Abstract
TP53 codon 72 polymorphism has been reported to affect regulatory networks central to glioma development. Although a number of published studies noted the association between TP53 codon 72 polymorphism and glioma risk, their conclusions were inconsistent. A meta-analysis was used to assess the possible association between TP53 codon 72 polymorphism and glioma risk. The PubMed databases were searched, relevant articles were identified and data were retrieved based on the inclusion criteria. The odds ratio (OR) and 95% confidence interval (95% CI) were determined on the pooled dataset. We retrieved eight different studies including 2,260 glioma cases and 3,506 controls. However, no association was found between the TP53 codon 72 polymorphism and glioma risk regarding the comparison between glioma cases and the controls. By further stratification based on criteria such as tumor grade, and the geographical location of the patients and the relevant controls, we found a significant association in the subgroup of patients with high-grade glioma in Europeans compared to controls in two models of TP53 codon 72 polymorphism, which include the dominant model [C/C + G/C vs. G/G: OR=1.35, 95% CI (1.14, 1.59), P=0.0005, Ph=0.13] and the additive model [C allele vs. G allele: OR=1.16, 95% CI (1.02, 1.33), P=0.03, Ph=0.37]. Our analysis suggests that TP53 codon 72 polymorphism is associated with an increased risk of high-grade glioma development in Europeans.
Keywords: meta-analysis, glioma, risk factors, TP53 codon 72 polymorphism
Introduction
Central nervous system tumors are the most common pediatric neoplasms, corresponding to approximately 20–23% of all cases of childhood cancer, and less than 2% of adult tumors (1). The prognosis for patients with primary central nervous system tumors, such as glioma, remains poor. The principal human tumor-suppressor gene TP53 encodes a protein, p53, activated by stresses such as DNA damage, aberrant growth signals and ultraviolet light. p53 acts as a nuclear transcription factor, binds to particular DNA sequences and activates the expression of adjacent genes, which directly or indirectly results in cell death or inhibition of cell divisions (2). Therefore, proper function of tumor-suppressor genes including TP53 is highly correlated with cancer risk. Approximately 25% of gliomas carry mutations in the TP53 gene (3).
A number of polymorphisms have been identified within the TP53 gene thus far, both in coding and non-coding regions (4). These polymorphisms include the serine 47 (5), the codon 72 (C→G, Pro→Arg) (6), intron 3 (+16 bp) and intron 6 (G→C) (7). Among these polymorphisms, the TP53 codon 72 polymorphism, which is located in a proline-rich region in exon 4, is the most frequently studied. The codon 72 polymorphism involves a guanine to cytosine nucleotide exchange, leading to a non-conservative change from arginine to proline (8,9). In a cell-based study, an increase in apoptosis rate by up to 15-fold was found for the Arg72 variant cells as compared to that in the Pro72 variant (10). In addition, an association between the Arg72 variant and increased risk of epithelial cancer (11,12) was reported. Certain authors found alternate correlations, i.e., an association between the Pro72 TP53 variant and increased cancer risk (13,14), whereas other authors (40,41) failed to confirm the link between TP53 codon 72 variants and the risk of cancer.
Studies exist regarding the association of the TP53 codon 72 polymorphism with susceptibility to the development of gliomas (15–21,42). However, the results obtained thus far were inconsistent. For example, Parhar et al (20) suggested a possible association between the codon 72 polymorphism and susceptibility to brain tumors, particularly high-grade astrocytomas. El Hallani et al (15) suggested that the codon 72 polymorphism was associated with the age-related onset of grade IV glioblastoma. Other investigators found that the codon 72 polymorphism is not correlated with susceptibility to glioma development (18,19,21,42). This discrepancy in the results may be due to different sample sizes, ethnicities or qualities including the genotyping method and study type among the various studies.
In this study, we performed a meta-analysis on the most current published reports in the region, to obtain the most precise estimation of the association between TP53 codon 72 polymorphism and the risk of human glioma.
Materials and methods
Eligibility of relevant studies
We searched the National Library of Medicine (PubMed) database, using the terms ‘(p53 OR TP53) AND ((brain tumor) OR glioma) AND polymorphism’ (the latest search was performed on June 3, 2011). In addition, we sent e-mails to the corresponding authors of the studies to retrieve the original data. The inclusion criteria were as follows: a) case-control studies with non-related subjects; b) sufficient data to calculate the odds ratio (OR); c) no deviation from the Hardy-Weinberg equilibrium (HWE) for the genotype distribution of the controls; and d) English articles. We excluded the following studies: a) studies that contained overlapping data; b) studies in which the number of wild-type genotypes could not be ascertained; and c) studies in which family members were studied.
Data extraction
All abstracts were read and articles were screened for suitability by two independent researchers (M.S. and R.H.). These investigators also read the full texts to extract data and reach a consensus on all of the eligible items: the first author, year of publication, country of study population, genotyping method, genotype frequency and source of the relevant control.
Meta- and statistical analysis
The meta-analysis evaluated the association between glioma risk and TP53 codon 72 polymorphism, which included the dominant model (G/G + G/C versus C/C), the recessive model (G/G versus G/C + C/C) and the additive model (G allele versus C allele). The strength of association was assessed by the OR with a corresponding 95% confidence interval (95% CI). The heterogeneity among these studies was checked by Q statistics, and was considered statistically significant when Ph<0.10 (22). Study heterogeneity was quantified by the I2 metric, which is independent of the number of studies in the meta-analysis (I2<25% no heterogeneity; 25≤I2≤50% moderate heterogeneity; I2>50% extreme heterogeneity) (23). The combined OR of each study was estimated by the fixed-effects (FE) model (Mantel-Haenszel) at Ph≥0.10. Otherwise, the random-effects (RE) model (DerSimonian and Laird) was applied (24). The studies were further stratified by glioma grade (high-grade gliomas, WHO classification III and IV; and low grade gliomas, WHO classification I and II) and the high-grade gliomas were subgrouped by geographical locations including Europe, America and Asia.
To assess the stability of the results, sensitivity and publication bias analysis were also performed. For the sensitivity analysis, one study was omitted each time to reflect the effect of the individual data-set to the pooled OR (25). Publication bias was investigated by Begg’s test (P<0.05 was considered to indicate statistical significance) (26) and Egger’s test (P<0.05 was considered to be statistically significant) (27). Deviations from HWE for controls were analyzed by the Chi-square goodness of fit test. Statistical analyses were performed using the Review Manager 5.0 software (The Cochrane Collaboration, Oxford, England) and STATA version 11 (STATA Corporation, College Station, TX, USA).
Results
Characteristics of the retrieved studies
The search terms used for the TP53 codon 72 polymorphism resulted in 268 articles, 22 of which were relevant upon further review. Three articles contained overlapping data (28–30), eight were no-control studies (31–37), and one was a non-glioma brain tumor study (38). In addition, deviation from HWE was found in one publication (39) and the glioma data could not be extracted from another (Fig. 1) (40). Therefore, eight studies published from 2004 to 2010, including 2,260 glioma cases and 3,506 controls, were eligible for the inclusion criteria in the meta-analysis. Table I shows the main characteristics of these studies, including the Jha study (39). The sample size in each study varies from 84 to 636. In addition, the original data of two studies were retrieved from the corresponding authors (18,41).
Figure 1.
The flow chart of study identification, including inclusion and exclusion criteria.
Table I.
Characteristics of studies included in the meta-analysis.
| References | Geographical location | Genotyping method | Number of cases (Age range, mean) | Number of controls (Age range, mean) | Source of controls | Matching |
|---|---|---|---|---|---|---|
| El Hallani et al (2009) | France | TaqMan | 254 (19.2–83.6, 56.5) | 238 (16–75, NA) | NA | Not matched |
| Idbaih et al (2007) | France | TaqMan | 293 (16–84, 43) | 175 (NA, NA) | NA | NA |
| Lima-Ramos et al (2008) | Europe | PCR-RFLP | 171 (NA, 49.5) | 526 (NA, 38.1) | Hospital-based | Gender |
| Malmer et al (2007) | Nordic-UK | TaqMan | 636 (18–69, 45) | 1461 (19–70, 50) | Population-based | Age, gender, geographical location |
| Parhar et al (2005) | USA | PCR-RFLP | 135 (0–79, NA) | 117 (NA, NA) | NA | Not matched |
| Pinto et al (2008) | Southeast Brazil | PCR-RFLP | 94 (1–75, 45) | 100 (18–72 45) | NA | Age, gender |
| Rajaraman et al (2007) | USA | TaqMan | 386 (>18, NA) | 547 (>18, NA) | Hospital-based | Age, gender, ethnicity, residential proximity to the hospital |
| Wang et al (2004) | USA | PCR-RFLP | 309 (20–60, NA) | 342 (20–60, NA) | Hospital-based | Age, gender, ethnicity |
| Jha et al (2010) | North India | Sequence analysis | 84 (NA, NA) | 112 (NA, NA) | NA | NA |
NA, data not available.
Gliomas from the eight eligible studies were classified according to the WHO classification criteria. Among them, six studies provided data on high-grade glioma genotype distribution, while the other two studies provided data on low-grade glioma genotype distribution (Table II). Therefore, these studies were treated as a mixed study at first, and then analyzed for high-grade and low-grade gliomas. The eight studies were conducted in different populations of various geographical locations: four were European populations (15,16,17,21), four were American (18,19,20,42) and only one was Asian (39). A stratified analysis for different geographical locations in high-grade gliomas was also conducted. However, such an analysis for geographical locations other than Europe was not performed due to insufficient data.
Table II.
Distribution of TP53 codon 72 genotype and allele frequencies.
| References | Genotype | Allele | HWE (P) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n) gliomas (high/low) | Controls (n) | Cases (n) gliomas (high/low) | Controls (n) | ||||||||
| GG | GC | CC | GG | GC | CC | G | C | G | C | ||
| El Hallani et al (2009) | 140 (140/NA) | 92 (92/NA) | 22 (22/NA) | 142 | 82 | 14 | 372 (372/NA) | 136 (136/NA) | 366 | 110 | 0.637956 |
| Idbaih et al (2007) | 149 (61/88) | 108 (45/63) | 18 (10/8) | 107 | 57 | 11 | 431 (178/253) | 155 (72/83) | 271 | 79 | 0.367309 |
| Lima-Ramos et al (2008) | 101 (47/NA) | 56 (24/NA) | 14 (4/NA) | 298 | 197 | 31 | 258 (118/NA) | 84 (32/NA) | 793 | 259 | 0.835726 |
| Malmer et al (2007) | 361 (159/NA) | 241 (194/NA) | 34 (10/NA) | 801 | 556 | 104 | 309 (214/NA) | 963 (512/NA) | 764 | 2158 | 0.576667 |
| Parhar et al (2005) | 38 (8/NA) | 94 (55/NA) | 3 (1/NA) | 72 | 42 | 3 | 170 (71/NA) | 100 (57/NA) | 186 | 48 | 0.275542 |
| Pinto et al (2008) | 53 (33/20) | 34 (14/20) | 7 (5/2) | 48 | 42 | 10 | 140 (80/60) | 48 (24/24) | 138 | 62 | 0.855325 |
| Rajaraman et al (2007) | 213 (NA/NA) | 146 (NA/NA) | 27 (NA/NA) | 300 | 209 | 38 | 572 (NA/NA) | 200 (NA/NA) | 809 | 285 | 0.84566 |
| Wang et al (2004) | 165 (NA/NA) | 126 (NA/NA) | 18 (NA/NA) | 194 | 128 | 20 | 456 (NA/NA) | 162 (NA/NA) | 516 | 168 | 0.853764 |
| Jha et al (2010) | 33 (13/NA) | 27 (17/NA) | 24 (13/NA) | 15 | 70 | 27 | 93 (43/NA) | 75 (43/NA) | 100 | 124 | 0.00512 |
Gliomas, all gliomas; high, high-grade gliomas; low, low-grade gliomas; NA, data not available. HWE, Hardy-Weinberg equilibrium.
Meta-analysis results
Primary meta-analysis results are shown in Table III. We adopted the random-effects model to test the association between TP53 codon 72 C allele and glioma risk. The overall OR for the C-allele was 1.10, its 95% CI was 0.93-1.30. Therefore, C allele was considered as a high-risk allele in this literature review of the data. The pooled estimates across all eight studies showed no association between TP53 codon 72 polymorphism and glioma risk within the three genotype models: [C/C + G/C vs. G/G: OR=1.17, 95% CI (0.91, 1.50), P=0.23, Ph<0.0001], [C/C vs. G/C+ G/G: OR=0.97, 95% CI (0.77, 1.21), P=0.77, Ph=0.64], [C allele vs. G allele: OR=1.10, 95% CI (0.93, 1.30), P=0.27, Ph=0.002].
Table III.
The odds ratios (ORs) of TP53 codon 72 polymorphism, glioma subtype and geographical location status with glioma.
| Allele and genotype | Outcome or subgroup | Studies | Cases/controls | Statistical method | Effect estimate | P | P (Heterogeneity) | I2(%) |
|---|---|---|---|---|---|---|---|---|
| C/C + G/C vs. G/G (dominant model) | ||||||||
| Gliomas | 8 | 2260/3506 | Odds ratio (M-H, Random, 95% CI) | 1.17 [0.91, 1.50] | 0.23 | <0.0001 | 78 | |
| High-grade gliomas | 6 | 924/2617 | Odds ratio (M-H, Random, 95% CI) | 1.45 [0.88, 2.39] | 0.15 | <0.00001 | 87 | |
| Low-grade gliomas | 2 | 201/275 | Odds ratio (M-H, Fixed, 95% CI) | 1.20 [0.82, 1.74] | 0.35 | 0.60 | 0 | |
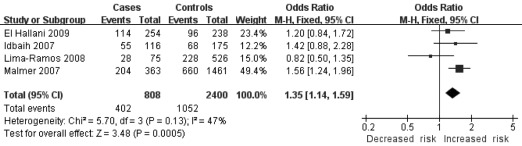
| High-grade gliomas in Europeans | 4 | 808/2400 | Odds ratio (M-H, Fixed, 95% CI) | 1.35 [1.14, 1.59] | 0.0005 | 0.13 | 47 | |
| C/C vs. G/C+ G/G (recessive model) | ||||||||
| Gliomas | 8 | 2260/3506 | Odds ratio (M-H, Fixed, 95% CI) | 0.97 [0.77, 1.21] | 0.77 | 0.64 | 0 | |
| High-grade gliomas | 6 | 924/2617 | Odds ratio (M-H, Random, 95% CI) | 0.88 [0.50, 1.56] | 0.67 | 0.07 | 52 | |
| Low-grade gliomas | 2 | 201/275 | Odds ratio (M-H, Fixed, 95% CI) | 0.67 [0.30, 1.47] | 0.32 | 0.54 | 0 | |
| High-grade gliomas in Europeans | 4 | 808/2400 | Odds ratio (M-H, Random, 95% CI) | 0.90 [0.43, 1.90] | 0.78 | 0.02 | 71 | |
| C allele vs. G allele (an additive model) | ||||||||
| Gliomas | 8 | 2260/3506 | Odds ratio (M-H, Random, 95% CI) | 1.10 [0.93, 1.30] | 0.27 | 0.002 | 68 | |
| High-grade gliomas | 6 | 924/2617 | Odds ratio (M-H, Random, 95% CI) | 1.23 [0.90, 1.66] | 0.19 | 0.0003 | 79 | |
| Low-grade gliomas | 2 | 201/275 | Odds ratio (M-H, Fixed, 95% CI) | 1.00 [0.74, 1.35] | 0.98 | 0.62 | 0 | |
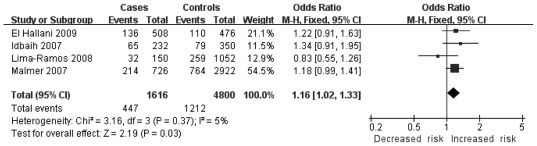
| High-grade gliomas in Europeans | 4 | 808/2400 | Odds ratio (M-H, Fixed, 95% CI) | 1.16 [1.02, 1.33] | 0.03 | 0.37 | 5 | |
In the stratified analysis for glioma grade, no association was found between high-grade and low-grade gliomas. However, heterogeneity was detected in high-grade gliomas [C/C + G/C vs. G/G: OR=1.45, 95% CI (0.88, 2.39), P=0.15, Ph<0.00001], [C allele vs. G allele: OR=1.23, 95% CI (0.90, 1.66), P=0.19, Ph=0.0003]. Therefore, we further sub-grouped the high-grade glioma group by geographical locations. Homogeneity and significant associations were found in two models: the dominant model [C/C + G/C vs. G/G: OR=1.35, 95% CI (1.14, 1.59), P=0.0005, Ph=0.13] (Fig. 2) and the additive model [C allele vs. G allele: OR=1.16, 95% CI (1.02, 1.33), P=0.03, Ph=0.37] (Fig. 3). No other significant association was found.
Figure 2.
Overall meta-analysis for the TP53 codon 72 polymorphism and glioma risk in the dominant model in high-grade gliomas in Europeans.
Figure 3.
Overall meta-analysis for the TP53 codon 72 polymorphism and glioma risk in the additive model in high-grade gliomas in Europeans.
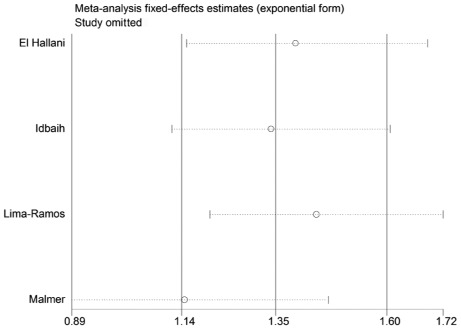
Sensitivity analysis
A single study in the meta-analysis was deleted to test for the effect of that individual data set on the pooled ORs. The corresponding pooled ORs were not significantly altered in the sensitivity analysis (Figs. 4 and 5).
Figure 4.
Sensitivity analysis for the TP53 codon 72 polymorphism in the dominant model in high-grade gliomas in Europeans.
Figure 5.
Sensitivity analysis for the TP53 codon 72 polymorphism in the additive model in high-grade gliomas in Europeans.
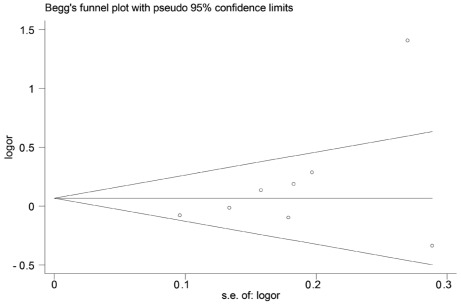
Publication bias
Both Begg’s and Egger’s test were performed to assess the publication bias. The results suggested no evidence of publication bias (C/C + G/C vs. G/G: Begg’s test P=0.174, Egger’s test P=0.194) (Figs. 6 and 7).
Figure 6.
Begg’s funnel plot of the TP53 codon 72 polymorphism and glioma risk.
Figure 7.
Begg’s test (P=0.174) and Egger’s test (P=0.194) of the TP53 codon 72 polymorphism and glioma risk.
Discussion
The TP53 codon 72 polymorphism was intensively studied and reported to affect the functions of the TP53 network, which is central to the development of gliomas, particularly high-grade gliomas. The previously published studies presented conflicting results over the association between TP53 codon 72 polymorphism and the risk of glioma. Therefore, we conducted this meta-analysis on data collected from the most up-to-date publications to evaluate this putative association.
The tumor suppressor gene TP53 is a core gene in the TP53 signaling pathway and is significant in tumor suppression. The mutation of the TP53 gene was recently defined as one of the most crucial factors in the development of malignant gliomas (43). Parhar et al (20) found a significant association between the G/C genotype and an increased risk for high-grade astrocytomas. However, the present results did not show any association between the type of single-nucleotide polymorphism (SNP) and the risk of high-grade gliomas. This inconsistency may be due to a number of reasons. First, our analysis stratified the data into high-grade and low-grade gliomas, whereas Parhar et al stratified the data into high-grade astrocytomas and non-astrocytomas. Since astrocytoma is a subtype of glioma, non-astrocytomas include other types of high-grade gliomas. Therefore, the tumor classification is different between the two studies. Second, the sample sizes were different. Parhar et al only included 252 subjects, whereas we included 2,145 subjects. The relatively small sample size in the study by Parhar et al may contribute to their conclusion. Our analysis, which combines data from all eight studies that included 2,145 subjects, should minimize the random error.
The genesis of glioma is closely associated with the interaction between environmental factors and genetic background. It was reported that the allele distribution at codon 72 of TP53 varies depending on geographical locations (44,45). Therefore, to clarify the association between codon 72 polymorphism and the glioma risk in different genetic backgrounds and remove heterogeneity, we analyzed the collected data by subgrouping the high-grade glioma group according to the geographical locations. The results have shown that the respective SNPs at codon 72 of TP53 are associated with an increased risk of high-grade gliomas in Europeans.
Based on the literature review of the available data, our results suggest that TP53 codon 72 C carriers (Pro) are associated with an increased risk of high-grade glioma in Europeans. Thomas et al (46) indicated that the Pro72 variant induced slower kinetics of apoptosis and suppressed transformation less efficiently than the Arg72 variant. In their study, Dumont et al (10) reported that the Pro72 variant bears only 1/15 apoptosis-inducing ability compared to the Arg72 variant in cells with endogenous p53, as well as in cell lines containing inducible alleles encoding the Pro72 or Arg72. Thus, our results are consistent with the data describing the biological functions of p53.
However, there are a number of limitations in our analysis. Although we carefully selected studies by performing a careful search, using strict study inclusion criteria, precise data extraction and statistical analysis, significant heterogeneity between studies still exists. E-mails were forwarded to the corresponding authors of each publication. However, only two of the original sets of data were retrieved; thus, we failed to adjust our meta-analysis by age and gender. Future studies should include other co-variants, such as age, gender, ethnicity, environmental factors and lifestyle for a more comprehensive understanding of the association between the TP53 codon 72 polymorphism and glioma risk.
In conclusion, our results confirm that TP53 codon 72 polymorphism may be associated with an increased risk of high-grade glioma development in Europeans.
Acknowledgements
The authors appreciate the assistance from G.R. Pinto (18), Ahmed Idbaih (41) and Guilherme Francisco (47) for sharing their original data. The research project was supported by the Key Project Science Foundation of Heilongjiang Province, China (Grant-ZD200804-01), the Chinese Postdoctoral Fellowship (Grant-2008043938) and the National Science Foundation of China (Grant NFSC-30227738), and Ministry of Education, Science and Technology Development Center College Research Foundation for the Doctoral Program (20092307110006).
References
- 1.Rickert CH, Paulus W. Epidemiology of central nervous system tumors in childhood and adolescence based on the new WHO classification. Childs Nerv Syst. 2001;17:503–511. doi: 10.1007/s003810100496. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Phatak P, Selvi SK, Divya T, Hegde AS, Hegde S, Somasundaram K. Alterations in tumour suppressor gene p53 in human gliomas from Indian patients. J Biosci. 2000;27:673–678. doi: 10.1007/BF02708375. [DOI] [PubMed] [Google Scholar]
- 4.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mut. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 5.Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet. 1993;53:752–759. [PMC free article] [PubMed] [Google Scholar]
- 6.Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotteri V. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol. 1986;6:4650–4656. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehman TA, Haffty BG, Carbone CJ, Bishop LR, Gumbs AA, Krishnan S, Shields PG, Modali R, Turner BC. Elevated frequency and functional activity of a specific germ-line p53 intron mutation in familial breast cancer. Cancer Res. 2000;60:1062–1069. [PubMed] [Google Scholar]
- 8.Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamuro D, Sabbatini P, White E, Prendergast GC. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 10.Dumont P, Leu JI, Pietra ACD, III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 11.Langerød A, Bukholm IRK, Bregård A, Lønning PE, Andersen TI, Rognum TO, Meling GI, Lothe RA, Børresen-Dale AL. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev. 2002;11:1684–1688. [PubMed] [Google Scholar]
- 12.Maarten TB, Struyk L, Tjong-A-Hung SP, Gruis N, Huurne J, Westendorp RGJ, Vermeer BJ, Bavinck JNB, Schegget J. Cutaneous squamous cell carcinoma and p53 codon 72 polymorphism: a need for screening? Mol Carcinog. 2001;30:56–61. doi: 10.1002/1098-2744(200101)30:1<56::aid-mc1013>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Granja F, Moraria J, Moraria EC, Correa LAC, Assumpcao LVM, Ward LS. Proline homozygosity in codon 72 of p53 is a factor of susceptibility for thyroid cancer. Cancer Lett. 2004;210:151–157. doi: 10.1016/j.canlet.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Tiwawech D, Srivatankul P, et al. The p53 codon 72 polymorphism in Thai nasopharyngeal carcinoma. Cancer Lett. 2003;198:69–75. doi: 10.1016/s0304-3835(03)00283-0. [DOI] [PubMed] [Google Scholar]
- 15.El Hallani S, Ducray F, Idbaih A, Marie Y, Boisselier B, Colin C, Laigle-Donadey F, Rodero M, Chinot O, Thillet J, Hoang-Xuan K, Delattre JY, Sanson M. TP53 codon 72 polymorphism is associated with age at onset of glioblastoma. Neurology. 2009;72:332–336. doi: 10.1212/01.wnl.0000341277.74885.ec. [DOI] [PubMed] [Google Scholar]
- 16.Idbaih A, Boisseliera B, Marie Y, Sanson M, El Hallani S, Crinière E, Fourtassi M, Paris S, Carpentier C, Rousseau A, et al. Influence of MDM2 SNP309 alone or in combination with the TP53 R72P polymorphism in oligodendroglial tumors. Brain Res. 2008;1198:16–20. doi: 10.1016/j.brainres.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Malmer BS, Feychting M, Lönn S, Lindström S, Grönberg H, Ahlbom A, Schwartzbaum J, Auvinen A, Collatz-Christensen H, Johansen C, Kiuru A, Mudie N, Salminen T, Schoemaker MJ, Swerdlow AJ, Henriksson R. Genetic variation in p53 and ATM haplotypes and risk of glioma and meningioma. J Neurooncol. 2007;82:229–237. doi: 10.1007/s11060-006-9275-1. [DOI] [PubMed] [Google Scholar]
- 18.Pinto GR, Yoshioka FKN, Silva RLL, Clara CA, Santos MJ, Almeida JRW, Burbano RR, Rey JA, Casartelli C. Prognostic value of TP53 Pro47Ser and Arg72Pro single nucleotide polymorphisms and the susceptibility to gliomas in individuals from Southeast Brazil. Genet Mol Res. 2008;7:207–216. doi: 10.4238/vol7-1gmr415. [DOI] [PubMed] [Google Scholar]
- 19.Rajaraman P, Wang SS, Rothman N, Brown MM, Black PM, Fine HA, Loeffler JS, Selker RG, Shapiro WR, Chanock SJ, Inskip PD. Polymorphisms in apoptosis and cell cycle control genes and risk of brain tumors in adults. Cancer Epidemiol Biomarkers Prev. 2007;16:1655–1661. doi: 10.1158/1055-9965.EPI-07-0314. [DOI] [PubMed] [Google Scholar]
- 20.Parhar P, Ezer R, Shao Y, Allen J, Miller D, Newcomb E. Possible association of p53 codon 72 polymorphism with susceptibility to adult and pediatric high-grade astrocytomas. Mol Brain Res. 2005;137:98–103. doi: 10.1016/j.molbrainres.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Lima-Ramos Vt, Pacheco-Figueiredo L, Costa S, Pardal F, Silva A, Amorim J, Lopes JM, Reis RM. TP53 codon 72 polymorphism in susceptibility, overall survival, and adjuvant therapy response of gliomas. Cancer Genet Cytogenet. 2008;180:14–19. doi: 10.1016/j.cancergencyto.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Zintzaras E, Ioannidis JP. Heterogeneity testing in metaanalysis of genome searches. Genet Epidemiol. 2005;28:123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 24.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 25.Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull. 1999;8:15–17. [Google Scholar]
- 26.Begg C, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 27.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Idbaiha A, Boisselier B, Sanson M, Criniere E, Livad S, Marie Y, Carpentier C, Paris S, Laigle-Donadey F, Mokhtari K, Kujas M, Hoang-Xuan K, Delattre O, Delattre J-Y. Tumor genomic profiling and TP53 germline mutation analysis of first-degree relative familial gliomas. Cancer Genet Cytogenet. 2007;176:121–126. doi: 10.1016/j.cancergencyto.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Malmer B, Gronberg H, Andersson U, Jonsson BA, Henriksson R. Microsatellite instability, PTEN and p53 germline mutations in glioma families. Acta Oncologica. 2001;40:633–637. doi: 10.1080/028418601750444196. [DOI] [PubMed] [Google Scholar]
- 30.Malmer B, Feychting M, Lonn S, Ahlbom A, Henriksson R. p53 Genotypes and risk of glioma and meningioma. Cancer Epidemiol Biomarkers Prev. 2005;14:2220–2223. doi: 10.1158/1055-9965.EPI-05-0234. [DOI] [PubMed] [Google Scholar]
- 31.Portwine C, Chilton-MacNeill S, Brown C, Sexsmith E, McLaughlin J, Malkin D. Absence of germline and somatic p53 alterations in children with sporadic brain tumors. J Neurooncol. 2001;52:227–235. doi: 10.1023/a:1010661831335. [DOI] [PubMed] [Google Scholar]
- 32.Zawlik I, Kita D, Vaccarella S, Mittelbronn M, Franceschi S, Ohgaki H. Common polymorphisms in the MDM2 and TP53 genes and the relationship between TP53 mutations and patient outcomes in glioblastomas. Brain Pathol. 2009;19:188–194. doi: 10.1111/j.1750-3639.2008.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aka K, Bruner JM, Bondy ML, Ligon K, Nishi K, Giglio AD, Moser RR, Levin VA, Saya H. Detection of p53 alterations in human astrocytomas using frozen tissue sections for the polymerase chain reaction. J Neuro Oncol. 1993;16:125–133. doi: 10.1007/BF01324699. [DOI] [PubMed] [Google Scholar]
- 34.Chang ZN, Guo CL, Ahronowitz I, Stemmer-Rachamimov RO, MacCollin M, Nunes FP. A role for the p53 pathway in the pathology of meningiomas with NF2 loss. J Neurooncol. 2008;91:265–270. doi: 10.1007/s11060-008-9721-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paunu N, Syrjakoski K, Sankila R, Simola KOJ, Helen P, Niemela M, Matikainen M, Isola J, Haapasalo H. Analysis of p53 tumor suppressor gene in families with multiple glioma patients. J Neurooncol. 2001;55:159–165. doi: 10.1023/a:1013890022041. [DOI] [PubMed] [Google Scholar]
- 36.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Patre PLD, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 37.Uno M, Oba-Shinjo SM, Wakamatsu A, Huang N, Avancini Ferreira Alves V, Rosemberg S, Pires de Aguiar PH, Leite C, Miura F, Marino Junior R, Scaff M, Nagahashi-Marie SK. Association of TP53 mutation, p53 overexpression, and p53 codon 72 polymorphism with susceptibility to apoptosis in adult patients with diffuse astrocytomas. Int J Biol Markers. 2006;21:50–57. doi: 10.1177/172460080602100108. [DOI] [PubMed] [Google Scholar]
- 38.Almeida LO, Custódio AC, Pinto GR, Santos MJ, Almeida JRW, Clara CA, Rey JA, Casartelli C. Polymorphisms and DNA methylation of gene TP53 associated with extra-axial brain tumors. Genet Mol Res. 2009;8:8–18. doi: 10.4238/vol8-1gmr518. [DOI] [PubMed] [Google Scholar]
- 39.Jha P, Jha P, Pathak P, Chosdol K, Suri V, Sharma MC, Kumar G, Singh M, Mahapatra AK, Chitra S. TP53 polymorphisms in gliomas from Indian patients: Study of codon 72 genotype, rs1642785, rs1800370 and 16 base pair insertion in intron-3. Exp Mol Pathol. 2011;90:167–172. doi: 10.1016/j.yexmp.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Biros E, Kalina I, Kohut A, Bogyiova E, Alagovic J, Ulla I. Allelic and haplotype frequencies of the p53 polymorphisms in brain tumor patients. Physiol Res. 2002;51:59–64. [PubMed] [Google Scholar]
- 41.Idbaih A, Boisselier B, Mariea Y, El Hallani S, Sanson M, Criniere E, Rodero M, Carpentier C, Paris S, Laigle-Donadey F, Ducray F, Hoang-Xuan K, Delattre JY. TP53 codon 72 polymorphism, p53 expression, and 1p/19q status in oligodendroglial tumors. Cancer Genet Cytogenet. 2007;177:103–107. doi: 10.1016/j.cancergencyto.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Wang LE. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res. 2004;64:5560–5563. doi: 10.1158/0008-5472.CAN-03-2181. [DOI] [PubMed] [Google Scholar]
- 43.Network TCGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjalander A, Birgander R, Saha N, Beckman L, Beckman G. p53 polymorphisms and haplotypes show distinct diff erences between major ethnic groups. Hum Hered. 1996;46:41–48. doi: 10.1159/000154324. [DOI] [PubMed] [Google Scholar]
- 45.Kashima T, Makino K, Soemantri A, Ishida T. TP53 codon 72 polymorphism in 12 populations of insular southeast Asia and Oceania. J Hum Genet. 2007;52:694–697. doi: 10.1007/s10038-007-0168-8. [DOI] [PubMed] [Google Scholar]
- 46.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashwski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francisco G, Menezes PR, Eluf-Neto J, Chammas R. Arg72Pro TP53 polymorphism and cancer susceptibility: A comprehensive meta-analysis of 302 case-control studies. Int J Cancer. 2011;129:920–30. doi: 10.1002/ijc.25710. [DOI] [PubMed] [Google Scholar]