Abstract
The purpose of this study was to detect the methylation of the RUNX3 gene promoter in non-small cell lung cancer (NSCLC) tissue and to explore the association of this methylation with clinical features of NSCLC. In 58 samples of NSCLC tissue and normal adjacent tissue, methylation of the RUNX3 gene promoter was measured by methylation-specific polymerase chain reaction. Correlation with clinicopathological characteristics was assessed. The results demonstrated that RUNX3 gene promoter methylation was present in 26/58 (44.8%) of NSCLC tissue samples and 10/58 (17.2%) of normal tissue samples, and that the difference was statistically significant between the two groups (χ2=10.311, p=0.001). Significantly, methylation of the RUNX3 gene promoter correlated with clinical stage, lymph node metastasis and the degree of differentiation (p<0.05) but not with age, gender, smoking history and pathological type (p>0.05). In conclusion, methylation of the RUNX3 gene promoter had a high relevance ratio in NSCLC tissue and correlated with clinical stage, lymph node metastasis and degree of differentiation; thus, this association may have clinical significance in NSCLC.
Keywords: non-small cell lung cancer, promoter, methylation, RUNX3
Introduction
Lung cancer is one of the most common types of malignant types of cancer worldwide, accounting for 14% of all malignant tumors. The morbidity and mortality rates of lung cancer are ranked the highest among various malignant tumors. Lung cancer may be divided into two categories by histopathology: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for 80 to 85% of all histological types of lung cancer. Although great progress has been made in the treatment of lung cancer in recent years, the five-year survival rate remains less than 15% (1). Postoperative, recurrent metastasis is a significant factor affecting prognosis. As such, a correct assessment of postoperative recurrent metastasis and appropriate standard adjuvant therapy following surgery are essential to improve the prognosis. At present, the main indicator that identifies and guides prognosis is TMN staging (1), but this approach is limited. Using TMN staging, many patients with stage I lung cancer (20–30%) fail to survive more than five years, while certain patients with more advanced stages (III or IV) live longer than expected. Therefore, the TMN staging assessment of prognosis cannot be broadly and accurately applied, and other prognostic markers are required. Currently, the majority of tumor markers used for clinical diagnosis are proteins or other biological macromolecules secreted into the extracellular portions by tumor cells; these markers usually appear in the advanced stages of cancer. However, the time of occurrence and the macromolecular level usually have no close association with the prognosis of NSCLC. Therefore, these tumor markers have limited importance or significance for early NSCLC prognosis.
In recent years, the emergence of epigenetics has defined another essential research direction. Modern tumor theory (2–5) considers that there are two types of mechanisms in tumor formation: i) genetic-level mutations or changes in the DNA nucleotide sequence, and ii) epigenetic mechanisms, in which the DNA nucleotide sequence is unchanged but the gene expression is affected at a transcriptional level through DNA chemical modification. Epigenetic change can be achieved by meiosis heredity. Epigenetic modifications include phenomena such as chromatin remodeling, DNA methylation, histone modifications and non-coding RNA regulation. Abnormalities in any of these four steps affect chromatin structure and gene expression and may lead to complex syndromes, such as multifactorial diseases and cancer. DNA methylation is one of the most common forms of DNA modification and is the most widely studied. Unlike DNA sequence changes, many epigenetic changes are reversible; this provides optimism for the treatment of disease through epigenetic modification.
The RUNX gene family has three members in mammals, RUNX1, RUNX2 and RUNX3 (6). The products of RUNX are essential for regulating growth signal transduction and gene expression and play significant roles in normal cell development and tumor formation. The RUNX3 gene was identified in 1994 by Levanon et al (7). It was originally termed acute myeloid leukemia gene 2 and was later known as core-binding factor α 3 gene (8) and polyomavirus enhancement factor- binding protein gene 2 (9). The RUNX3 gene is located at 1p36.1 of the short arm chromosome 1 in humans and chromosome 4 in mice. The gene length is approximately 67 kb and it contains two promoters, P1 and P2, and six exons. The RUNX3 protein is a crucial regulatory factor in the TGF-β signaling pathway. RUNX3 deletion results in the limited function of Smad proteins and the promotion of TGF-β signaling, which leads to tumor development (10).
Decreased RUNX3 expression or deletion are mainly due to methylation or allelic loss. For example, the high methylation of the RUNX3 CpG island is closely correlated with cancer incidence and may be a useful diagnostic biomarker (11). On the other hand, a decreased RUNX3 expression or deletion is correlated to the prognosis of breast cancer (12), bladder cancer (13), pancreatic cancer (14) and other types of cancer. Currently, the association of RUNX3 gene promoter methylation in lung cancer and its clinical features is unknown. In this study, the methylation-specific polymerase chain reaction (MSP) method was used to detect RUNX3 gene promoter methylation in NSCLC and healthy adjacent tissues. In addition, we analyzed its association with clinical and pathological features of lung cancer to reveal the potential clinical significance of RUNX3 gene promoter methylation in the diagnosis of NSCLC.
Materials and methods
Clinical data
Specimens were collected from 58 patients with NSCLC by surgical resection between January 2008 and December 2010 at The Affiliated Jangyin People’s Hospital of Southeast University Medical College, China. All specimens were pathologically diagnosed as NSCLC by biopsy, and none of the subjects received pre-operative radiotherapy, chemotherapy or other cancer treatments. The subjects comprised 36 males and 22 females, aged 38–72 years, with a median age of 57 years and a mean age of 56.6 (±8.7) years. Other characteristics included: squamous cell carcinoma (n=26), adenocarcinoma (n=32); clinical stages I and II (n=25), clinical stages III and IV (n=33); lymph node metastasis (n=34), no lymph node metastasis (n=24); highly differentiated tumor (n=35), poorly differentiated tumor (n=23); smoking history (n=42), and no smoking history (n=16). Each subject specimen contained distinct NSCLC tissue and corresponding normal adjacent tissue. The specimens were frozen and stored in liquid nitrogen immediately after sampling.
Experimental methods
Reagents and primers
DNA extraction kits were purchased from Beijing Tiangen Biochemistry Technology Limited Company (Beijing, China), and the methylation kits were purchased from Zymo Research Corporation (Irvine, California, USA). Primers were synthetized by Takara Biotech Co. (Dalian, China).
RUNX3 gene promoter methylation analysis
Genomic DNA was extracted from fresh tissue according to kit instructions. Genomic DNA was modified with sulfite. The methylation modification was performed for the genomic DNA according to kit instructions, and the modified genomic DNA was stored at −20°C until further use. DNA was purified and recovered for MSP analysis. The primers for RUNX3 gene promoter methylation and demethylation are available in the literature (15). The methylated primer sequence was: forward: 5′-ATAATAGCGGTCGTTAGGGCGTCG-3′; and reverse: 5′-GCTTCTACTTTCCCACTTCTCACA-3′. The non-methylated primer sequence was: forward: 5′-ATAATAGTTGTT GTTAGGGTGTTG-3′; and reverse: 5′-ACTTCTACTTTC CCACTTCTCACA-3′. The PCR reaction system was 25 μl, including 10X PCR buffer, 1.5 mmol/l MgCl2, 10 mmol/l dNTP, 0.5 mmol/l forward and reverse primers and 1 unit Taq DNA polymerase. PCR conditions were as follows: denaturation at 94°C for 3 min, then at 94°C for 30 sec, at 55°C (methylated) or 56°C (non-methylated) for 90 sec, and at 72°C for 60 sec, 35 cycles in total, and finally at 72°C for 10 min. PCR products were analyzed by agarose electrophoresis (1.0%) and visualized with ImageMaster® VDS.
Statistical methods
SPSS 13.0 software was used to process the data. The four-cell table χ2 test was used to compare the methylation differences between the RUNX3 gene promoter groups. The above hypothesis test was two-sided with a test level (α) of 0.05; p<0.05 was considered to be statistically significant.
Results
Test results of RUNX3 gene promoter methylation
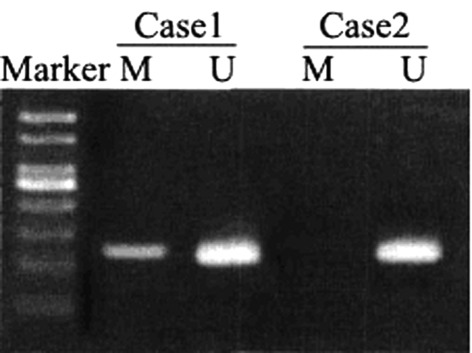
Among the 58 cases of NSCLC, 26 had RUNX3 gene promoter methylation, giving a relevance ratio of 44.8%; whereas 10 samples from the normal adjacent tissues had RUNX3 gene promoter methylation, giving a relevance ratio of 17.2%. The methylation rate of RUNX3 gene promoter in the NSCLC tissue was significantly higher than that in the normal adjacent tissue (χ2=10.311, p=0.001). The PCR electrophoresis results are shown in Fig. 1; Case 1 is methylated RUNX3 and Case 2 is non-methylated RUNX3.
Figure 1.
Electrophoretogram on the methylation of RUNX3 gene promoter. Lane M, methylated; Lane U, unmethylated.
The correlation between RUNX3 gene promoter methylation and clinicopathological characteristics
Table I shows that RUNX3 gene promoter methylation was correlated to the clinical stage, lymph node metastasis and degree of differentiation. Patients in clinical stages III and IV had a significantly higher methylation relevance ratio (57.6%) than patients in clinical stages I and II (28.0%, p=0.025). Patients with lymph node metastasis had a significantly higher methylation relevance ratio (55.9%) than those without lymph node metastasis (29.2%, p=0.044). Patients with poorly differentiated tumors also had a significantly higher methylation relevance ratio (69.6%) than those with moderately- or highly-differentiated tumors (28.6%, p=0.002). However, the RUNX3 gene promoter methylation relevance ratio was not significantly different by gender, age, pathological type and smoking history (p>0.05).
Table I.
Correlation between methylation of RUNX3 gene promoter and clinicopathological characteristics [n (%)].
| Clinical data | N | Methylation | Non-methylation | χ2 | P-value |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 36 | 14 (38.9) | 22 (61.1) | ||
| Female | 22 | 12 (54.5) | 10 (45.5) | 1.353 | 0.245 |
| Age | |||||
| <60 years | 34 | 15 (41.1) | 18 (55.9) | ||
| ≥60 years | 24 | 11 (45.8) | 14 (54.2) | 0.017 | 0.897 |
| Pathological type | |||||
| Squamous cell carcinoma | 26 | 13 (50.0) | 13 (50.0) | ||
| Adenocarcinoma | 32 | 13 (40.6) | 19 (59.4) | 0.510 | 0.475 |
| Clinical staging | |||||
| Stage I+II | 25 | 7 (28.0) | 18 (72.0) | ||
| Stage III+IV | 33 | 19 (57.6) | 14 (42.4) | 5.031 | 0.025 |
| Lymph node metastasis | |||||
| No | 34 | 7 (29.2) | 17 (70.8) | ||
| Yes | 24 | 19 (55.9) | 15 (44.1) | 4.060 | 0.044 |
| Differentiation degree | |||||
| Poorly differentiated | 23 | 16 (69.6) | 7 (30.4) | ||
| Moderately and highly differentiated | 35 | 10 (28.6) | 25 (71.4) | 9.431 | 0.002 |
| Smoking history | |||||
| No | 16 | 10 (62.5) | 6 (37.5) | ||
| Yes | 42 | 16 (38.1) | 26 (61.9) | 2.790 | 0.095 |
Discussion
The RUNX3 gene is a recently discovered tumor suppressor gene that regulates cell growth and apoptosis and plays a significant regulatory role in transcription and cell signal transduction. RUNX3 functions impact both normal physiological development and tumor development, and a reduced function of RUNX3 plays a role in the development of various human malignancies (16). Numerous studies have shown that a decreased RUNX3 expression, RUNX3 deletion, or high methylation of the RUNX3 CpG island are common in gastric, liver, lung, breast, pancreatic and colorectal cancer. It has also been confirmed that RUNX3 methylation is tumor-specific. Tan et al (17) examined the presence of methylated RUNX3, p16, RASSF1A and CDH1 in serum from 70 cases of cancer (metastatic breast cancer, NSCLC, gastric cancer, pancreatic cancer, colorectal cancer and hepatocellular carcinoma). Serum specimens from 10 healthy volunteers were used as controls. RUNX3 methylation was present in 44 patients. Of the four genes screened, at least one methylated gene was present in 62 patients, while a 3-gene combined detection (without RUNX3) was present in 50 cases. No methylation was detected in serum specimens from the 10 healthy volunteers. Therefore, as serum test markers, methylation of RUNX3 is more sensitive, and the combined detection approach of multiple genes (including the RUNX3 gene) can improve cancer diagnosis. The results from the current study demonstrated that the methylation rate of the RUNX3 gene promoter in NSCLC tissues was significantly higher than that in the normal adjacent lung tissues (44.8 versus 17.2%), indicating that RUNX3 promoter methylation was common in NSCLC.
The study performed by Sato et al (18) indicated that the RUNX3 methylation rate in NSCLC patients without a smoking history was significantly higher than that in patients with a smoking history, and that the RUNX3 methylation rate in adenocarcinoma patients was higher than that in squamous cell carcinoma patients. For adenocarcinoma, abnormal RUNX3 methylation was the essential mechanism for the inactivation of RUNX3 expression, and gene silencing was essential for the occurrence of lung cancer. Yanagawa et al (19)demonstrated that RUNX3 methylation in NSCLC tissues was not significantly different between groups by age (<69 versus >70 years), gender, smoking history or presence of pleural invasion (19,20). RUNX3 methylation was higher in patients in stage I as compared to stages II to III, but the difference was not statistically significant. The RUNX3 methylation rate in adenocarcinoma was significantly higher than that in squamous cell carcinoma. We speculated that RUNX3 methylation has a greater impact on NSCLC progression during earlier stages by exhibiting distant metastasis beyond stage H/III. Li et al (21) demonstrated that a decreased RUNX3 mRNA expression correlated with abnormal methylation status. The detected methylation rate of the RUNX3 gene promoter was 28% in adenocarcinoma, 12% in squamous cell carcinoma and 58% in large cell carcinoma. No methylation was detected in normal lung tissues, and RUNX3 methylation increased with increasing clinical stage.
The results of the current study have shown that RUNX3 gene promoter methylation was correlated to clinical stage, lymph node metastasis and degree of differentiation. Patients in clinical stages III and IV had a significantly higher methylation relevance ratio (57.6%) than patients in stages I and II (28.0%), and patients with lymph node metastasis had a significantly higher methylation relevance ratio (62.5%) than those without lymph node metastasis (32.4%). Patients with poorly-differentiated tumors also had a significantly higher methylation relevance ratio (65.2%) than those who with moderately- or highly-differentiated tumors (31.4%). However, the RUNX3 methylation relevance ratio was not significantly different by gender, age, pathological type and smoking history. Therefore, this study demonstrated that RUNX3 methylation had no significant correlation with pathological type or smoking history. The results may have been affected by study factors such as sample size and geographic and population differences. Further research on these associations is required.
In conclusion, RUNX3 methylation plays a significant role in the occurrence, development and postoperative recurrent metastasis of NSCLC. RUNX3 methylation may be an essential postoperative prognostic biomarker for NSCLC. Since changes in methylation epigenetic inheritance are reversible, clinical treatment through demethylation (22,23) may be useful to delay recurrent metastasis and improve prognosis. This approach brings new direction and hope for cancer treatment through gene-targeted therapy.
References
- 1.Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH. Cohypermethylation of p16 and FHIT promoters as a prognostic factor of recurrence in surgically resected stage I non-small cell lung cancer. Cancer Res. 2006;66:4049–4054. doi: 10.1158/0008-5472.CAN-05-3813. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carraway H, Herman J. Montoring methylation changes in caner. Methods Mol Biol. 2007;383:187–202. doi: 10.1007/978-1-59745-335-6_12. [DOI] [PubMed] [Google Scholar]
- 5.Chim CS, Liang R, Leung MH, Kwong YL. Aberrant gene methylation implicated in the progression of monoclonal gammopathy of undetermined significance to multiple myeloma. J Clin Pathol. 2007;60:104–106. doi: 10.1136/jcp.2006.036715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- 7.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 8.Ito Y. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 1999;4:685–696. doi: 10.1046/j.1365-2443.1999.00298.x. [DOI] [PubMed] [Google Scholar]
- 9.Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene. 2004;23:4336–4340. doi: 10.1038/sj.onc.1207286. [DOI] [PubMed] [Google Scholar]
- 10.Miyazono K, Suzuki H, Imamura T. Regulation of TGF-beta signaling and its roles in progression of tumors. Cancer Sci. 2003;94:230–234. doi: 10.1111/j.1349-7006.2003.tb01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244–251. doi: 10.1159/000132385. [DOI] [PubMed] [Google Scholar]
- 13.Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC, Kim WJ. Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J Urol. 2008;180:1141–1145. doi: 10.1016/j.juro.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Nomoto S, Kinoshita T, Mori T, Kato K, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Adverse prognosis of epigenetic inactivation in RUNX3 gene at 1p36 in human pancreatic cancer. Br J Cancer. 2008;98:1690–1695. doi: 10.1038/sj.bjc.6604333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waki T, Tamura G, Sato M, Terashima M, Nishizuka S, Motoyama T. Promoter methylation status of DAP-kinase and RUNX3 genes in neoplastic and non-neoplastic gastric epithelia. Cancer Sci. 2003;94:360–364. doi: 10.1111/j.1349-7006.2003.tb01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich MJ, Rad R, Langer R, Voland P, Hoefler H, Schmid RM, Prinz C, Gerhard M. Lack of RUNX3 regulation in human gastric cancer. J Pathol. 2006;210:141–146. doi: 10.1002/path.2042. [DOI] [PubMed] [Google Scholar]
- 17.Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18:1225–1230. [PubMed] [Google Scholar]
- 18.Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, Mori M. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep. 2006;15:129–135. [PubMed] [Google Scholar]
- 19.Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;5:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li QL, Kim HR, Kim WJ, Choi JK, Lee YH, Kim HM, Li LS, Kim H, Chang J, Ito Y, Youl Lee K, Bae SC. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun. 2004;314:223–228. doi: 10.1016/j.bbrc.2003.12.079. [DOI] [PubMed] [Google Scholar]
- 22.Herranz M, Esteller M. DNA methylation and histone modifications in patients with cancer: potential prognostic and therapeutic targets. Methods Mol Biol. 2007;361:25–62. doi: 10.1385/1-59745-208-4:25. [DOI] [PubMed] [Google Scholar]
- 23.Fojtova M, Piskala A, Votruba I, Otmar M, Bartova E, Kovarik A. Efficacy of DNA hypomethylating capacities of 5-aza-2′-deoxycytidine and its alpha anomer. Pharmacol Res. 2007;55:16–22. doi: 10.1016/j.phrs.2006.09.008. [DOI] [PubMed] [Google Scholar]