Abstract
Chronic myelogenous leukemia (CML) has a typical progressive course with transition from a chronic phase to a terminal blast crisis phase. The mechanisms that lead to disease progression remain to be elucidated. To understand the role of aberrant methylation in the progression of CML, DNA methylation patterns in 16 patients with CML blast crisis were analyzed. Methylation status was analyzed by methylation-specific PCR (MSP) for 13 genes, including cell cycle regulating genes, DNA repair genes, apoptosis-related genes, a differentiation-associated gene and a cytokine signaling gene. The frequency of samples with methylation in each of the following genes were: p15, 18%; MGMT, 12%; RARβ, 12%; p16, 6%; DAPK, 6% and FHIT, 6%. In total, four (25%) cases showed methylation of at least one gene. None of the 16 cases showed hypermethylation of the hMLH1 or hMSH2 genes. These results suggest that hypermethylation of cell cycle control genes, but not DNA mismatch repair genes, play a significant role in the progression of CML.
Keywords: chronic myelogenous leukemia, blast crisis, promoter hypermethylation, cell cycle control genes, DNA repair genes, epigenetics
Introduction
Chronic myelogenous leukemia (CML) has a typical progressive course with transition from the chronic phase to the terminal blast crisis phase. The mechanisms that lead to disease progression have yet to be elucidated. Cytogenetic and genetic changes occur in the majority of patients during disease progression. Approximately 70–80% of patients with CML blast crisis show additional chromosomal changes involving chromosomes 7, 8, 17, 19, 21 and 22, sometimes with duplication of the Ph chromosome (1). Genetic changes occurring in the progression to blast crisis include mutation of the p53 (20–30%), amplification of the c-myc (20%), deletion of the p16 (15%) and mutation of the Ras (6%) gene (2).
DNA methylation at CpG sites in promoter regions is a frequent, acquired epigenetic event involved in the pathogenesis of various types of human malignancies. Methylation in the promoter region is capable of causing gene silencing, which may provide an alternative pathway to gene inactivation, in addition to deletions or mutations. The ABL1, calcitonin, ER and HIC1 genes were found to be frequently methylated in CML (3). Moreover, methylation of the ABL1 gene is associated with the progression of CML (4). These methylation phenotypes in CML provided a rationale for using demethylating agents such as 5-azacytidine and decitabine in a clinical setting, and preliminary clinical results were reported (3,5). To determine the role of aberrant methylation in the progression of CML, we analyzed DNA methylation patterns in CML blast crisis.
Materials and methods
Bone marrow cells were obtained from 16 patients who developed blast crisis during the follow-up of CML. Genomic DNA was extracted from low density mononuclear cells after the bone marrow cells were centrifuged in the presence of TRIzol reagent (Life Technologies Inc., Rockville, MD, USA). Control DNA was extracted from the peripheral blood of 10 healthy individuals. Methylation-specific PCR (MSP) was performed as previously described (6,7). Briefly, 1.0 μg genomic DNA was modified by bisulfite, and PCR was performed using specific primers for each of the genes. Following amplification, 10 μl PCR product was separated on 2% agarose gel containing 0.3 mg/ml ethidium bromide. A total of 13 genes, including cell cycle regulating genes (p14, p15, p16, Rb, APC and FHIT), DNA repair genes (hMLH1, hMSH2 and MGMT), apoptosis-related genes (DAPK and RIZ1), a differentiation-associated gene (RARβ) and a cytokine signaling gene (SOCS-1) were analyzed in this study.
Results and Discussion
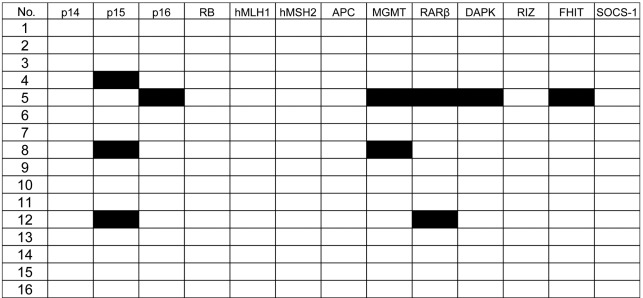
The methylation status of the 13 genes was analyzed in 16 patients in the blast crisis phase of CML. The frequency of samples with methylation in each of the following genes was: p15, 18%; MGMT, 12%; RARβ, 12%; p16, 6%; DAPK, 6%; FHIT, 6% (Figs. 1 and 2). The p14, Rb, hMLH1, hMSH2, APC, RIZ or SOCS-1 genes were not methylated in any of the patients. Moreover, none of these genes were methylated in white blood cell DNA from 10 healthy individuals.
Figure 1.
The methylation profile of 13 genes in 16 CML blast crisis patients. We analyzed the methylation status of the 13 genes in 16 patients with CML blast crisis. Black boxes denote methylated cases and white boxes denote non-methylated cases.
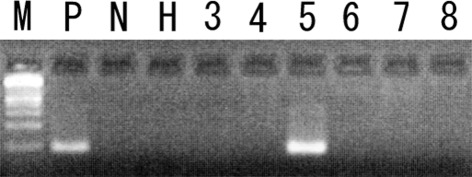
Figure 2.
Methylation analysis of the DAPK gene in CML blast crisis patients. Examples are provided for patients 3–8 in the M primer. Patient 5 shows methylation in the DAPK promoter. Patients 3, 4 and 6–8 show no methylation in the DAPK promoter. P, positive control (universally methylated DNA); N, negative control (peripheral blood DNA from a healthy individual); H, H2O blank (PCR negative control); M, 100 bp DNA ladder.
In total, 4 (25%) of the 16 CML blast crisis patients had methylation of at least one of these genes; 1 case (6%) had methylation of five target genes; 2 (12%) had two genes methylated, and the remaining case (6%) had methylation in one gene. Although the number of cases that were analyzed is limited, no significant correlation was found between methylation status and the clinical characteristics of CML.
The most frequently methylated gene was p15 (3 of 16 patients: 18%). This is in accordance with previous reports (8,9). The other cell cycle control gene, p16, was methylated in another patient. Taken together, 4 (25%) of the 16 patients had methylation in cell cycle control genes, suggesting that inactivation of cell cycle control genes by promoter hypermethylation plays a significant role in the progression of CML.
Microsatellite instability (MSI) is caused by defects of the DNA mismatch repair system, and inactivation of the hMLH1 and hMSH2 genes by promoter hypermethylation is frequently associated with MSI (10). In this study, none of the 16 cases showed hypermethylation of the hMLH1 and hMSH2 genes. This is in accordance with our previous findings that MSI is infrequent in CML blast crisis patients (11). Taken together, the deficiency of the DNA mismatch repair system does not contribute to the disease progression of CML. We conclude that hypermethylation of the cell cycle control genes, and not DNA mismatch repair genes, plays a significant role in the progression of CML.
Acknowledgements
This study was supported in part by grants from the Uehara Memorial Foundation and a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Shet AS, Jahagirdar BN, Verfaillie CM. Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia. 2002;16:1402–1411. doi: 10.1038/sj.leu.2402577. [DOI] [PubMed] [Google Scholar]
- 2.Randhawa GS, Cui H, Barletta JA, et al. Loss of imprinting in disease progression in chronic myelogenous leukemia. Blood. 1998;91:3144–3147. [PubMed] [Google Scholar]
- 3.Santini V, Kantarjian HM, Issa JP. Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Ann Intern Med. 2001;134:573–86. doi: 10.7326/0003-4819-134-7-200104030-00011. [DOI] [PubMed] [Google Scholar]
- 4.Asimakopoulos FA, Shteper PJ, Krichevsky S, et al. ABL1 methylation is a distinct molecular event associated with clonal evolution of chronic myeloid leukemia. Blood. 1999;94:2452–2460. [PubMed] [Google Scholar]
- 5.Sacchi S, Kantarjian HM, O'Brien S, et al. Chronic myelogenous leukemia in nonlymphoid blastic phase: analysis of the results of first salvage therapy with three different treatment approaches for 162 patients. Cancer. 1999;86:2632–2641. doi: 10.1002/(sici)1097-0142(19991215)86:12<2632::aid-cncr7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann WK, Tsukasaki K, Takeuchi N, Takeuchi S, Koeffler HP. Methylation analysis of cell cycle control genes in adult T-cell leukemia/lymphoma. Leuk Lymphoma. 2001;42:1107–1109. doi: 10.3109/10428190109097731. [DOI] [PubMed] [Google Scholar]
- 7.Uehara E, Takeuchi S, Tasaka T, et al. Aberrant methylation in promoter-associated CpG islands of multiple genes in therapy-related leukemia. Int J Oncol. 2003;23:693–696. [PubMed] [Google Scholar]
- 8.Nguyen TT, Mohrbacher AF, Tsai YC, et al. Quantitative measure of c-abl and p15 methylation in chronic myelogenous leukemia: biological implications. Blood. 2000;95:2990–2992. [PubMed] [Google Scholar]
- 9.Kusy S, Cividin M, Sorel N, et al. p14ARF, p15INK4b, and p16INK4a methylation status in chronic myelogenous leukemia. Blood. 2003;101:374–375. doi: 10.1182/blood-2002-09-2834. [DOI] [PubMed] [Google Scholar]
- 10.Sheikhha MH, Tobal K, Liu Yin JA. High level of microsatellite instability but not hypermethylation of mismatch repair genes in therapy-related and secondary acute myeloid leukaemia and myelodysplastic syndrome. Br J Haematol. 2002;117:359–365. doi: 10.1046/j.1365-2141.2002.03458.x. [DOI] [PubMed] [Google Scholar]
- 11.Mori N, Takeuchi S, Tasaka T, et al. Absence of microsatellite instability during the progression of chronic myelocytic leukemia. Leukemia. 1997;11:151–152. doi: 10.1038/sj.leu.2400524. [DOI] [PubMed] [Google Scholar]