Introduction
The essence of the mitochondria lies within their ability to generate energy, maintain metabolisms, and regulate signaling cascades. As fundamental constituents of numerous rudimentary cellular processes, the molecular architecture and the proteome of the mitochondria are critical to intracellular biochemistry. The behavior of the mitochondria is subjugated to the environment of this organelle; perturbation of mitochondrial homeostasis can prompt pathophysiological conditions to arise.
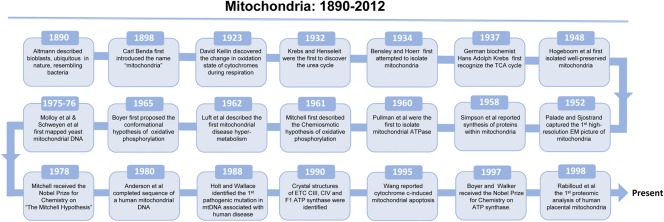
The history of mitochondrial research can be traced back to Rudolf Albrecht von Kolliker in 1857 when he first described the mitochondrion. In 1890, Richard Altmann termed the mitochondria “bioblasts,” noted their ubiquitous nature, and showed exceptional foresight when he explained that these bioblasts were living inside cells and were responsible for “elementary functions.” The endosymbiotic theory was first articulated by Konstantin Mereschkowski in 1905. Although several hypotheses were put forth to explain the merger between mitochondria and cells, it was Lynn Margulis who tied the endosymbiotic theory to biochemical and cytological evidence. Mitochondria are of α-protobacterial origin, engulfed by cells through endosymbiosis about two billion years ago. The name “mitochondrion” was first introduced in 1898 by Carl Benda from the Greek words “mitos” (thread) and “chondros” (granule). The first well-preserved mitochondria were isolated by Hogeboom et al. (1948). This approach proved to be significantly advantageous to mitochondrial research for several decades, in particular, on the characterization of mitochondrial proteome biology and function which we focus today. A key result from the remarkable hypothesis of chemiosmosis by Mitchell (Ernster and Schatz, 1981) is an organized subproteome, i.e., the electron transport complexes, with a dedicated role, supporting the oxidation and phosphorylation function in the mitochondria. The significance of the respiratory complexes to biology has been highly recognized, but that to the proteome design was much less appreciated. The complexity of this apparatus was further realized with the introduction of innovative methods to visualize three-dimensional structures and the role of the mitochondria in cell death signaling (Ernster and Schatz, 1981). These findings inspired the research efforts to link biological pathways with their subproteome-based molecular participants in the regulation of mitochondrial function. Fig. 1 summarizes the milestones of the long and fruitful history of mitochondrial research.
Figure 1.
Milestones in mitochondrial research. Representative milestones in mitochondrial research, including mitochondrial function, structure, and mitochondrial medicine, are listed in a chronological sequence (1890–current). Our apologies to those not represented in this timeline.
The relationship between mitochondria and disease has been firmly established, with one of the first reported mitochondrial diseases by Rolf Luft and colleagues at Karolinska University in Stockholm, Sweden. A patient was presented with hypermetabolism unrelated to thyroid dysfunction. Through morphological (increased number of mitochondria) and biochemical (loosely coupled mitochondrial oxidative phosphorylation [OXPHOS]) evidence, this group of investigators concluded that mitochondria were the source of these symptoms, marking the beginning of an era of mitochondrial medicine. Throughout the 1970s, other landmark discoveries singled out specific metabolic as well as enzymatic deficiencies, such as pyruvate dehydrogenase and carnitine palmitoyltransferase deficiencies. Over 20 years passed between the discovery of mitochondrial DNA (mtDNA) and its mapping. In 1988, for the first time, mutations in mtDNA-associated diseases were reported (Wallace, 2010). As the detection of mitochondrial dysfunctions became more prevalent, the identification of mitochondrial components, as well as the characterization of mitochondrial proteome dynamics, increased in relevance. This Perspective will provide current knowledge of mitochondrial biology and an update on the design of the mitochondrial proteome, with an effort to contextualize mitochondrial proteomes to their biological pathways and the consequences of the proteome compositions to clinical phenotypes.
Proteins supporting mitochondrial structure and biology
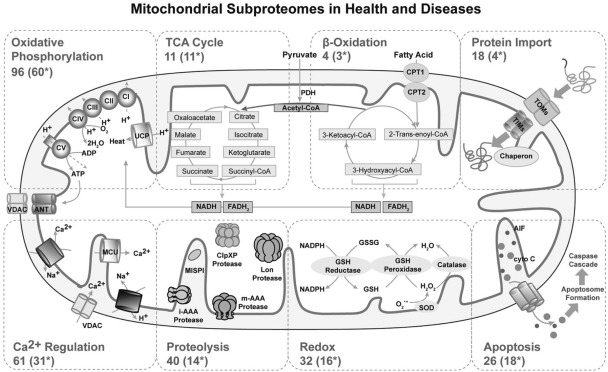
The mitochondrion is an organelle comprised of four distinct compartments—the outer mitochondrial membrane (OMM), the intermembrane space, the inner mitochondrial membrane (IMM), and the matrix—which are bordered by its unique double-membrane structure. Production and transportation of electrolytes are facilitated by proteins embedded within the membranes. Approximately 1,000 proteins have been identified by several proteomic investigations (Mootha et al., 2003; Taylor et al., 2003; Forner et al., 2006; Kislinger et al., 2006; Pagliarini et al., 2008; Zhang et al., 2008) to support mitochondrial structure and function. Fig. 2 summarizes the major mitochondrial cellular processes, their subproteomes, and their involvements in metabolic diseases.
Figure 2.
Schematic overview of mitochondrial subproteomes in health and disease. This figure contextualizes mitochondrial proteomes to their biological pathways and the consequences of the proteome compositions to clinical phenotypes. Mitochondrial pathways responsible for fundamental cellular functions within the cell, including OXPHOS, TCA cycle, β oxidation, apoptosis, proteolysis, redox, protein import, and calcium regulation, are detailed. Other mitochondrial functions related to specific cell types are not listed. Within each mitochondrial subproteome, the first number represents the total number of proteins associated with its function (the number of proteins was collected in our previous study; Zhang et al., 2008); the next number inside the parentheses (*) signifies the portion of the total proteins in this particular subproteome reported to be implicated in metabolic diseases.
The majority of proteins comprising the mitochondria are encoded by nuclear DNA. However, unlike their surrounding organelles, mitochondria possess their own DNA, which acts as scripts for the 13 proteins constituting seven proteins for complex I, one for complex III, three for complex IV, and two for complex V of the electron transport chain (ETC), also known as the respiratory complexes. In most multicellular organisms, mtDNA are inherited maternally. Because there are many copies of mtDNA and there are many mitochondria in all cell types, most mitochondrial mutations often have few detrimental effects on mitochondrial function until they reach sufficient number, a “threshold effect.” Not surprisingly, this threshold effect is lower in highly aerobic tissues such as the eye, brain, and heart (DiMauro and Schon, 2003). Therefore, under pathogenic mitochondrial conditions, these tissues are vulnerable and may be affected, indicative of mitochondrial dysfunction. To date, proteomic approaches have been successful in characterizing the subproteome supporting OXPHOS function, including the identification of 88 proteins from the human heart (Taylor et al., 2003) and 96 proteins from the mouse heart (Zhang et al., 2008). These proteins represent 90% or more of the entire OXPHOS subproteome (nuclear and mDNA combined). The remaining 10% of the OXPHOS proteins are known only by their genetic information, but are undetected by proteomic methods. They may have exceptionally low abundance or may carry unique biochemical properties (e.g., hydrophobic proteins); their protein characterization may require approaches with higher sensitivity (e.g., antibodies).
As the key energy producer of the cell, the mitochondrion is fundamental for many metabolic processes. The products of glycolysis enter the mitochondrial matrix to continue their conversion to energy in the tricarboxylic acid (TCA) cycle, which possesses a subproteome composed of 11 proteins in the mouse cardiac mitochondria (Zhang et al., 2008). Subsequently, NADH and FADH2 proceed to donate electrons to the ETC components, eventually reducing oxygen to water. An electrochemical gradient is established and maintained to harness the energy produced by the subsequent flow of protons back into the matrix to synthesize ATP from ADP and phosphate.
Calcium plays a multifaceted role in mitochondrial function. Mitochondrial calcium regulation can be denoted by influx, matrix buffering, and efflux (Weiss et al., 2003; Glancy and Balaban, 2012). A total of 61 proteins from the mouse heart (Zhang et al., 2008) has been reported to support calcium regulation in mitochondria. Mitochondria have been implicated as fundamental in the induction of cell death; two differential processes have been shown to lyse the cells: necrosis and apoptosis. The protein identities defining both the necrosis and apoptosis pathways are only partially understood. Release of apoptotic factors such as cytochrome c, Smac/Diablo, and apoptosis-induced factors into the cytosol attracts caspase-9 and apoptotic protease-activating factor 1 to form the apoptosome with ATP. The Bcl-2 family proteins are also activated during the apoptotic process. A total of 26 proteins in the mouse heart (Zhang et al., 2008) has been identified to be related to apoptosis. In parallel, mitochondria produce free radicals by leaking electrons to oxygen in the process of transferring electrons through the ETC, in particular complexes I and III. To counteract this, endogenous scavenging enzymes and antioxidants are activated to eliminate reactive oxygen species (ROS); this defense system includes superoxide dismutases, catalase, peroxidredoxin, glutathione peroxidase, and reduced glutathione. Currently, 29 proteins in the human heart are affiliated with redox functions (Taylor et al., 2003), whereas 32 proteins were reported in the mouse heart. However, when the system is challenged to rapidly convert ROS to water, oxidative damage will accumulate, leading to mitochondrial dysfunction.
Mitochondrial proteases participate not only in protein proteolysis (Lau et al., 2012), but they also are emerging as crucial regulators of mitochondrial function. The many types of mitochondrial proteases include the PIM1/Lon protease, the mitochondrial intermembrane space protease I and the ClpXP protease in the matrix, and the i-AAA protease and the m-AAA protease residing in the IMM. Proteases are shown to play essential roles in mitochondrial morphology maintenance, mitochondrial biogenesis, and mitochondrial metabolism regulation (Bulteau and Bayot, 2011). Mitochondrial proteases are evolutionarily conserved from yeast to humans in support of the evolutionary prokaryotic ancestors. Disturbances of the mitochondrial proteolytic system affect mitochondrial homeostasis. Thus far, 40 proteases were reported from the mouse heart (Zhang et al., 2008) in the mitochondria.
Proteome profiling of mitochondria
The mitochondrial proteome is unique, complex, and dynamically regulated as it adapts to the needs of the tissues or disease states (Johnson et al., 2007b; Balaban, 2010). Over the past decade, tremendous efforts have been made to explore mitochondrial subproteomes and their posttranslational modifications (PTMs) in both physiological and pathological environments. Indeed, mitochondrial proteomes in various organisms and tissues, including yeast (Sickmann et al., 2003), mouse (Mootha et al., 2003; Kislinger et al., 2006; Pagliarini et al., 2008; Zhang et al., 2008), human (Taylor et al., 2003; Gaucher et al., 2004), rat (Forner et al., 2006; Reifschneider et al., 2006; Johnson et al., 2007b), and drosophila (Alonso et al., 2005) have been investigated. Fig. 3 deciphers the state-of-the-art experimental workflow used in mitochondrial proteomic studies, including mitochondrial isolation, purification, mass spectrometry (MS) identification, and data analyses.
Figure 3.
Current state-of-the-art technology platform to characterize mitochondrial proteomes. This figure depicts the current technology platform used to characterize mitochondrial proteomes from model systems, including mitochondrial preparation, protein/peptide fractionations, MS instrumentation, protein identification/PTM analyses, protein quantification, and data validation.
Preparation of pure and functionally viable mitochondria is an important first step to achieving reliable and reproducible proteomic outputs. Hogeboom et al. (1948) developed the first protocol to isolate rat liver mitochondria based on a differential centrifugation. Thereafter, isolation methods have been modified and tailored based on different tissues and species. Mitochondria could be further purified by free-flow electrophoresis or density gradient centrifugation with either percoll or other dense materials such as metrizamide, sucrose, and nycodenz. Mitochondrial purity and intactness could be validated by immunoblottings using specific organelle protein markers, membrane potential measurements, and respiratory control index assessments, as well as electron microscopy morphology detection (Zhang et al., 2008). If a particular mitochondrial subproteome, such as OMM, IMM, or OXPHOS, is the subject of interest, isolation of these subcompartments is desirable. Sample fractionation has been frequently applied to reduce the complexity before the sample is subjected to MS analyses. Different approaches have been used, including gel-based approaches such as one-dimensional SDS-PAGE, two-dimensional PAGE (2-DE), and blue native PAGE, as well as gel-free–based approaches such as immunoprecipitation, liquid chromatography (LC), and free-flow electrophoresis. Different separation approaches are complementary. 2-DE is preferred to separate soluble and high-abundance proteins but poorly resolves hydrophobic proteins, low-abundance proteins, and proteins with extreme isoelectric point values. The combination of a blue native PAGE and a SDS-PAGE could be used to enrich the identification of OXPHOS protein complexes (Reifschneider et al., 2006). Gel-free–based shotgun proteomic analyses, combining a strong cation exchange LC and a reversed-phased LC, are beneficial for the detection of proteins with low abundance and high hydrophobicity.
An early study on mitochondrial proteomes by Rabilloud et al. (1998) was conducted on human placental mitochondria using 2-DE followed by MS analyses; they reported a total of 46 proteins. Subsequently, Taylor and his collaborators (Taylor et al., 2003; Gaucher et al., 2004) characterized the human cardiac mitochondrial proteome using SDS-PAGE separation and multidimensional LC coupled with MS analyses, where 722 proteins were reported. Furthermore, Mootha et al. (2003) characterized mitochondrial proteomes from the mouse brain, heart, liver, and kidney samples. 399 proteins were identified from proteomic studies and 428 proteins were reported from gene annotation analyses, combining for a total of 591 distinct proteins from different tissues. Among these 399 proteins, 107 proteins were conserved across tissues. Later, Kislinger et al. (2006) performed a global proteomic survey of four organellar compartments (cytosol, membranes, mitochondria, and nuclei) in six mouse organs. 4,768 proteins were identified, of which 1,075 proteins were localized in the brain mitochondria, 667 in the heart mitochondria, 789 in the kidney mitochondria, 775 in the liver mitochondria, 1,072 in the lung mitochondria, and 901 in the placenta mitochondria. Among these mitochondrial proteins, only 132 proteins were conserved across tissues. Later on, studies by Zhang et al. (2008) identified 940 distinct proteins from mouse cardiac mitochondria using functionally validated mitochondria, among which 480 proteins were not identified by the aforementioned major proteomic profiling studies (Mootha et al., 2003; Taylor et al., 2003; Kislinger et al., 2006). Additionally, Pagliarini et al. (2008) combined MS analyses with GFP tagging, as well as machine learning, and created a mitochondrial compendium of 1,098 genes and their protein expressions across 14 different mouse tissues. By linking the characterized proteins in this inventory to known mitochondrial pathways, 19 proteins were predicted to be important for the function of ETC complex I, of which one protein (C8orf38) was further validated. Many methodological issues may contribute to these mitochondrial proteome differences as reported by their MS-based identifications. There are issues using mitochondrial samples from different tissues, including sample preparations, contaminants from abundant proteins, the overall molecular compositions of the targeted mitochondrial proteome, the dynamic ranges of protein abundance in the particular mitochondrial proteome of interest, etc. Apart from these technical factors, it is highly possible that the reported diversity in mitochondrial proteomes is a result of their tissue specificity. Despite the variability of MS-based proteomic approaches, the proteomic results from different research groups indicate the heterogeneity of mitochondrial proteomes among the rat/mouse tissues (Forner et al., 2006; Kislinger et al., 2006; Johnson et al., 2007a,b). These inter-tissue comparisons recognize that the mitochondrial proteomes are tuned to meet the metabolic and signaling requirements of their environment. For example, heart mitochondria require a constant and stable supply of ATP to maintain cardiac function, whereas the liver mitochondria are orientated toward a more biosynthetic role conducive for metabolic function. In addition, using a combination of comparative genomics and computational algorithms, the interspecies comparisons support the notion of the existence of conserved and heterogeneous proteins in the mitochondrial proteome (Richly et al., 2003).
The four subcompartments of the mitochondria are comprised of different protein contents. The matrix incorporates approximately two thirds of the mitochondrial proteins, with the IMM containing 21%, the intermembrane space occupying ∼6%, and the OMM encompassing a mere 4% of the overall proteins of the mitochondrion (Distler et al., 2008). These values are still under investigation as low-abundance proteins are difficult to detect with the current technology. Using organic acid, Da Cruz and Martinou (2008) extracted the IMM fractions of hepatocytes and found 182 proteins using a 2-DE-LC-MS/MS. McDonald et al. (2006) further identified 348 proteins through three separation techniques (2D-LC with ProteomeLab PF 2D Protein Fractionation System, 2-DE, and RP-HPLC). A two-step digestion with trypsin and proteinase K facilitated the detection of OMM proteins previously unidentified (Distler et al., 2008). Another study of Saccharomyces cerevisiae by Zahedi et al. (2006) encountered 112 OMM proteins, including integral and peripheral membrane proteins.
In parallel to protein identification, two main strategies have been applied to the quantitative mitochondrial proteomic analyses: an MS-based label-free approach and differential labeling quantitative techniques. The label-free quantification is based on either the measurement of the peptide precursor intensity ions of a protein or the number of fragmented spectra peptides of a protein. This approach has been successfully used in the comparison of mitochondrial proteome changes under physiological and pathological conditions in various animal models (Forner et al., 2006; Johnson et al., 2007a). Isotope-labeling experiments include two-dimensional difference in-gel electrophoresis (2D-DIGE), isotope-coded affinity tag, isobaric tags for relative and absolute quantitation (iTRAQ), and stable isotopic labeling by amino acids in cell culture (SILAC), as well as stable isotopic labeling in mammals (SILAM). These approaches are more accurate, but the reagents are costly and require specialized bioinformatics tools.
In recent years, several databases of mitochondrial proteins have been created, including MitoP2 (Elstner et al., 2008), Mitoproteome (Cotter et al., 2004), MitoCarta (Pagliarini et al., 2008), MitoMiner (Smith and Robinson, 2009), as well as COPaKB, i.e., the Cardiac Organellar Protein Atlas Knowledgebase (http://www.heartproteome.org). These databases list mitochondrial proteins identified via multiple approaches, including MS/MS analyses, literature curations, and bioinformatics evaluations. COPaKB is an integrated resource of proteome biology configured to specifically focus on cardiovascular biology and medicine. Its first release includes proteomic data from large-scale proteomic surveys of cardiac mitochondrial proteins.
Defining the mitochondrial proteome is a challenge because of the dynamics of this organelle. Approximately 293 out of 940 proteins identified were found to contain mitochondrial targeting sequences (Zhang et al., 2008). In contrast, a majority of proteins do not bear the mitochondrial targeting sequences; however, they can travel to multiple subcellular localizations in parallel to their mitochondrial residency. Some proteins anchor onto the outer membrane of mitochondria with a loose attachment; they are called mitochondrial-associated proteins. Targeting sequence prediction cannot be used as a sole strategy to validate the mitochondrial localization of proteins caused by a higher false-positive prediction rate as well as multiple protein import mechanisms. Some mitochondrial proteins are only expressed in certain developmental stages or specific species. Furthermore, some mitochondrial proteins are expressed in very low abundances and do not meet the detection threshold criteria of MS. Collectively, the above issues (the nature of mitochondrial proteins and the technological limitations) contribute to the current discrepancy of the existing mitochondrial proteome datasets.
PTM of mitochondrial proteins
Major protein PTMs including phosphorylation, acetylation, S-nitrosylation (SNO), succinylation, and O-GlcNAcylation are reported to take place in the mitochondrial proteome (Deng et al., 2011; Hart et al., 2011; Wu et al., 2011; Zhang et al., 2011). Together, these PTMs orchestrate the functions of mitochondrial proteins by modulating their subcellular localizations, protein–protein interactions, and stability. For example, phosphorylation has been shown to regulate essential metabolic flux into the TCA cycle (Boja et al., 2009) and the assembly of the ETC complexes (Kane et al., 2010; Phillips et al., 2012).
Understanding the molecular and physiological roles of PTMs in mitochondria is a pivotal step toward advancing our knowledge in mitochondrial biology. As in proteome profiling, the literature reflects the indispensability of MS technology in PTM analyses. Numerous mitochondrial PTMs have now been unequivocally discerned by MS-based proteomics experiments (Boja et al., 2009). Tables 1 and 2 list representative human and mouse cardiac mitochondrial proteins from the TCA cycle and ETC complexes, and their respective phosphorylation and acetylation sites identified through large-scale MS screening, as indicated in UniProt (The UniProt Consortium, 2012). The functions and regulations of these PTMs are currently under intense investigations.
Table 1.
Known phosphorylation sites on UniProt among ETC and TCA proteins in the mouse and human heart
| Protein name | Species | UniProt accession | Phosphorylation site(s) |
| ATP synthase subunit e | Mouse | Q06185 | Y32 |
| ATP synthase subunit γ | Mouse | Q91VR2 | S146 |
| ATP synthase subunit α | Mouse | Q03265 | S76 |
| ATP synthase subunit β | Mouse | P56480 | S529 |
| Cytochrome b-c1 complex subunit 1 | Human | P31930 | T381 |
| Cytochrome b-c1 complex subunit 2 | Human | P22695 | T86 |
| Cytochrome b-c1 complex subunit 6 | Human | P07919 | S61 |
| Cytochrome b-c1 complex subunit 6 | Mouse | P99028 | T61 |
| Fumarate hydratase | Human | P07954 | Y491 |
| Isocitrate dehydrogenase subunit γ | Mouse | P70404 | T363 |
| Malate dehydrogenase | Human | P40926 | Y56 |
| NADH-ubiquinone oxidoreductase 18-kD subunit | Human | O43181 | T32, S34 |
| NADH-ubiquinone oxidoreductase B22 subunit | Human | Q9Y6M9 | S85 |
| NADH-ubiquinone oxidoreductase PDSW subunit | Human | O96000 | Y56, Y143 |
| NADH-ubiquinone oxidoreductase subunit 2 | Human | P03891 | T258 |
| NADH-ubiquinone oxidoreductase subunit B14.5a | Human | O95182 | S63 |
| Succinate dehydrogenase flavoprotein subunit | Mouse | Q8K2B3 | T252 |
| Succinyl-CoA synthetase β-A chain | Mouse | Q9Z2I9 | S279 |
| Succinyl-CoA synthetase β-A chain | Human | Q9P2R7 | Y84 |
Proteins from ETC complexes and the TCA cycle were experimentally determined from mouse and human heart mitochondria. The PTM information of the protein was retrieved from UniProt. The referenced phosphorylation sites were identified from their respective species but may have originated from different tissues other than the heart (e.g. mouse liver, human HeLa cells). Tissue-specific PTM distribution is a matter of intense investigation.
Table 2.
Known acetylation sites on UniProt among ETC and TCA proteins in the mouse and human heart
| Protein name | Species | UniProt accession | Acetylation site(s) |
| Aconitase | Human | Q99798 | K50, K573, K605 |
| Mouse | Q99KI0 | K50 | |
| Acyl carrier protein | Human | O14561 | K92 |
| ATP synthase protein 8 | Mouse | P03930 | K54 |
| ATP synthase subunit b | Human | P24539 | K221, K233 |
| Mouse | Q9CQQ7 | K115, K131, K162, K188, K221, K225, K233 | |
| ATP synthase subunit d | Human | O75947 | K85, K95, K117, K149 |
| Mouse | Q9DCX2 | K63, K78, K85, K99, K117, K149 | |
| ATP synthase subunit f | Human | P56134 | A2 |
| ATP synthase subunit g | Human | O75964 | A2, K24 |
| Mouse | Q9CPQ8 | K54, K66 | |
| ATP synthase subunit γ | Human | P36542 | K55, K154, K197 |
| Mouse | Q91VR2 | K79, K90, K115 | |
| ATP synthase subunit O | Human | P48047 | K54, K60, K70, K162, K172, K192 |
| Mouse | Q9DB20 | K60, K70, K158, K162, K172, K176, K192 | |
| ATP synthase subunit α | Human | P25705 | K161, K261, K305, K434, K498, K506, K539 |
| Mouse | Q03265 | K132, K230, K239, K261, K305, K427, K498, K531, K539 | |
| ATP synthase subunit β | Human | P06576 | K133, K198, K426, K485 |
| Mouse | P56480 | K133, K259, K522 | |
| ATP synthase-coupling factor 6 | Human | P18859 | K41, K46, K99, K105 |
| Mouse | P97450 | K84, K99 | |
| Citrate synthase | Human | O75390 | K327, K366, K375, K382, K393 |
| Cytochrome b-c1 complex subunit 1 | Human | P31930 | K111 |
| Cytochrome b-c1 complex subunit 2 | Mouse | Q9DB77 | K159, K250 |
| Cytochrome b-c1 complex subunit 6 | Human | P07919 | K85 |
| Mouse | P99028 | K40 | |
| Cytochrome b-c1 complex subunit 7 | Human | P14927 | K78 |
| Cytochrome c oxidase polypeptide VIc | Mouse | Q9CPQ1 | K61 |
| Cytochrome c oxidase subunit 4 isoform 1 | Human | P13073 | K53, K60 |
| Cytochrome c oxidase subunit 5B | Human | P10606 | K121 |
| Cytochrome c oxidase subunit 7A-related | Human | O14548 | K69 |
| Dihydrolipoyl dehydrogenase | Human | P09622 | K143, K267, K320, K410, K417, K420 |
| Mouse | O08749 | K127 | |
| Fumarate hydratase | Human | P07954 | K66, K80, K94, K256, K292 |
| Mouse | P97807 | K63, K112, K474 | |
| Isocitrate dehydrogenase subunit β | Human | O43837 | K146, K199, K374 |
| Isocitric dehydrogenase subunit α | Human | P50213 | K343 |
| Malate dehydrogenase | Human | P40926 | K165, K185, K301, K307, K314, K329, K335 |
| Mouse | P08249 | K157, K239, K314 | |
| NADH-ubiquinone oxidoreductase 13-kD A subunit | Mouse | P52503 | K112 |
| NADH-ubiquinone oxidoreductase 13-kD B subunit | Mouse | Q9CPP6 | K36, K46, K98 |
| NADH-ubiquinone oxidoreductase 39-kD subunit | Mouse | Q9DC69 | K370 |
| NADH-ubiquinone oxidoreductase 42-kD subunit | Mouse | Q99LC3 | K122, K181, K242, K350 |
| NADH-ubiquinone oxidoreductase 51-kD subunit | Mouse | Q91YT0 | K81, K104, K375 |
| NADH-ubiquinone oxidoreductase 75-kD subunit | Mouse | Q91VD9 | K84 |
| NADH-ubiquinone oxidoreductase B12 subunit | Mouse | Q9CQZ6 | K29, K40 |
| NADH-ubiquinone oxidoreductase B22 subunit | Human | Q9Y6M9 | A2 |
| NADH-ubiquinone oxidoreductase B8 subunit | Human | O43678 | A2 |
| Mouse | Q9CQ75 | K64, K75 | |
| NADH-ubiquinone oxidoreductase MLRQ subunit | Mouse | Q62425 | K56 |
| NADH-ubiquinone oxidoreductase subunit B14.5b | Human | O95298 | K114 |
| NADH-ubiquinone oxidoreductase subunit B17.2 | Mouse | Q7TMF3 | K43, K47 |
| Succinate dehydrogenase flavoprotein subunit | Human | P31040 | K179, K335, K541, K547, K608 |
| Mouse | Q8K2B3 | K179, K423, K485, K498, K538, K547 | |
| Succinyl-CoA synthetase β A chain | Human | Q9P2R7 | K78, K143 |
| Succinyl-CoA synthetase β G chain | Human | Q96I99 | K73, K227, K291, K338 |
| Succinyl-CoA synthetase subunit α | Human | P53597 | K54 |
Proteins from ETC complexes and the TCA cycle were experimentally determined from mouse and human heart mitochondria. The PTM information of the protein was retrieved from UniProt. The referenced acetylation sites were identified from their respective species but may have originated from different tissues other than the heart (e.g. mouse liver, human HeLa cells). Tissue-specific PTM distribution is a matter of intense investigation.
MS technologies used in PTM studies often parallel those of proteome profiling. MS ionization techniques such as electrospray ionization and matrix-assisted laser desorption/ionization (Kane et al., 2010) have been extensively used to study mitochondrial PTMs, with electrospray ionization commonly coupled to upstream LC separation and matrix-assisted laser desorption/ionization coupled with gel-based separation. For tandem MS (MS/MS) experiments, various fragmentation mechanisms have been used; because of the benefit afforded by electron transfer dissociation in preserving peptide backbone information for PTM analyses, it has been applied complementarily with conventional collision-induced dissociation to maximize the revelation of phosphorylation sites (Deng et al., 2011).
Characterization of PTM functions and their regulations in disease additionally requires the degree of modifications to be measured and compared across samples. In this regard, traditional 2D-DIGE (Hopper et al., 2006) in conjunction with spot identification by MS has been applied to quantify mitochondrial PTMs; iTRAQ has been performed with high-resolution Orbitrap mass spectrometers through the fragmentation mechanism known as high-energy collision dissociation (Boja et al., 2009).
An emerging MS quantification technology for mitochondrial PTM peptides is targeted multiple reaction monitoring (MRM) using a triple-quadrupole mass spectrometer. An MRM assay can specifically identify peptides through their signature transition ions and LC retention times and is tremendously valuable in demarcating modification sites in proximity. Although present transition prediction algorithms for PTM peptides remain immature and thus the attainment of assay transitions for a specific PTM demands lapidary experimentation and validation, the transitions of PTM sites become a valuable resource for all researchers once deduced. When incorporated with stable isotope internal standards, MRM assays are capable of sensitive and site-specific determination of the absolute quantity of many distinct modification sites in one experiment (Lam et al., 2012).
Finally, the cardinal challenge in the identification and quantification of PTMs pertains to the transiency and low stoichiometry of protein modifications. The use of PTM-specific enrichments before MS acquisition is thus virtually obligatory for any analysis to reduce sample complexity and enhance the sensitivity of detection. Several methods have been exploited to selectively enrich phosphopeptides from the proteome, such as strong cation exchange (Boja et al., 2009), titanium dioxide (Deng et al., 2011; Lam et al., 2012), and immobilized metal ion affinity chromatography (Lee et al., 2007). Immunoprecipitation-based extraction has been harnessed to enrich a wide range of modifications, including acetylated proteins through anti-acetyllysine antibodies (Kim et al., 2006; Choudhary et al., 2009), whereas biotin-switch assay with avidin affinity column has enabled S-nitrosylated peptides to be distinguished and isolated (Sun et al., 2007). Lastly, O-GlcNAcylated peptides can be enriched by enzymatic chemical tagging (Wang et al., 2010) or antibodies. The ensuing yield, rich in peptides containing the modifications of interest, could be identified and quantified by the aforementioned techniques.
Mitochondrial proteomes in pathogenesis and pathophysiology
As the central hubs of energy production and other important signaling pathways, mitochondria are predisposed to be entangled in numerous human pathological phenotypes. Dysfunction of mitochondrial proteins caused by either environmental changes or genetic mutations has been shown to be directly associated with various diseases including cardiomyopathies, diabetes, and neurodegenerative disorders.
Mitochondrial proteomic analyses in heart failure and cardioprotection.
Mitochondria comprise ∼35–40% of the volume of cardiomyocytes (Neubauer, 2007); it is not surprising that many mitochondrial proteomic studies focus on heart tissues to gain insight into the alterations of the mitochondrial proteome under different cardiac phenotypes including heart failure (Gucek and Murphy, 2010; Hollander et al., 2011). As Hollander et al. (2011) reported, a commonality among patients with end-stage heart failure revealed a down-regulation of four mitochondrial enzymes involved in the metabolic production of energy including VDAC1, PDH E3-binding protein, ATP synthase D chain, and malate dehydrogenase; these changes were detected by using 2-DE accompanied by protein identification with LC-MS/MS. However, proteins of the ETC were dichotomous with the majority of the proteins appearing in lower abundances and greater abundances present in only 10 ETC proteins in complexes I, II, IV, and V. Intermittent hypoxia (IH) training improves cardioprotection by developing an adjustment to reduced oxygen. After IH training in rats, mitochondrial aconitase, malate dehydrogenase, and the electron transfer flavoprotein α subunit used in lipid metabolism were found to be up-regulated (Rosca and Hoppel, 2010). In contrast, two proteins, aspartate aminotransferase and the ubiquinol cytochrome c reductase iron sulfur subunit, were down-regulated upon exposure to IH (Rosca and Hoppel, 2010). Before the introduction of myocardial ischemia, IH treatment promoted more prominent ATP reserves (Lesnefsky et al., 2001; Rosca and Hoppel, 2010). Although myocardial ischemia typically impairs cardiac and metabolic function, a comparison of myocardial ischemic–induced normoxic rats and IH-treated rats resulted in significantly higher ATP levels in IH-treated rats both after 30 min of ischemia as well as after 30 min of reperfusion; the IH-treated rats not only showed a greater tolerance for ischemic insult but also exhibited significantly enhanced ATP reserves both before and after myocardial ischemia/reperfusion (Lin et al., 2008; Milano et al., 2011; Zhu et al., 2012). Generation of ROS has been shown to be detrimental to mitochondria; however, peroxiredoxin 5 can inhibit the production of ROS and remove hydrogen peroxide from the system. Previous studies have revealed that IH treatment elevated levels of antioxidant enzymes such as thioredoxin and antioxidant glutathione (Lin et al., 2008). Increased expression of peroxiredoxin 5, a thioredoxin peroxidase, has been reported to be an adjunct to IH training (Gucek and Murphy, 2010; Zhu et al., 2012). In addition, heat shock protein 60 has been found to alter the ratio of Bcl-2/Bax in favor of preventing ischemia–reperfusion apoptosis, protecting the cell from rupturing (Gucek and Murphy, 2010; Zhu et al., 2012).
Mounting evidence has shown that alterations of mitochondrial PTMs also contribute to cardioprotection (Chen et al., 2008; Das et al., 2008; Lin et al., 2009; Rasola et al., 2010). PKCε has been reported to translocate to the mitochondria and phosphorylate proteins such as aldehyde dehydrogenase 2 and α-ketoglutarate dehydrogenase. This molecular modification leads to a decrease in ROS production, resulting in cardioprotection (Chen et al., 2008; Lagranha et al., 2010). Inhibition of glycogen synthase kinase moderates the transportation of ATP, whereas activation of glycogen synthase kinase phosphorylates cyclophillin D, thereby triggering the activation of the mitochondrial permeability transition pore. In addition, nitric oxide indirectly acts as an inhibitory mechanism in the generation of ROS (Sun et al., 2007; Lin et al., 2009); it promotes increases in SNO of complex I, consequently reducing ROS production. Furthermore, SNO causes ATP synthase to reverse its function, prompting a depletion of ATP. SNO has also been ascertained to inhibit irreversible oxidation of proteins. Modifications of mitochondrial proteins and PTMs can lead to alterations in cardiac function. A comprehensive understanding of the mitochondrial proteome will continue to support the advancement of our knowledge in cardiac physiology and cardiac dysfunction.
Mitochondrial proteomic analyses in metabolic disorder–related diseases.
The development of diabetes mellitus typically accompanies the dysfunction of insulin production and absorption by the body (Baseler et al., 2011). Using 2-DE followed by LC-MS/MS analyses, recent studies from Taurino et al. (2012) observed the decreased expression of the Ndufs3 protein subunit of complex I in steptozotocin-induced type 1 diabetic rats. In combination with genetic (a decreased mRNA level) as well as biochemical (impaired catalytic activity of complex I) evidence, this group of investigators concluded that Ndufs3 is a critical contributor to the onset of diabetic encephalopathy in type 1 diabetes (Taurino et al., 2012). Through the use of iTRAQ and 2D-DIGE, subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) were analyzed to determine whether type 1 diabetes influenced the proteomic makeup of these two mitochondrial subpopulations in the heart (Baseler et al., 2011). The proteomic makeup of IFM was affected to a greater extent than SSM, as exemplified by a decrease in fatty acid oxidation and OXPHOS proteins. Compared with diabetic SSM, the expression levels of adenine nucleotide translocator, mitochondrial phosphate carrier, mitofilin, inner membrane translocases, and mitochondrial heat shock protein 70 were decreased in diabetic IFM. The levels of mitochondrial protein import were unchanged in diabetic SSM, whereas the levels of mitochondrial protein import were substantially decreased in diabetic IFM (Baseler et al., 2011; Davidson, 2011). PTMs, specifically protein oxidations and deamidations, were more prevalent within IFM. Therefore, proteomic alterations were linked to the dysfunction of mitochondrial protein import such as mitochondrial heat shock protein 70. This evidence underscores the role of the mitochondrial proteomes underlying pathophysiology of diabetes.
Many mitochondrial disease–related phenotypes are associated with excessive ROS, including aging. The body typically counteracts the effects of low amounts of ROS with antioxidant defenses (e.g., MnSOD); as it ages, the antioxidative ability decreases, resulting in enhanced oxidant production. Mitochondrial ETC activity diminishes with aging in humans and other primates; this decline is associated with an increase of mtDNA mutations. An accumulation of mtDNA mutations aggravates the defects in OXPHOS machinery, eventually resulting in energetic failures. Mitochondrial calcium levels also contribute to the aging process as mitochondria become more sensitive to intracellular calcium overload. During aging, the expression and activity of calcium regulating proteins are reduced, leading to a disturbed calcium extrusion (Lesnefsky and Hoppel, 2006). This intracellular calcium overload directly damages the activation of proteases, nucleases, and calcium-dependent phospholipases, as well as indirectly hindering the OXPHOS machinery, diminishing the supply of ATP and triggering additional ROS release. As part of the aging process, additional roles of mitochondria remain to be elucidated.
Respiratory chain diseases (RCDs) occupy a large subcategory of mitochondrial disorders and are considered among the most common genetic metabolic disorders. Approximately 20% of RCDs are caused by mtDNA mutations, with the remaining RCDs resulting from nuclear anomalies (DiMauro and Schon, 2003; Calvo and Mootha, 2010; Wallace, 2010). Defects of proteins within the five complexes of the ETC have been implicated in certain diseases. All proteins encoded by mtDNA (referred to as MT) are linked to diseases such as Leber’s hereditary optic neuropathy, including MT-ND1, MT-ND2, MT-ND4, MT-ND4L, MT-ND6, and MT-CYB. In parallel, cardiomyopathies are affiliated with mtDNA mutations in MT-ND2, MT-CYB, and MT-CO1. Additional examples include myoclonic seizures affiliated with mutations in MT-ND3, Leigh’s syndrome (LS) (MT-ND5 and MT-CO3), mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes or MELAS (MT-ND5, MT-ND6, MT-CYB, and MT-CO2), Parkisonism (MT-CYB), and myoglobinuria (MT-CO1 and MT-CO3). In addition, six particular nuclear-encoded subunits of complex I (NDUFS1, NDUFS2, NDUFS4, NDUFS7, NDUFS8, and NDUFV1) have been associated with LS and leukodystrophy. Each individual subunit in complex II is involved in the synthesis of diseases including LS, paraganglioma, and pheochromocytoma. The complex III mitochondrial chaperone BCS1 is also linked to GRACILE syndrome, in addition to LS. The occurrence of these insults can arise during disruptions of mutations or deletions in oogenesis, early embryogenesis, or transcription/translation. In addition, the differential expression of the OXPHOS subproteomes within various tissues can entail a tissue-specific phenotype of particular RCDs because of the different metabolic needs of diverse tissue types (Johnson et al., 2007b; Balaban, 2010). Complete characterization of the OXPHOS subproteome, its biochemical properties, and its biological functions will further our understanding of RCDs in terms of providing prospective diagnostic biomarkers and therapeutic targets in the detection of RCDs.
Conclusion and future perspectives
Contextualization of mitochondrial proteins to their biological pathways and their corresponding linkages to clinical phenotypes affords great opportunities to advance our understanding regarding the fundamentals of mitochondrial diseases. Proteomics investigations in the past 15 years have paved a foundation for future studies to establish a comprehensive mitochondrial proteome map. Several challenges remain. At the technology front, these include the identification of proteins in low abundance, proteins that are associated with mitochondria, or proteins with unique biochemical features. Future tasks detailing comprehensive and quantitative characterization of the mitochondrial protein PTMs are also daunting. In the biology arena, functional information regarding individual components within many mitochondrial subproteomes is far from completion. Characterization of these proteins may lead to the discovery of novel mitochondrial functions, of which we are not yet aware. Integrating MS strategies with other approaches such as computational biology, protein arrays, and biochemical analyses will facilitate the advancement and completion of a mitochondrial proteome knowledgebase. The ultimate goal of mitochondrial proteome research is to bridge the knowledge gap between mitochondrial compositions and their functionalities, therefore providing potential diagnostic and prognostic targets for mitochondrial-associated diseases.
This Perspectives series includes articles by Sheu et al., Balaban, Santo-Domingo and Demaurex, Wei and Dirksen, O-Uchi et al., Nowikovsky et al., and Galloway and Yoon.
Acknowledgments
We apologize to those whose relevant work was not cited because of a space limitation of the references. We thank members of our laboratory for helpful discussions.
The authors are supported by a National Institutes of Health (NIH) Heart, Lung and Blood Institute Proteomics Center Award (HHSN268201000035C to P. Ping) as well as by an NIH Multiple Principle Investigators R01 Award (HL01228 to P. Ping, J. Weiss, and H. Cai).
Shey-Shing Sheu served as guest editor.
Footnotes
Abbreviations used in this paper:
- 2-DE
- two-dimensional PAGE
- 2D-DIGE
- two-dimensional difference in-gel electrophoresis
- ETC
- electron transport chain
- IFM
- interfibrillar mitochondria
- IH
- intermittent hypoxia
- IMM
- inner mitochondrial membrane
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LC
- liquid chromatography
- LS
- Leigh’s syndrome
- MRM
- multiple reaction monitoring
- MS
- mass spectrometry
- mtDNA
- mitochondrial DNA
- OMM
- outer mitochondrial membrane
- OXPHOS
- oxidative phosphorylation
- PTM
- posttranslational modification
- RCD
- respiratory chain disease
- ROS
- reactive oxygen species
- SNO
- S-nitrosylation
- SSM
- subsarcolemmal mitochondria
- TCA
- tricarboxylic acid
References
- Alonso J., Rodriguez J.M., Baena-López L.A., Santarén J.F. 2005. Characterization of the Drosophila melanogaster mitochondrial proteome. J. Proteome Res. 4:1636–1645 10.1021/pr050130c [DOI] [PubMed] [Google Scholar]
- Balaban R.S. 2010. The mitochondrial proteome: a dynamic functional program in tissues and disease states. Environ. Mol. Mutagen. 51:352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler W.A., Dabkowski E.R., Williamson C.L., Croston T.L., Thapa D., Powell M.J., Razunguzwa T.T., Hollander J.M. 2011. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300:R186–R200 10.1152/ajpregu.00423.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boja E.S., Phillips D., French S.A., Harris R.A., Balaban R.S. 2009. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J. Proteome Res. 8:4665–4675 10.1021/pr900387b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau A.L., Bayot A. 2011. Mitochondrial proteases and cancer. Biochim. Biophys. Acta. 1807:595–601 10.1016/j.bbabio.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Calvo S.E., Mootha V.K. 2010. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11:25–44 10.1146/annurev-genom-082509-141720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Budas G.R., Churchill E.N., Disatnik M.H., Hurley T.D., Mochly-Rosen D. 2008. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 321:1493–1495 10.1126/science.1158554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325:834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Cotter D., Guda P., Fahy E., Subramaniam S. 2004. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 32(Database issue):D463–D467 10.1093/nar/gkh048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S., Martinou J.C. 2008. Purification and proteomic analysis of the mouse liver mitochondrial inner membrane. Methods Mol. Biol. 432:101–116 10.1007/978-1-59745-028-7_7 [DOI] [PubMed] [Google Scholar]
- Das S., Wong R., Rajapakse N., Murphy E., Steenbergen C. 2008. Glycogen synthase kinase 3 inhibition slows mitochondrial adenine nucleotide transport and regulates voltage-dependent anion channel phosphorylation. Circ. Res. 103:983–991 10.1161/CIRCRESAHA.108.178970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S.M. 2011. A needle in a haystack: focus on “Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart”. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300:R183–R185 10.1152/ajpregu.00751.2010 [DOI] [PubMed] [Google Scholar]
- Deng N., Zhang J., Zong C., Wang Y., Lu H., Yang P., Wang W., Young G.W., Wang Y., Korge P., et al. 2011. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol. Cell. Proteomics. 10:M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S., Schon E.A. 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348:2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- Distler A.M., Kerner J., Hoppel C.L. 2008. Proteomics of mitochondrial inner and outer membranes. Proteomics. 8:4066–4082 10.1002/pmic.200800102 [DOI] [PubMed] [Google Scholar]
- Elstner M., Andreoli C., Ahting U., Tetko I., Klopstock T., Meitinger T., Prokisch H. 2008. MitoP2: an integrative tool for the analysis of the mitochondrial proteome. Mol. Biotechnol. 40:306–315 10.1007/s12033-008-9100-5 [DOI] [PubMed] [Google Scholar]
- Ernster L., Schatz G. 1981. Mitochondria: a historical review. J. Cell Biol. 91:227s–255s 10.1083/jcb.91.3.227s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F., Foster L.J., Campanaro S., Valle G., Mann M. 2006. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics. 5:608–619 [DOI] [PubMed] [Google Scholar]
- Gaucher S.P., Taylor S.W., Fahy E., Zhang B., Warnock D.E., Ghosh S.S., Gibson B.W. 2004. Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J. Proteome Res. 3:495–505 10.1021/pr034102a [DOI] [PubMed] [Google Scholar]
- Glancy B., Balaban R.S. 2012. Role of mitochondrial ca(2+) in the regulation of cellular energetics. Biochemistry. 51:2959–2973 10.1021/bi2018909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gucek M., Murphy E. 2010. What can we learn about cardioprotection from the cardiac mitochondrial proteome? Cardiovasc. Res. 88:211–218 10.1093/cvr/cvq277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G.W., Slawson C., Ramirez-Correa G., Lagerlof O. 2011. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80:825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeboom G.H., Schneider W.C., Pallade G.E. 1948. Cytochemical studies of mammalian tissues; isolation of intact mitochondria from rat liver; some biochemical properties of mitochondria and submicroscopic particulate material. J. Biol. Chem. 172:619–635 [PubMed] [Google Scholar]
- Hollander J.M., Baseler W.A., Dabkowski E.R. 2011. Proteomic remodeling of mitochondria in heart failure. Congest. Heart Fail. 17:262–268 10.1111/j.1751-7133.2011.00254.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper R.K., Carroll S., Aponte A.M., Johnson D.T., French S., Shen R.F., Witzmann F.A., Harris R.A., Balaban R.S. 2006. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 45:2524–2536 10.1021/bi052475e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.T., Harris R.A., Blair P.V., Balaban R.S. 2007a. Functional consequences of mitochondrial proteome heterogeneity. Am. J. Physiol. Cell Physiol. 292:C698–C707 10.1152/ajpcell.00109.2006 [DOI] [PubMed] [Google Scholar]
- Johnson D.T., Harris R.A., French S., Blair P.V., You J., Bemis K.G., Wang M., Balaban R.S. 2007b. Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Physiol. Cell Physiol. 292:C689–C697 10.1152/ajpcell.00108.2006 [DOI] [PubMed] [Google Scholar]
- Kane L.A., Youngman M.J., Jensen R.E., Van Eyk J.E. 2010. Phosphorylation of the F(1)F(o) ATP synthase beta subunit: functional and structural consequences assessed in a model system. Circ. Res. 106:504–513 10.1161/CIRCRESAHA.109.214155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 23:607–618 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., et al. 2006. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 125:173–186 10.1016/j.cell.2006.01.044 [DOI] [PubMed] [Google Scholar]
- Lagranha C.J., Deschamps A., Aponte A., Steenbergen C., Murphy E. 2010. Sex differences in the phosphorylation of mitochondrial proteins result in reduced production of reactive oxygen species and cardioprotection in females. Circ. Res. 106:1681–1691 10.1161/CIRCRESAHA.109.213645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.P., Scruggs S.B., Kim T.Y., Zong C., Lau E., Wang D., Ryan C.M., Faull K.F., Ping P. 2012. An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. J. Proteomics. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E., Wang D., Zhang J., Yu H., Lam M.P., Liang X., Zong N., Kim T.Y., Ping P. 2012. Substrate- and isoform-specific proteome stability in normal and stressed cardiac mitochondria. Circ. Res. 110:1174–1178 10.1161/CIRCRESAHA.112.268359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Xu Y., Chen Y., Sprung R., Kim S.C., Xie S., Zhao Y. 2007. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics. 6:669–676 10.1074/mcp.M600218-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnefsky E.J., Hoppel C.L. 2006. Oxidative phosphorylation and aging. Ageing Res. Rev. 5:402–433 10.1016/j.arr.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Lesnefsky E.J., Moghaddas S., Tandler B., Kerner J., Hoppel C.L. 2001. Mitochondrial dysfunction in cardiac disease: ischemia—reperfusion, aging, and heart failure. J. Mol. Cell. Cardiol. 33:1065–1089 10.1006/jmcc.2001.1378 [DOI] [PubMed] [Google Scholar]
- Lin J.S., Chen Y.S., Chiang H.S., Ma M.C. 2008. Hypoxic preconditioning protects rat hearts against ischaemia-reperfusion injury: role of erythropoietin on progenitor cell mobilization. J. Physiol. 586:5757–5769 10.1113/jphysiol.2008.160887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Steenbergen C., Murphy E., Sun J. 2009. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 120:245–254 10.1161/CIRCULATIONAHA.109.868729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald T., Sheng S., Stanley B., Chen D., Ko Y., Cole R.N., Pedersen P., Van Eyk J.E. 2006. Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell. Proteomics. 5:2392–2411 10.1074/mcp.T500036-MCP200 [DOI] [PubMed] [Google Scholar]
- Milano G., Bianciardi P., Rochemont V., Vassalli G., Segesser L.K., Corno A.F., Guazzi M., Samaja M. 2011. Phosphodiesterase-5 inhibition mimics intermittent reoxygenation and improves cardioprotection in the hypoxic myocardium. PLoS ONE. 6:e27910 10.1371/journal.pone.0027910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M., et al. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 115:629–640 10.1016/S0092-8674(03)00926-7 [DOI] [PubMed] [Google Scholar]
- Neubauer S. 2007. The failing heart—an engine out of fuel. N. Engl. J. Med. 356:1140–1151 10.1056/NEJMra063052 [DOI] [PubMed] [Google Scholar]
- Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 134:112–123 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D., Covian R., Aponte A.M., Glancy B., Taylor J.F., Chess D.J., Balaban R.S. 2012. Regulation of oxidative phosphorylation complex activity: effects of tissue-specific metabolic stress within an allometric series and acute changes in workload. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302:R1034–R1048 10.1152/ajpregu.00596.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T., Kieffer S., Procaccio V., Louwagie M., Courchesne P.L., Patterson S.D., Martinez P., Garin J., Lunardi J. 1998. Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: toward a human mitochondrial proteome. Electrophoresis. 19:1006–1014 10.1002/elps.1150190616 [DOI] [PubMed] [Google Scholar]
- Rasola A., Sciacovelli M., Chiara F., Pantic B., Brusilow W.S., Bernardi P. 2010. Activation of mitochondrial ERK protects cancer cells from death through inhibition of the permeability transition. Proc. Natl. Acad. Sci. USA. 107:726–731 10.1073/pnas.0912742107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifschneider N.H., Goto S., Nakamoto H., Takahashi R., Sugawa M., Dencher N.A., Krause F. 2006. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J. Proteome Res. 5:1117–1132 10.1021/pr0504440 [DOI] [PubMed] [Google Scholar]
- Richly E., Chinnery P.F., Leister D. 2003. Evolutionary diversification of mitochondrial proteomes: implications for human disease. Trends Genet. 19:356–362 10.1016/S0168-9525(03)00137-9 [DOI] [PubMed] [Google Scholar]
- Rosca M.G., Hoppel C.L. 2010. Mitochondria in heart failure. Cardiovasc. Res. 88:40–50 10.1093/cvr/cvq240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H.E., Schönfisch B., Perschil I., Chacinska A., Guiard B., et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 100:13207–13212 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.C., Robinson A.J. 2009. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol. Cell. Proteomics. 8:1324–1337 10.1074/mcp.M800373-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Morgan M., Shen R.F., Steenbergen C., Murphy E. 2007. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ. Res. 101:1155–1163 10.1161/CIRCRESAHA.107.155879 [DOI] [PubMed] [Google Scholar]
- Taurino F., Stanca E., Siculella L., Trentadue R., Papa S., Zanotti F., Gnoni A. 2012. Mitochondrial proteome analysis reveals depression of the Ndufs3 subunit and activity of complex I in diabetic rat brain. J. Proteomics. 75:2331–2341 10.1016/j.jprot.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Taylor S.W., Fahy E., Zhang B., Glenn G.M., Warnock D.E., Wiley S., Murphy A.N., Gaucher S.P., Capaldi R.A., Gibson B.W., Ghosh S.S. 2003. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21:281–286 10.1038/nbt793 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40:D71–D75 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. 2010. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51:440–450 [DOI] [PubMed] [Google Scholar]
- Wang Z., Udeshi N.D., O’Malley M., Shabanowitz J., Hunt D.F., Hart G.W. 2010. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics. 9:153–160 10.1074/mcp.M900268-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.N., Korge P., Honda H.M., Ping P. 2003. Role of the mitochondrial permeability transition in myocardial disease. Circ. Res. 93:292–301 10.1161/01.RES.0000087542.26971.D4 [DOI] [PubMed] [Google Scholar]
- Wu R., Haas W., Dephoure N., Huttlin E.L., Zhai B., Sowa M.E., Gygi S.P. 2011. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods. 8:677–683 10.1038/nmeth.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi R.P., Sickmann A., Boehm A.M., Winkler C., Zufall N., Schönfisch B., Guiard B., Pfanner N., Meisinger C. 2006. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell. 17:1436–1450 10.1091/mbc.E05-08-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li X., Mueller M., Wang Y., Zong C., Deng N., Vondriska T.M., Liem D.A., Yang J.I., Korge P., et al. 2008. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 8:1564–1575 10.1002/pmic.200700851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. 2011. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7:58–63 10.1038/nchembio.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.Z., Wu X.F., Zhang Y., Zhou Z.N. 2012. Proteomic analysis of mitochondrial proteins in cardiomyocytes from rats subjected to intermittent hypoxia. Eur. J. Appl. Physiol. 112:1037–1046 10.1007/s00421-011-2050-9 [DOI] [PubMed] [Google Scholar]