacks PG, Schantz SP, Chaganti RS. Homozygous deletions and loss of expression of the CDKN2 gene occur frequently in head and neck squamous cell carcinoma cell lines but infrequently in primary tumors. Genes Chromosomes Cancer. 1995;13:94–98. doi: 10.1002/gcc.2870130204. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, Merlo A, Mao L, Lapidus RG, Issa J-PJ, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 23.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]