Originally identified as a key element of mitochondrial volume homeostasis through regulation of K+–H+ exchange (KHE), the LETM1 protein family is also involved in respiratory chain biogenesis and in the pathogenesis of seizures in the Wolf–Hirschhorn syndrome (WHS). To add further complexity, LETM1 has been recently proposed to catalyze mitochondrial H+–Ca2+ exchange, which would imply a role in mitochondrial Ca2+ homeostasis as well. In the following paragraphs, we summarize the current state of the art about the functions of LETM1 and its role in pathophysiology, with some emphasis on whether it is a feasible candidate for regulation of mitochondrial Ca2+ homeostasis.
The LETM1 protein family
LETM1 (leucine–zipper–EF hand-containing transmembrane region) encodes a mitochondrial protein conserved in all lower eukaryotes, animals, and plants. All members of the LETM1 family share the same protein features and architecture, with a hydrophobic N-terminal portion spanning the inner membrane and a large hydrophilic portion including the C terminus located in the matrix. The primary amino acid sequence of the C-terminal part predicts at least two coiled-coil domains and two Ca2+-binding EF hand–like motifs (Nowikovsky et al., 2004). However, the EF hands differ from canonical EF hand motifs by several residues, and the α helices are not directly flanking the Ca2+-binding loop. The amino acid sequence of yeast LETM1 proteins lacks these putative Ca2+-binding EF hand motifs. The N-terminal region of the LETM1 protein superfamily displays a mitochondrial targeting sequence and a single transmembrane domain. The latter is highly conserved in all orthologues (Nowikovsky et al., 2004; Schlickum et al., 2004) and rich in proline residues, as usually observed in ion channels. Yeast Saccharomyces cerevisiae encodes two LETM1 orthologues, YOL027c and YPR125w. YOL027c codes for proteins of 65 kD and has been referred to as the MDM38 gene because its deletion alters mitochondrial distribution and morphology (Dimmer et al., 2002). YPR125w is also named MRS7, as it was initially identified in a genetic screen for multicopy suppressors of petite yeast strains lacking the mitochondrial Mg2+ transporter MRS2 (Waldherr et al., 1993) or YLH47 for yeast LETM1 homologue of 47 kD (Frazier et al., 2006).
Besides the LETM1 protein of 83.4 kD, the human genome also encodes the LETM1-like protein LETM2, the product of a related open reading frame that originated by gene duplication. Studies on the rat LETM1 homologue revealed that it is ubiquitously expressed, whereas LETM2 is only expressed in testis and sperm (Tamai et al., 2008). The Caenorhabditis elegans LETM1 is encoded by the F58G11.1 gene and shares 79% amino acid identity with human LETM1 (Hasegawa and van der Bliek, 2007). The Drosophila melanogaster homologue is the largest member of the LETM1 protein family with 113.6 kD and is encoded by the open reading frame annotated as CG4589. This protein shares 42% sequence identity with human LETM1 (McQuibban et al., 2010).
Biochemical analyses of Mdm38, Mrs7, and human LETM1 proteins indicated that they are components of high molecular weight complexes (Schlickum et al., 2004; Hasegawa and van der Bliek, 2007; Dimmer et al., 2008; Tamai et al., 2008; Zotova et al., 2010). In blue native PAGE, Mrs7p appeared within two major protein complexes of ∼130 and 250 kD, respectively, as well as in a minor complex of ∼500 kD; Mdm38 appeared within complexes of ∼140, 230, and 500 kD; and LETM1 appeared within a major complex of 300 and a minor complex of ∼500 kD (Hasegawa and van der Bliek, 2007; Dimmer et al., 2008; Zotova et al., 2010). In fact, Mdm38p and LETM1 were found as homodimers (Hasegawa and van der Bliek, 2007; Zotova et al., 2010) and may exist in homomultimers or interact with other proteins. In one study, LETM1 and BCS1L, an AAA-ATPase chaperone with a function in assembling the respiratory complex III, were both identified within a protein complex of ∼300 kD (Tamai et al., 2008). BCS1L is associated with several human disorders including the GRACILE and the Björnstad syndromes (Morán et al., 2010; DiMauro and Garone, 2011). Interestingly, in BCS1L mutant fibroblasts, the LETM1 protein levels were increased but shifted from the 300-kD complex toward higher molecular weight species (>500 kD) (Tamai et al., 2008), suggesting that LETM1 may require wild-type BCS1L to be inserted into the 300-kD complex. Although there is a general consensus on the molecular size of the Mdm38/LETM1 protein complexes, the identity of the interaction partners of the LETM1 protein family members is still a matter of debate, likely reflecting the different pull-down and protein-tagging approaches (Frazier et al., 2006; Zotova et al., 2010) and the discrete Mdm38p/LETM1-containing complexes.
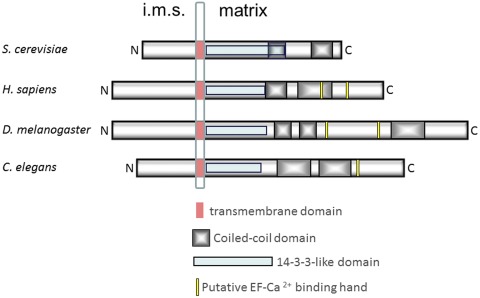
Recently, structural analyses were performed to solve the crystal structure of the soluble part of Mdm38p from the C terminus of the transmembrane domain to the C terminus of the protein residues 159–573 (Lupo et al., 2011). Although the first highly conserved 23 amino acids could not be resolved, data revealed that the amino acid stretch between residues 182 and 408 formed a 14-3-3–like domain. Classical 14-3-3 proteins share nine antiparallel α helices, with helices 1 and 2 forming the dimerization domain and helices 3, 5, 7, and 9 forming the concave substrate site. These proteins are involved in a large number of regulatory cellular processes (Lupo et al., 2011). As revealed by the structural analysis, Mdm38 exposes a hydrophobic cavity within the 14-3-3–like domain reminiscent of the substrate-binding site of the canonical 14-3-3 proteins and conserved in the LETM1 protein family. However, the 14-3-3–like domain of Mdm38 lacks the first two α helices (Lupo et al., 2011). From the available structural evidence and membrane topology (Dimmer et al., 2008), the Mdm38/LETM1 superfamily thus comprises an N-terminal domain facing the intermembrane space, a transmembrane domain that is likely to regulate dimerization, and a C-terminal domain facing the mitochondrial matrix, which bears a substrate-binding site, coiled-coil domains, and, with the exception of yeast, putative Ca2+-binding EF hand sites (Fig. 1).
Figure 1.
The LETM1 protein superfamily. Schematic representation of selected LETM1 family members aligned on the highly conserved transmembrane region. Specific domains are represented, and bar lengths are proportional to the length of the corresponding amino acid sequences. i.m.s., intermembrane space.
LETM1 and mitochondrial potassium homeostasis
In the process of characterizing yeast genes encoding putative mitochondrial cation transporters, Nowikovsky et al. (2004) identified Mdm38p as an essential element both for mitochondrial KHE activity and for growth of yeast cells on nonfermentable substrates. Passive swelling experiments with isolated mitochondria in K+ acetate (a sensitive method to detect electroneutral KHE; see Bernardi, 1999) demonstrated that Mdm38p/Yol027cp is essential for mitochondrial KHE in yeast, an activity that was lacking in mdm38 knockout mutants (Nowikovsky et al., 2004; Froschauer et al., 2005). Direct KHE assays in submitochondrial particles with entrapped K+- and H+-sensitive fluorescent dyes PBFI and BCECF, respectively, which allow continuous recording of changes in free K+ and H+ concentrations over time, indicated that Mdm38p-mediated K+ and H+ fluxes were obligatorily coupled and electroneutral (Froschauer et al., 2005). These results are compelling evidence that Mdm38p/Yol027cp is an essential component of the mitochondrial KHE (Nowikovsky et al., 2004; Froschauer et al., 2005), a hypothesis that is strongly supported by the finding that nigericin (an ionophore that catalyzes electroneutral KHE) restored aerobic growth of Mdm38p/Yol027cp-null yeast strains and reverted mitochondrial swelling in situ (Nowikovsky et al., 2007). In contrast to MDM38, deletion of MRS7 had no severe effect on growth or KHE activity (Frazier et al., 2006; Zotova et al., 2010). However, reversion to normal K+ homeostasis and growth features could also be achieved by overexpression of the Mdm38p yeast homologues Ypr125wp/Mrs7p and LETM1 or DmLETM1, the human or Drosophila orthologues, confirming the functional homology. As mentioned above, the LETM1 protein family bears only one transmembrane domain, suggesting that its members may not constitute the KHE itself (at least as a monomer) but rather an essential component that allows the exchange activity to take place. Biochemical data confirming that these proteins are part of a higher molecular weight complex suggest that the KHE might consist of discrete subunits. To identify other components of the KHE, a genetic screen for multi-copy suppressors of the petite phenotype of mdm38Δ cells was performed and led to the identification of the unknown gene YDL183c. Overexpression of YDL183c was able to restore the KHE activity, confirming a functional homology to MDM38 and MRS7. YDL183c encodes a 37-kD mitochondrial membrane protein with a single predicted transmembrane domain that contains several proline residues but otherwise appears not to be related to Mdm38p. Disruption of YDL183c had a weak petite phenotype at high growth temperatures. However, the triple deletion of MDM38, MRS7, and YDL183c exhibited a complete negative phenotype on nonfermentable substrates, strongly reduced growth on glucose, and complete block of residual K+ and H+ fluxes. All synthetic phenotypes of the triple mutant were fully restored by nigericin in vitro and in vivo (Zotova et al., 2010).
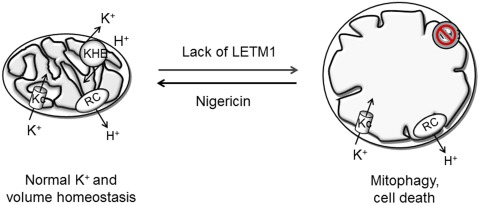
Atomic absorption measurements revealed a significant increase of mitochondrial K+ in the absence of Mdm38p and, conversely, mitochondrial K+ depletion under overexpression of Mdm38p (Nowikovsky et al., 2004). Consistent with a role of K+ in controlling mitochondrial volume and morphology, Mdm38p or LETM1 depletion resulted in extreme matrix swelling and cristae loss, whereas Mdm38p or LETM1 overexpression caused mitochondrial contraction and cristae swelling (Hasegawa and van der Bliek, 2007; Nowikovsky et al., 2007). Interestingly, nigericin fully reverted the phenotype of LETM1 ablation in HeLa cells (Dimmer et al., 2008). Moreover, considerable mitochondrial depolarization was caused upon MDM38 or LETM1 deletion (Nowikovsky et al., 2004; Frazier et al., 2006; McQuibban et al., 2010). Surprisingly, studies performed in yeast and mammalian cell cultures led to the observation of ongoing selective autophagy (mitophagy) as a result of shutoff of KHE activity (Nowikovsky et al., 2007; McQuibban et al., 2010). To dissect the phenotypes by depleting Mdm38p in a controlled fashion as a function of time, a conditional MDM38 shutoff system was generated (Nowikovsky et al., 2007). The primary effect of MDM38 down-regulation was the absence of mitochondrial KHE activity, confirming the essential function of Mdm38 in regulating K+ homeostasis. Electron microscopy revealed the presence of swollen and fragmented mitochondria closely associated with and/or ingested by vacuoles. These data provided the first evidence that mitochondrial dysfunction and osmotic swelling caused by excess matrix K+ leads to mitophagy. Importantly, the addition of nigericin reverted mitochondrial swelling and ongoing mitophagy, confirming the causal link between the two events (Nowikovsky et al., 2007). Down-regulation of DmLETM1 in Drosophila cell lines also led to increased mitophagy, as revealed under confocal or electron microscopy (McQuibban et al., 2010). A schematic representation of the role of LETM1 in mitochondrial K+ (hence volume) homeostasis and of the consequences of its absence is presented in Fig. 2, which highlights how K+ influx via K+ channels (KC) must be counterbalanced by activity of the KHE to maintain volume homeostasis; in the absence of LETM1, mitochondria cannot compensate electrophoretic K+, resulting in matrix swelling causing, in turn, increased mitophagy and/or cell death.
Figure 2.
Mitochondrial K+ and volume homeostasis. RC, respiratory chain; KC, potassium channels; KHE, K+–H+ exchange. For further explanation, see text.
LETM1 in respiratory chain biogenesis
Frazier et al. (2006) reported that lack of Mdm38p caused reduced steady-state levels of the mitochondrially encoded proteins and decreased insertion of cytochrome b and Atp6 into the respiratory complexes. Destabilized interaction of Mdm38 with mitochondrial ribosomes was proposed to account for the defective assembly of the respiratory chain (Frazier et al., 2006). However, time course studies revealed that these defects were secondary to the Mdm38 deletion, impaired KHE, and inner membrane depolarization, as they were rescued by the addition of low concentrations of nigericin to the growth media (Nowikovsky et al., 2007). Hence, mdm38Δ-induced K+ overload or membrane depolarization may destabilize the protein assembly of respiratory complexes.
To further explore the function of LETM1 as a ribosome-associated protein, a double deletion of MDM38 and of the ribosome receptor MBA1 was performed that led to a synthetic translational defect causing the complete loss of Cox1 and Cytb. This synthetic defect could not be reverted by nigericin (Bauerschmitt et al., 2010), similarly to the damage caused by deletion of the soluble part of Mdm38 (Lupo et al., 2011), thus supporting the notion that LETM1 has a role in mitochondrial biogenesis in addition to that in ion homeostasis. In human cells, lack of LETM1 was associated with a decreased number of cristae, depolarization, and impaired assembly of respiratory chain supercomplexes (Tamai et al., 2008). In another study, LETM1 was shown to bind the mitochondrial ribosome protein L36 and cause an L36-dependent drop of ATP production (Piao et al., 2009a). However, silencing of LETM1 in HeLa cells failed to affect defects in complex III assembly, or to cause overt or latent mitochondrial dysfunction (Dimmer et al., 2008), further complicating the picture. The role of Mdm38/LETM1 in the biogenesis of respiratory complexes is essentially based on binding to ribosomal proteins. Furthermore, the extreme mitochondrial swelling observed in the absence of Mdm38/LETM1 was never seen in cells with inactivated components of the respiratory complexes or in ρ0 cells (Hasegawa and van der Bliek, 2007; Tamai et al., 2008).
LETM1 in mitochondrial calcium transport
In a genome-wide RNAi screening of Drosophila for proteins involved in Ca2+ uptake in mitochondria in situ, a Drosophila homologue of the human LETM1 gene was identified as strongly affecting mitochondrial Ca2+ and H+ fluxes. A drastic reduction of mitochondrial Ca2+ uptake was observed when protein expression was suppressed; purification and reconstitution of LETM1 in liposomes allowed detection of coupled Ca2+ and H+ fluxes in opposite directions, which were inhibited by ruthenium red (RR) (Jiang et al., 2009). These findings led to the conclusion that LETM1 is a H+–Ca2+ exchanger mediating RR-sensitive Ca2+ uptake in energized mitochondria (Jiang et al., 2009). We note that the finding that Ca2+ fluxes are mirrored by H+ fluxes in liposomes reconstituted with LETM1 is certainly compatible with an obligatory H+–Ca2+ antiporter mechanism but does not prove it; indeed, Ca2+ flux could be electrophoretic (as is the case for transport via mitochondrial Ca2+ uniporter [MCU]), and H+ movement could occur independently of LETM1 to provide charge compensation.
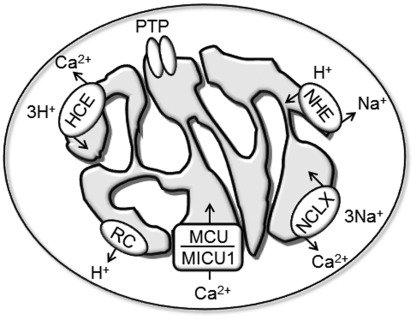
The mechanisms through which energized mitochondria take up and release Ca2+ have been the subject of a large number of studies over the years (Rizzuto et al., 2000; Drago et al., 2011). The pathways characterized so far (Fig. 3) are: (a) the MCU, which in energized mitochondria mediates Ca2+ uptake via electrophoretic transport across the inner membrane (Baughman et al., 2011; De Stefani et al., 2011) through a specific channel (Kirichok et al., 2004) that is inhibited by RR (Moore, 1971) and is modulated by a protein component with a single transmembrane domain, mitochondrial Ca2+ uptake 1 (Perocchi et al., 2010); (b) the Na+–Ca2+ exchanger, an RR-insensitive system mediated by NCLX (Palty et al., 2010) that in energized mitochondria catalyzes Ca2+ release (Carafoli et al., 1974; Crompton et al., 1976); (c) the RR-insensitive putative H+–Ca2+ exchanger, which mediates Ca2+ release and whose activity is favored by the membrane potential (Bernardi and Azzone, 1982, 1983) suggesting a stoichiometry of more than 2H+–Ca2+; and (d) the permeability transition pore (PTP), a high conductance channel (Kinnally et al., 1989; Petronilli et al., 1989) that plays an important role in cell death (Rasola et al., 2010); owing to its large size allowing depolarization, the PTP could provide mitochondria with a fast Ca2+ release channel preventing matrix Ca2+ overload (Bernardi and Petronilli, 1996; Bernardi, 1999), an idea supported by some studies (Altschuld et al., 1992; Elrod et al., 2010; Barsukova et al., 2011) and by the recent identification in Drosophila of a H+-permeant Ca2+ release channel with features intermediate between the PTP of yeast and mammals (von Stockum et al., 2011).
Figure 3.
Pathways for Ca2+ transport in mammalian mitochondria. RC, respiratory chain; MCU, mitochondrial Ca2+ uniporter; MICU1, mitochondrial Ca2+ uptake 1; NCLX, Na+–Ca2+ exchanger; HCE, H+–Ca2+ exchanger; PTP, permeability transition pore. For explanation, see text.
As already mentioned, LETM1 is a single transmembrane domain protein, a structure that is unlikely to directly mediate Ca2+ transport either as a channel or by mediating H+–Ca2+ antiport. Indeed, all known exchangers and channels possess multiple transmembrane domains. Are the features reported for LETM1-mediated Ca2+ transport consistent with its modulation of any of the known Ca2+ influx/efflux pathways? We tend to exclude both a mechanism involving the Na+–Ca2+ exchanger, because this mechanism can only catalyze Ca2+ efflux in coupled respiring mitochondria, and the PTP, because its opening causes depolarization, which prevents Ca2+ uptake and favors Ca2+ release (Bernardi, 1999). A modulation of the MCU, on the other hand, is not fully consistent with the reconstitution experiments of Jiang et al. (2009), where only LETM1 was apparently present. Although the possibility that LETM1 modulates previously unidentified Ca2+ transport systems cannot be ruled out, its proposed role as a H+–Ca2+ exchanger is discussed below.
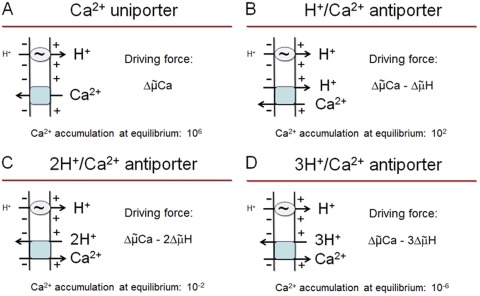
If LETM1-mediated (or modulated) Ca2+ transport is obligatorily coupled to H+ translocation via an exchange mechanism, assessment of the H+–Ca2+ stoichiometry is essential for predictions to be made about the direction of Ca2+ flux it mediates in energized mitochondria. If the H+–Ca2+ stoichiometry is 1, Ca2+ uptake is favored by the and opposed by the ; net charge translocation is 1, and the predicted Ca2+ accumulation at equilibrium is 102 if the ΔpH is 1 unit, alkaline inside (Fig. 4 B and Appendix). This mode of Ca2+ uptake would have to compete with the MCU, whose driving force is 10,000-fold larger (Fig. 4 A and Appendix). Jiang et al. (2009) have suggested that LETM1 catalyzes Ca2+ accumulation in energized mitochondria only at low cytoplasmic Ca2+ concentrations, whereas MCU predominates at higher Ca2+ levels. The possibility that Ca2+ is taken up with a single charge by energized mitochondria (either in symport with anions, e.g., Pi or OH−, or via a H+–Ca2+ antiporter) has been discussed in the field (Heaton and Nicholls, 1976; Azzone et al., 1977; Moyle and Mitchell, 1977) but is inconsistent with all the measurements of net charge translocation during Ca2+ transport, which has consistently been found to be 2 (Scarpa and Azzone, 1970; Rottenberg and Scarpa, 1974; Wingrove et al., 1984). Furthermore, mitochondrial Ca2+ uptake is almost completely suppressed by MCU-specific siRNAs (Baughman et al., 2011; De Stefani et al., 2011), findings that strongly argue against the existence of a H+–Ca2+ antiporter mediating mitochondrial Ca2+ uptake.
Figure 4.
Predicted equilibrium Ca2+ accumulation depending on mode of Ca2+ transport in energized mitochondria. The scheme highlights the net direction of Ca2+ flux for transport via a Ca2+ uniporter (A), a H+–Ca2+ exchanger (B), a 2H+–Ca2+ exchanger (C), and a 3H+–Ca2+ exchanger (D) in respiring mitochondria. The predicted equilibrium Ca2+ accumulation was calculated as described in the Appendix and refers to the matrix/cytosol concentration ratio so that a negative figure means that Ca2+ is more concentrated in the cytosol. The squiggle denotes the redox-coupled H+ pumps. For further explanation, see text and Appendix.
Transport mediated by a 2H+–Ca2+ antiporter is electroneutral, and therefore the direction of Ca2+ flux will depend on the proton and Ca2+ chemical gradients; given the existence of an alkaline-inside pH difference, such a mechanism cannot mediate Ca2+ uptake unless cytosolic [Ca2+] becomes more than 100-fold higher than matrix [Ca2+], with a predicted equilibrium accumulation of 10−2 (Fig. 4 C and Appendix). Thus, in respiring coupled mitochondria under physiological conditions, a 2H+–Ca2+ exchanger should catalyze Ca2+ efflux.
If the transport is mediated by a 3H+–Ca2+ antiporter, Ca2+ efflux is greatly favored, as easily seen by the predicted equilibrium Ca2+ distribution, which would be 10−6 (i.e., 1,000,000 higher in the cytosol than in the matrix) (Fig. 4 D and Appendix). This is indeed a likely mechanism for mitochondrial Na+-independent Ca2+ release, which is insensitive to RR and slowed down by depolarization (Bernardi and Azzone, 1982, 1983).
Whatever the mechanism of LETM1-catalyzed Ca2+ transport, it should be mentioned that the Ca2+-transporting activity of LETM1 in reconstituted systems has so far been studied in K+-free media (Nowikovsky et al., 2004; Jiang et al., 2009), and that the relevant question of whether Ca2+ is actually transported at physiological K+ concentrations has not been addressed. A change of selectivity of channels and transporters is often observed in the absence of the physiologically transported species. For example, in the absence of divalent cations, the MCU conducts large Na+ currents that can be inhibited by RR and disappear as soon as Mg2+ or Ca2+ is added (Kirichok et al., 2004). Although the experiments of Jiang et al. (2009) in living and permeabilized cells were performed at physiological K+ and Mg2+ concentrations, in our view, whether the uptake of Ca2+ under those conditions was actually mediated by LETM1 remains to be established.
A recent study addressed the putative role of LETM1 and UCP2/3 in mitochondrial Ca2+ transport in endothelial cells (Waldeck-Weiermair et al., 2011). The role of UCP2/3 as MCUs (Trenker et al., 2007) has been a matter of debate (Brookes et al., 2008; Trenker et al., 2008), and although this topic in not within the scope of our review, we feel that such a role for UCP2/3 has been strongly undermined by the subsequent discovery of mitochondrial Ca2+ uptake 1 (Perocchi et al., 2010) and MCU (Baughman et al., 2011; De Stefani et al., 2011). As for LETM1, it has been reported that its knockdown strongly diminished the transfer of Ca2+ into mitochondria, resulting in subsequent reduction of store-operated Ca2+ entry (Waldeck-Weiermair et al., 2011). Although knockdown of LETM1 was reported not to impact cellular ATP levels and membrane potential, its impact on K+ homeostasis has not been considered or discussed in this study (Waldeck-Weiermair et al., 2011), which in our view will need to be confirmed. We wish to stress that in most other investigations with comparable levels of LETM1 suppression, significant changes of mitochondrial membrane potential were observed (Nowikovsky et al., 2004; Frazier et al., 2006; McQuibban et al., 2010).
In summary, we think that the occurrence and mode of LETM1-dependent Ca2+ transport need further study to establish whether, besides its undisputable role in KHE (Nowikovsky et al., 2009), LETM1 can also support or modulate Ca2+ uptake in energized mitochondria.
LETM1 and the WHS
The WHS (MIM 194190) is a complex disease involving the central nervous system (Hirschhorn et al., 1965; Wolf et al., 1965) caused by partial heterozygous deletion of the terminal portion of the short arm of chromosome 4 involving the 4p16.3 region (Endele et al., 1999; Zollino et al., 2003). A critical region of 165 kb has been defined (WHSCR-1), whose deletion causes the clinical hallmarks of the disease, i.e., severe growth and mental retardation, hypotonia, midline fusion defects, and typical facial dysmorphism (Johnson et al., 1976; Wilson et al., 1981). A subset of cases of WHS is characterized by seizures, which start during the first year of life and are a frequent cause of death (Zollino et al., 2000). The LETM1 gene has been localized less than 80 kb distal to the WHSCR-1 region, and it is invariably deleted in WHS patients with seizures and preserved in those without epilepsy (Schlickum et al., 2004) regardless of the WHSCR-1 deletion. Indeed, haploinsufficiency of the WHSCR-1 region results in an atypical WHS phenotype with no seizures, whereas involvement of the WHSCR-2, which includes LETM1, causes the typical WHS phenotype with psychomotor retardation and epilepsy. Thus, LETM1 haploinsufficiency is a potential causative event for WHS with seizures and possibly also in other forms of epilepsy. Given the key role of LETM1 in mitochondrial K+ homeostasis, it may be predicted that reduced activity of the KHE is followed by osmotic matrix swelling, outer membrane rupture, cytochrome c release, and impaired ATP production. In keeping with this prediction, depletion of LETM1 in wild-type cells caused matrix swelling and fragmentation that, like in yeast, could be rescued by proper amounts of nigericin (Hasegawa and van der Bliek, 2007; Nowikovsky et al., 2007; Dimmer et al., 2008). Overexpression of LETM1 instead resulted in mitochondrial matrix contraction (Dimmer et al., 2008). Collectively, these findings critically support a role of the mitochondrial K+ cycle in pathophysiological swelling–contraction events of mitochondria. Impaired mitochondrial ATP production after mitochondrial swelling may affect the neurons’ ability to maintain the plasma membrane potential and thus increase excitability to the threshold required to trigger seizures, a hypothesis that has not been tested so far. Fibroblastoid cells from one WHS patient showed a detectable decrease in LETM1 protein content, but there was no obvious phenotypic effect on mitochondria. Only stronger reduction in LETM1 expression caused mitochondrial changes and finally cell death (Dimmer et al., 2008). Yet it is well possible that in other types of cells, e.g., neurons, the LETM1 protein contents of WHS patients is below the threshold required to maintain mitochondrial cation homeostasis, and that the basis for cell dysfunction in WHS may be lowered KHE activity, which could be treated with pharmacological agents.
To understand the function of LETM1 in development, Drosophila was used as an animal model. Drosophila is a powerful model organism, and the availability of the genome-wide library of Drosophila RNAi transgenes combined with a wide range of GAL4 drivers allows for the targeting of conditional inactivation of a gene function in a tissue-specific manner (Dietzl et al., 2007). DmLETM1 knockdown recapitulated several important hallmarks of WHS, as ubiquitous or muscle-specific down-regulation significantly delayed development and resulted in small body size and early death. Down-regulation of DmLetm1 in the ommatidia caused deformations of the receptor cells and cell death. Down-regulation in the neuronal system resulted in reduced locomotor activity, and electrophysiological analyses revealed reduced synaptic neurotransmitter release (McQuibban et al., 2010).
A growing list of functions for LETM1
A role for LETM1 as a regulator of the mitochondrial-shaping protein OPA1 has been proposed based on LETM1 overexpression studies, where it was found that mitochondria fragment and OPA1 are differently cleaved, s-OPA1 rather than l-OPA1 being detected after overexpression of LETM1 (Piao et al., 2009b). It is worth recalling that MDM38, the yeast homologue of LETM1, was originally identified together with a series of other genes in a screen for mutants affecting mitochondrial morphology (Dimmer et al., 2002). Some of these genes did turn out to encode proteins required for mitochondrial fusion or fission, yet a large number of these genes encode proteins involved in different functions, including lipid metabolism and cation transport (Dimmer et al., 2002; Altmann and Westermann, 2005). Because LETM1 has complex effects on mitochondrial volume, membrane potential, and ATP levels (Nowikovsky et al., 2009), it is presently hard to tell whether the reported effects on OPA1 cleavage are primary or secondary to other events discussed in the preceding paragraphs.
It has recently been found that ER Ca2+ uptake in permeabilized neurons and cardiomyocytes was strongly inhibited by replacing K+ with Na+ or tetraethylammonium, or by treatment with quinine or propranolol (Kuum et al., 2012), well-known inhibitors of the mitochondrial KHE (Nakashima and Garlid, 1982). The inhibitory effects of propranolol were relieved by nigericin, indicating that proton–potassium exchange is essential for ER Ca2+ uptake; strikingly, fluorescence microscopy and Western blot analysis revealed the presence of LETM1 in the ER (Kuum et al., 2012). These data are consistent with a key role of LETM1 in maintaining ER pH homeostasis and exerting an indirect regulatory role on Ca2+ uptake in specific cell types (Kuum et al., 2012). An ER retention signal is found at the C terminus of LETM1 in some organisms; it will be interesting to assess the mechanisms through which LETM1 can be sorted to the ER or to the mitochondria, and whether this involves posttranscriptional modifications, as in the case of NADH–cytochrome b5 reductase (Borgese et al., 1996; Colombo et al., 2005).
Recent work has explored the potential role of LETM1 in cell death as related to carcinogenesis. LETM1 overexpression induced necrotic cell death, which was matched by decreased mitochondrial biogenesis and ATP production, and by AMPK activation (Piao et al., 2009a; Hwang et al., 2010); yet the expression levels of LETM1 were found to be significantly increased in multiple human cancer tissues (Piao et al., 2009a). It is not clear what mechanism(s) mediates the effects of enforced LETM1 overexpression, however, and we think that this issue must be addressed before safe conclusions can be made on the role of LETM1 in tumorigenesis.
Conclusions and perspectives
In summary, studies in yeast, human cell cultures, and Drosophila underscore the prominent role of LETM1 in maintaining mitochondrial K+ and volume homeostasis and respiratory chain assembly; genetic studies on WHS patients identify the deletion of LETM1 as a most likely causative event of seizures associated with the disease. Understanding the pathophysiology of LETM1 deficiency in vivo may thus lead to important advances toward the therapy of WHS and possibly of other forms of central nervous system diseases with seizures, which are often linked to mitochondrial defects (DiMauro and Schon, 2008). On the other hand, in our view, the potential role of LETM1 in mitochondrial Ca2+ homeostasis demands further scrutiny, with specific emphasis on the H+–Ca2+ stoichiometry and on assessing whether this protein can catalyze Ca2+ transport at physiological K+ concentrations. However, it is easy to forecast that the growing interest in LETM1 and the discovery of additional functions for this protein will rapidly lead to substantial advances in our understanding of its roles in pathophysiology.
Appendix
Predicted equilibrium Ca2+ accumulation ratio according to the mechanism of transport
For each of the four transport mechanisms depicted in Fig. 4, assuming that ψm − ψc (Δψ) is −180 mV, 2.303 RT/F = 60 mV, and [H+]m/[H+]c = 0.1, the ratio between the concentrations of Ca2+ in the mitochondrial matrix (Ca2+m) and in the cytoplasm (Ca2+c) at equilibrium can be calculated by considering the corresponding driving forces (is the electrochemical, and µ is the chemical potential) equal to zero as follows:
Ca2+ uniporter
H+–Ca2+ antiporter
2H+–Ca2+ antiporter
3H+–Ca2+ antiporter
This Perspectives series includes articles by Sheu et al., Zhang et al., Balaban, Santo-Domingo and Demaurex, Wei and Dirksen, O-Uchi et al., and Galloway and Yoon.
Acknowledgments
We would like to thank Prof. Daniela Pietrobon for helpful discussions on the thermodynamics of mitochondrial Ca2+ transport.
Our work is supported by grants from Schweizerische Nationalfonds and Associazione Italiana per la Ricerca sul Cancro, Fondazione Cariparo, MIUR-Italy, Italian Institute of Technology, Telethon-Italy, the University of Padova, Fonds zur Förderung der wissenschaftlichen Forschung (FWF), and Genomforschung Austria (GEN-AU). L. Scorrano is a Senior Telethon Scientist.
Shey-Shing Sheu served as guest editor.
Footnotes
Abbreviations used in this paper:
- KHE
- K+–H+ exchange
- MCU
- mitochondrial Ca2+ uniporter
- PTP
- permeability transition pore
- RR
- ruthenium red
- WHS
- Wolf–Hirschhorn syndrome
References
- Altmann K., Westermann B. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 16:5410–5417 10.1091/mbc.E05-07-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuld R.A., Hohl C.M., Castillo L.C., Garleb A.A., Starling R.C., Brierley G.P. 1992. Cyclosporin inhibits mitochondrial calcium efflux in isolated adult rat ventricular cardiomyocytes. Am. J. Physiol. 262:H1699–H1704 [DOI] [PubMed] [Google Scholar]
- Azzone G.F., Pozzan T., Massari S., Bragadin M., Dell’Antone P. 1977. H+/site ratio and steady state distribution of divalent cations in mitochondria. FEBS Lett. 78:21–24 10.1016/0014-5793(77)80264-0 [DOI] [PubMed] [Google Scholar]
- Barsukova A., Komarov A., Hajnóczky G., Bernardi P., Bourdette D., Forte M. 2011. Activation of the mitochondrial permeability transition pore modulates Ca2+ responses to physiological stimuli in adult neurons. Eur. J. Neurosci. 33:831–842 10.1111/j.1460-9568.2010.07576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmitt H., Mick D.U., Deckers M., Vollmer C., Funes S., Kehrein K., Ott M., Rehling P., Herrmann J.M. 2010. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol. Biol. Cell. 21:1937–1944 10.1091/mbc.E10-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L., et al. 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 476:341–345 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P. 1999. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol. Rev. 79:1127–1155 [DOI] [PubMed] [Google Scholar]
- Bernardi P., Azzone G.F. 1982. A membrane potential-modulated pathway for Ca2+ efflux in rat liver mitochondria. FEBS Lett. 139:13–16 10.1016/0014-5793(82)80476-6 [DOI] [PubMed] [Google Scholar]
- Bernardi P., Azzone G.F. 1983. Regulation of Ca2+ efflux in rat liver mitochondria. Role of membrane potential. Eur. J. Biochem. 134:377–383 10.1111/j.1432-1033.1983.tb07578.x [DOI] [PubMed] [Google Scholar]
- Bernardi P., Petronilli V. 1996. The permeability transition pore as a mitochondrial calcium release channel: a critical appraisal. J. Bioenerg. Biomembr. 28:131–138 10.1007/BF02110643 [DOI] [PubMed] [Google Scholar]
- Borgese N., Aggujaro D., Carrera P., Pietrini G., Bassetti M. 1996. A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J. Cell Biol. 135:1501–1513 10.1083/jcb.135.6.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes P.S., Parker N., Buckingham J.A., Vidal-Puig A., Halestrap A.P., Gunter T.E., Nicholls D.G., Bernardi P., Lemasters J.J., Brand M.D. 2008. UCPs—unlikely calcium porters. Nat. Cell Biol. 10:1235–1237 10.1038/ncb1108-1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E., Tiozzo R., Lugli G., Crovetti F., Kratzing C. 1974. The release of calcium from heart mitochondria by sodium. J. Mol. Cell. Cardiol. 6:361–371 10.1016/0022-2828(74)90077-7 [DOI] [PubMed] [Google Scholar]
- Colombo S., Longhi R., Alcaro S., Ortuso F., Sprocati T., Flora A., Borgese N. 2005. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 168:735–745 10.1083/jcb.200407082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M., Capano M., Carafoli E. 1976. The sodium-induced efflux of calcium from heart mitochondria. A possible mechanism for the regulation of mitochondrial calcium. Eur. J. Biochem. 69:453–462 10.1111/j.1432-1033.1976.tb10930.x [DOI] [Google Scholar]
- De Stefani D., Raffaello A., Teardo E., Szabò I., Rizzuto R. 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 476:336–340 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- DiMauro S., Garone C. 2011. Metabolic disorders of fetal life: glycogenoses and mitochondrial defects of the mitochondrial respiratory chain. Semin. Fetal Neonatal Med. 16:181–189 10.1016/j.siny.2011.04.010 [DOI] [PubMed] [Google Scholar]
- DiMauro S., Schon E.A. 2008. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 31:91–123 10.1146/annurev.neuro.30.051606.094302 [DOI] [PubMed] [Google Scholar]
- Dimmer K.S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:847–853 10.1091/mbc.01-12-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer K.S., Navoni F., Casarin A., Trevisson E., Endele S., Winterpacht A., Salviati L., Scorrano L. 2008. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum. Mol. Genet. 17:201–214 10.1093/hmg/ddm297 [DOI] [PubMed] [Google Scholar]
- Drago I., Pizzo P., Pozzan T. 2011. After half a century mitochondrial calcium in- and efflux machineries reveal themselves. EMBO J. 30:4119–4125 10.1038/emboj.2011.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod J.W., Wong R., Mishra S., Vagnozzi R.J., Sakthievel B., Goonasekera S.A., Karch J., Gabel S., Farber J., Force T., et al. 2010. Cyclophilin D controls mitochondrial pore-dependent Ca2+ exchange, metabolic flexibility, and propensity for heart failure in mice. J. Clin. Invest. 120:3680–3687 10.1172/JCI43171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S., Fuhry M., Pak S.J., Zabel B.U., Winterpacht A. 1999. LETM1, a novel gene encoding a putative EF-hand Ca2+-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics. 60:218–225 10.1006/geno.1999.5881 [DOI] [PubMed] [Google Scholar]
- Frazier A.E., Taylor R.D., Mick D.U., Warscheid B., Stoepel N., Meyer H.E., Ryan M.T., Guiard B., Rehling P. 2006. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172:553–564 10.1083/jcb.200505060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froschauer E., Nowikovsky K., Schweyen R.J. 2005. Electroneutral K+/H+ exchange in mitochondrial membrane vesicles involves Yol027/Letm1 proteins. Biochim. Biophys. Acta. 1711:41–48 10.1016/j.bbamem.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Hasegawa A., van der Bliek A.M. 2007. Inverse correlation between expression of the Wolfs Hirschhorn candidate gene Letm1 and mitochondrial volume in C. elegans and in mammalian cells. Hum. Mol. Genet. 16:2061–2071 10.1093/hmg/ddm154 [DOI] [PubMed] [Google Scholar]
- Heaton G.M., Nicholls D.G. 1976. The calcium conductance of the inner membrane of rat liver mitochondria and the determination of the calcium electrochemical gradient. Biochem. J. 156:635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn K., Cooper H.L., Firschein I.L. 1965. Deletion of short arms of chromosome 4-5 in a child with defects of midline fusion. Humangenetik. 1:479–482 [DOI] [PubMed] [Google Scholar]
- Hwang S.K., Piao L., Lim H.T., Minai-Tehrani A., Yu K.N., Ha Y.C., Chae C.H., Lee K.H., Beck G.R., Park J., Cho M.H. 2010. Suppression of lung tumorigenesis by leucine zipper/EF hand-containing transmembrane-1. PLoS ONE. 5:e12535 10.1371/journal.pone.0012535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Zhao L., Clapham D.E. 2009. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 326:144–147 10.1126/science.1175145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.P., Mulder R.D., Hosen R. 1976. The Wolf-Hirschhorn (4p-) syndrome. Clin. Genet. 10:104–112 10.1111/j.1399-0004.1976.tb00021.x [DOI] [PubMed] [Google Scholar]
- Kinnally K.W., Campo M.L., Tedeschi H. 1989. Mitochondrial channel activity studied by patch-clamping mitoplasts. J. Bioenerg. Biomembr. 21:497–506 10.1007/BF00762521 [DOI] [PubMed] [Google Scholar]
- Kirichok Y., Krapivinsky G., Clapham D.E. 2004. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 427:360–364 10.1038/nature02246 [DOI] [PubMed] [Google Scholar]
- Kuum M., Veksler V., Liiv J., Ventura-Clapier R., Kaasik A. 2012. Endoplasmic reticulum potassium-hydrogen exchanger and small conductance calcium-activated potassium channel activities are essential for ER calcium uptake in neurons and cardiomyocytes. J. Cell Sci. 125:625–633 10.1242/jcs.090126 [DOI] [PubMed] [Google Scholar]
- Lupo D., Vollmer C., Deckers M., Mick D.U., Tews I., Sinning I., Rehling P. 2011. Mdm38 is a 14-3-3-like receptor and associates with the protein synthesis machinery at the inner mitochondrial membrane. Traffic. 12:1457–1466 10.1111/j.1600-0854.2011.01239.x [DOI] [PubMed] [Google Scholar]
- McQuibban A.G., Joza N., Megighian A., Scorzeto M., Zanini D., Reipert S., Richter C., Schweyen R.J., Nowikovsky K. 2010. A Drosophila mutant of LETM1, a candidate gene for seizures in Wolf-Hirschhorn syndrome. Hum. Mol. Genet. 19:987–1000 10.1093/hmg/ddp563 [DOI] [PubMed] [Google Scholar]
- Moore C.L. 1971. Specific inhibition of mitochondrial Ca++ transport by ruthenium red. Biochem. Biophys. Res. Commun. 42:298–305 10.1016/0006-291X(71)90102-1 [DOI] [PubMed] [Google Scholar]
- Morán M., Marín-Buera L., Gil-Borlado M.C., Rivera H., Blázquez A., Seneca S., Vázquez-López M., Arenas J., Martín M.A., Ugalde C. 2010. Cellular pathophysiological consequences of BCS1L mutations in mitochondrial complex III enzyme deficiency. Hum. Mutat. 31:930–941 10.1002/humu.21294 [DOI] [PubMed] [Google Scholar]
- Moyle J., Mitchell P. 1977. Electric charge stoicheiometry of calcium translocation in rat liver mitochondria. FEBS Lett. 73:131–136 10.1016/0014-5793(77)80964-2 [DOI] [PubMed] [Google Scholar]
- Nakashima R.A., Garlid K.D. 1982. Quinine inhibition of Na+ and K+ transport provides evidence for two cation/H+ exchangers in rat liver mitochondria. J. Biol. Chem. 257:9252–9254 [PubMed] [Google Scholar]
- Nowikovsky K., Froschauer E.M., Zsurka G., Samaj J., Reipert S., Kolisek M., Wiesenberger G., Schweyen R.J. 2004. The LETM1/YOL027 gene family encodes a factor of the mitochondrial K+ homeostasis with a potential role in the Wolf-Hirschhorn syndrome. J. Biol. Chem. 279:30307–30315 10.1074/jbc.M403607200 [DOI] [PubMed] [Google Scholar]
- Nowikovsky K., Reipert S., Devenish R.J., Schweyen R.J. 2007. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 14:1647–1656 10.1038/sj.cdd.4402167 [DOI] [PubMed] [Google Scholar]
- Nowikovsky K., Schweyen R.J., Bernardi P. 2009. Pathophysiology of mitochondrial volume homeostasis: potassium transport and permeability transition. Biochim. Biophys. Acta. 1787:345–350 10.1016/j.bbabio.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Palty R., Silverman W.F., Hershfinkel M., Caporale T., Sensi S.L., Parnis J., Nolte C., Fishman D., Shoshan-Barmatz V., Herrmann S., et al. 2010. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. USA. 107:436–441 10.1073/pnas.0908099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F., Gohil V.M., Girgis H.S., Bao X.R., McCombs J.E., Palmer A.E., Mootha V.K. 2010. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature. 467:291–296 10.1038/nature09358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronilli V., Szabò I., Zoratti M. 1989. The inner mitochondrial membrane contains ion-conducting channels similar to those found in bacteria. FEBS Lett. 259:137–143 10.1016/0014-5793(89)81513-3 [DOI] [PubMed] [Google Scholar]
- Piao L., Li Y., Kim S.J., Byun H.S., Huang S.M., Hwang S.K., Yang K.J., Park K.A., Won M., Hong J., et al. 2009a. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 69:3397–3404 10.1158/0008-5472.CAN-08-3235 [DOI] [PubMed] [Google Scholar]
- Piao L., Li Y., Kim S.J., Sohn K.C., Yang K.J., Park K.A., Byun H.S., Won M., Hong J., Hur G.M., et al. 2009b. Regulation of OPA1-mediated mitochondrial fusion by leucine zipper/EF-hand-containing transmembrane protein-1 plays a role in apoptosis. Cell. Signal. 21:767–777 10.1016/j.cellsig.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Rasola A., Sciacovelli M., Pantic B., Bernardi P. 2010. Signal transduction to the permeability transition pore. FEBS Lett. 584:1989–1996 10.1016/j.febslet.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Bernardi P., Pozzan T. 2000. Mitochondria as all-round players of the calcium game. J. Physiol. 529:37–47 10.1111/j.1469-7793.2000.00037.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg H., Scarpa A. 1974. Calcium uptake and membrane potential in mitochondria. Biochemistry. 13:4811–4817 10.1021/bi00720a020 [DOI] [PubMed] [Google Scholar]
- Scarpa A., Azzone G.F. 1970. The mechanism of ion translocation in mitochondria. 4. Coupling of K+ efflux with Ca2+ uptake. Eur. J. Biochem. 12:328–335 10.1111/j.1432-1033.1970.tb00854.x [DOI] [PubMed] [Google Scholar]
- Schlickum S., Moghekar A., Simpson J.C., Steglich C., O’Brien R.J., Winterpacht A., Endele S.U. 2004. LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics. 83:254–261 10.1016/j.ygeno.2003.08.013 [DOI] [PubMed] [Google Scholar]
- Tamai S., Iida H., Yokota S., Sayano T., Kiguchiya S., Ishihara N., Hayashi J., Mihara K., Oka T. 2008. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J. Cell Sci. 121:2588–2600 10.1242/jcs.026625 [DOI] [PubMed] [Google Scholar]
- Trenker M., Malli R., Fertschai I., Levak-Frank S., Graier W.F. 2007. Uncoupling proteins 2 and 3 are fundamental for mitochondrial Ca2+ uniport. Nat. Cell Biol. 9:445–452 10.1038/ncb1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenker M., Fertschai I., Malli R., Graier W.F. 2008. UCP2/3-likely to be fundamental for mitochondrial Ca2+ uniport. Nat. Cell Biol. 10:1237–1240 10.1038/ncb1108-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stockum S., Basso E., Petronilli V., Sabatelli P., Forte M.A., Bernardi P. 2011. Properties of Ca2+ transport in mitochondria of Drosophila melanogaster. J. Biol. Chem. 286:41163–41170 10.1074/jbc.M111.268375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldeck-Weiermair M., Jean-Quartier C., Rost R., Khan M.J., Vishnu N., Bondarenko A.I., Imamura H., Malli R., Graier W.F. 2011. Leucine zipper EF hand-containing transmembrane protein 1 (Letm1) and uncoupling proteins 2 and 3 (UCP2/3) contribute to two distinct mitochondrial Ca2+ uptake pathways. J. Biol. Chem. 286:28444–28455 10.1074/jbc.M111.244517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldherr M., Ragnini A., Jank B., Teply R., Wiesenberger G., Schweyen R.J. 1993. A multitude of suppressors of group II intron-splicing defects in yeast. Curr. Genet. 24:301–306 10.1007/BF00336780 [DOI] [PubMed] [Google Scholar]
- Wilson M.G., Towner J.W., Coffin G.S., Ebbin A.J., Siris E., Brager P. 1981. Genetic and clinical studies in 13 patients with the Wolf-Hirschhorn syndrome [del(4p)]. Hum. Genet. 59:297–307 (del(4p)). 10.1007/BF00295461 [DOI] [PubMed] [Google Scholar]
- Wingrove D.E., Amatruda J.M., Gunter T.E. 1984. Glucagon effects on the membrane potential and calcium uptake rate of rat liver mitochondria. J. Biol. Chem. 259:9390–9394 [PubMed] [Google Scholar]
- Wolf U., Porsch R., Baitsch H., Reinwein H. 1965. Deletion on short arms of a B-chromosome without “Cri du Chat” syndrome. Lancet. 1:769 10.1016/S0140-6736(65)92136-7 [DOI] [PubMed] [Google Scholar]
- Zollino M., Di Stefano C., Zampino G., Mastroiacovo P., Wright T.J., Sorge G., Selicorni A., Tenconi R., Zappalà A., Battaglia A., et al. 2000. Genotype-phenotype correlations and clinical diagnostic criteria in Wolf-Hirschhorn syndrome. Am. J. Med. Genet. 94:254–261 [DOI] [PubMed] [Google Scholar]
- Zollino M., Lecce R., Fischetto R., Murdolo M., Faravelli F., Selicorni A., Buttè C., Memo L., Capovilla G., Neri G. 2003. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am. J. Hum. Genet. 72:590–597 10.1086/367925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotova L., Aleschko M., Sponder G., Baumgartner R., Reipert S., Prinz M., Schweyen R.J., Nowikovsky K. 2010. Novel components of an active mitochondrial K+/H+ exchange. J. Biol. Chem. 285:14399–14414 10.1074/jbc.M109.059956 [DOI] [PMC free article] [PubMed] [Google Scholar]