Abstract
The human Val66Met single nucleotide polymorphism in the brain-derived neurotrophic factor (BDNF) gene impacts BDNF signaling at the cellular level. At the neural-systems level, it is associated with differences in prefrontal cortex (PFC) and hippocampal function during performance of cognitive and affective tasks. Because the impact of this variant on basal prefrontal and hippocampal activity is not known but may be relevant to understanding the function of this gene in health and disease, we studied 94 healthy individuals with H215O PET to assess regional cerebral blood flow (rCBF) during rest and tested for between-genotype differences. Because BDNF and gonadal steroid hormones conjointly influence neuronal growth, survival, and plasticity in hippocampus and PFC, we also tested for sex × genotype interactions. Finally, in light of the known impact of BDNF on plasticity and dendritic arborization, we complimented direct rCBF comparisons with connectivity analyses to determine how activity in hippocampal and prefrontal regions showing between-genotype group differences covaries with rCBF in other nodes throughout the brain in a genotype- or sex-dependent manner. Compared with Val homozygotes, Met carriers had higher rCBF in prefrontal (BA25 extending into BA10) and hippocampal/parahippocampal regions. Moreover, there were significant sex × genotype interactions in regions (including frontal, parahippocampal, and lateral temporal cortex) in which Val homozygotes showed higher rCBF in females than males, but Met carriers showed the opposite relationship. Functional connectivity analysis demonstrated that correlations of BA25, hippocampus, and parahippocampus with frontal and temporal networks were positive for Val homozygotes and negative for Met carriers. In addition, sex × genotype analysis of functional connectivity revealed that genotype affected directionality of the inter-regional correlations differentially in men versus women. Our data indicate that BDNF allelic variation and sex interactively affect basal prefrontal and hippocampal function.
Introduction
Brain-derived neurotrophic factor (BDNF), the most abundant neurotrophin in the human CNS, is robustly expressed in the hippocampus (Conner et al., 1997) and cortical regions, including prefrontal cortex (PFC) (Hofer et al., 1990; Huntley et al., 1992; Pezawas et al., 2004; Weickert et al., 2005), and is important in regulating synaptic plasticity as well as neuronal survival, migration, differentiation, and dendritic growth (Huang and Reichardt, 2001). In rodents, inhibition of BDNF signaling impairs spatial learning and memory (Linnarsson et al., 1997; Mizuno et al., 2000). Because of its critical involvement in brain regions and cognitive operations important for higher-order human function and because of its role in the mesolimbic dopamine circuit (Berton et al., 2006), BDNF is of interest in the pathogenesis of a number of psychiatric disorders and may have implications for their treatment.
A human functional single nucleotide polymorphism (SNP) with a valine (Val) substituted by a methionine (Met) at codon 66 (rs6265) in the 5′ pro-region of the BDNF gene on chromosome 11p13 is observed with a population frequency of 20–30% in Caucasians (Shimizu et al., 2004). This SNP affects intracellular processing and activity-dependent modulation of BDNF (Egan et al., 2003; Chen et al., 2004), which, in turn, influences long-term changes in hippocampal synapses (Lu, 2003). The BDNF Met allele is associated with poorer episodic memory (Hariri et al., 2003), abnormal cognition- and affect-related hippocampal activation, and reduced levels of hippocampal n-acetyl aspartate, an intracellular marker of neuronal integrity (Egan et al., 2003). The Val66Met polymorphism has also been associated with susceptibility to neuropsychiatric disorders, particularly schizophrenia (Neves-Pereira et al., 2005), Alzheimer's disease (Ventriglia et al., 2002), bipolar disorder (Sklar et al., 2002), anxiety (Soliman et al., 2010), and depression (Gatt et al., 2009), all of which have sexually dimorphic expression of disease onset, severity, and/or course (World Health Organization, 2008).
Sex specificity in illnesses in which BDNF may play a role could occur because of common effects of ovarian steroids and BDNF on molecular and neural systems (Miranda et al., 1993; Murphy et al., 1998). Estrogen and BDNF activate similar signal transduction pathways through estrogen receptors and trkB, respectively (Scharfman and MacLusky, 2006), and conjointly influence neural growth, survival, and plasticity in hippocampus and PFC (Henderson, 1996; Benraiss et al., 2001). Additionally, the BDNF gene contains a functional estrogen-response element, and estrogen-ligand complexes can bind this sequence, inducing BDNF expression (Sohrabji et al., 1995). Because of these BDNF and gonadal hormone interactions, we hypothesized that the Val66Met polymorphism would affect brain function and circuit-level activity differently in men and women.
To elucidate the effect of this SNP, and its interaction with sex, on basal brain function, we used the H215O positron emission tomography (PET) technique, a gold-standard method for measuring resting regional cerebral blood flow (rCBF), an indicator of local cerebral metabolism and a parameter that is as yet unstudied in this context. We assessed so-called resting rCBF as a function of BDNF genotype in men versus women, with particular focus on hippocampal/parahippocampal and prefrontal BDNF-rich brain regions. In addition, because BDNF mediates synaptic plasticity (Yang et al., 2009; Ninan et al., 2010) and dendritic arborization (Tyler and Pozzo-Miller, 2003; Ji et al., 2010), we hypothesized that functional interactions between BDNF-rich regions and other parts of the brain would vary with BDNF genotype and sex.
Materials and Methods
Participants
Ninety-four healthy, right-handed Caucasians, aged 18 to 50 years, were recruited and screened with the Structured Clinical Interview for DSM-IV Axis I Disorders and DSM-IV Axis II Disorders Research Version (First et al., 1996) to ensure that they were free of confounding past or present psychiatric illness. In addition, board-certified physicians, including radiologists, assessed each subject with physical examination, medical history review, a battery of laboratory tests, and a structural MRI scan to rule out significant medical illness, substance abuse, centrally active medications, or neurostructural abnormalities. Study procedures were approved by the National Institutes of Health Institutional Review Board and Radiation Safety Committee, and all participants provided written informed consent.
Each of the 94 participants underwent two resting PET scans and was genotyped. There were 66 Val homozygotes (mean ± SD age, 31.1 ± 8.5 years) consisting of 33 males and 33 females (aged 30.1 ± 8.1 and 30.0 ± 8.8 years, respectively), as well as 28 Met carriers (mean ± SD age, 32.1 ± 9.1 years), consisting of 14 males and 14 females (aged 31.1 ± 8.6 and 33.0 ± 9.8 years).
BDNF Val66Met genotyping
Blood was collected from all subjects, and DNA was extracted using standard methods. BDNF genotyping and allelic discrimination were determined by TaqMan 5′ exonuclease assay using a custom-designed assay (Applied Biosystems). BDNF genotype frequencies were in Hardy–Weinberg equilibrium (X2 = 0.09). To examine occult genetic stratification, a common functional polymorphism in catechol-O-methyltransferase Val158Met was genotyped, and no significant variation in allele frequency was found.
PET data acquisition
Subjects were instructed to refrain from alcohol, nicotine, and caffeine for 4 hours before the scan and were told not to ingest over-the-counter medications that could affect rCBF for the preceding 24 h. On the day of the study, participants were instructed to lie still in the scanner and rest with their eyes closed. An intravenous bolus of 10 mCi of H215O was administered before each of two 60 s scans performed 6 min apart. PET data were collected with a GE Advance three-dimensional PET scanner (septa retracted, 4.25 mm slice separation, 35 slices, axial field of view of 15.3 cm; GE Healthcare). Scans were corrected for attenuation and were reconstructed into 32 axial planes (6.5 mm full-width at half-maximum).
PET data analysis
Preprocessing.
The reconstructed PET data were processed and analyzed with SPM5 (Wellcome Department of Cognitive Neurology, University College London, London, UK; http://www.fil.ion.ucl.ac.uk/spm/). Images were coregistered, corrected for background activity, anatomically normalized to an average template, scaled proportionally to remove variations in global blood flow, and smoothed using a 10 mm Gaussian kernel. After preprocessing, the two resting rCBF scans of each subject were averaged and entered into group-level analyses.
rCBF analyses.
To test for rCBF differences between genotype groups across the entire brain, a voxelwise random effects analysis was performed. In light of our a priori, region-specific hypotheses, small volume correction (SVC) for familywise error (FWE) was additionally applied to results within the hippocampal/parahippocampal complex and PFC; anatomical regions of interest were created with the Wake Forest Pick-Atlas tool in SPM for left and right hippocampi, parahippocampal gyri, and PFC (composed of BA9, BA10, BA11, BA25, and BA46, combined). To test for male–female differences in the effect of the Val66Met polymorphism on resting rCBF, a voxelwise sex × genotype interaction analysis was performed.
Functional connectivity analyses.
To test for between-genotype differences in functional connectivity, rCBF was extracted from spheres with 5 mm radii drawn around seed voxels within regions of interest hypothesized a priori (hippocampus/parahippocampal gyrus and PFC) that best distinguished the Val homozygotes from the Met carriers as delineated by the between-genotype analysis (shown in Table 1). First, to determine a search area for the between-genotype and the sex × genotype correlational analyses, rCBF from these spheres was entered into a series of voxelwise regression analyses that were run on the entire cohort, regardless of sex or genotype. Next, the rCBF values for each of these spheres were entered into voxelwise regression analyses for each genotype group separately to determine group-specific correlational patterns within the search area. Finally, between-genotype analyses were performed on the resulting correlational maps for each seed region.
Table 1.
Regions showing increased rCBF for BDNF Met carriers compared with Val homozygotes
| Brain region | MNI coordinates |
Cluster size | t values | p values | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| L subgenual cingulate (BA25) | −4 | 28 | −12 | 176 | 4.04 | 0.00006* |
| L insula | −42 | −10 | −2 | 168 | 3.97 | 0.00007 |
| R inferior temporal gyrus | 50 | −60 | −2 | 43 | 3.71 | 0.0002 |
| L parahippocampal gyrus | −14 | 6 | −28 | 41 | 3.58 | 0.0003* |
| R middle temporal gyrus | 60 | 4 | −18 | 60 | 3.53 | 0.0003 |
| R hippocampus | 32 | −10 | −12 | 13 | 3.30 | 0.0007 |
No regions showed the opposite effect.
* FWE corrected at p < 0.05 for SVC. Cluster size reported at p < 0.001. L, Left; R, right.
To test for sex × genotype effects on functional connectivity, we used the same search areas as defined above: connectivity maps for the hippocampus/parahippocampal gyrus and PFC seed regions that best distinguished rCBF in the Val homozygotes from rCBF in the Met carriers for the entire group, regardless of genotype or sex. Within these whole-group connectivity maps, sex × genotype interaction analyses were performed on the correlational data.
Results significant at the p < 0.001 level with cluster size >10 voxels are reported. Peak voxel p values are given, with FWE correction for multiple comparisons indicated when SVC was applied.
Results
BDNF genotype and rCBF
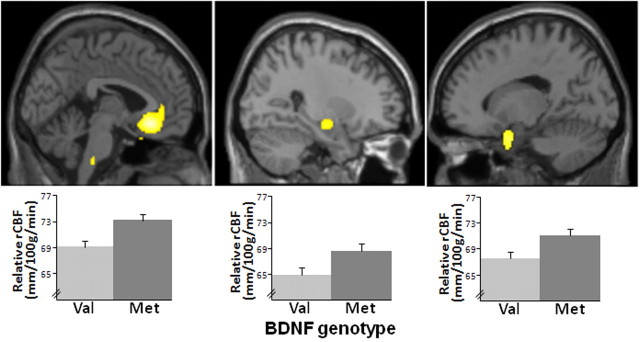
Whole-brain voxelwise analysis examining the main effect of genotype revealed no regions in which rCBF of Val homozygotes exceeded that of Met carriers. In contrast, compared with Val homozygotes, Met carriers demonstrated increased resting rCBF in regions hypothesized a priori (Fig. 1), specifically in ventromedial PFC (BA25 extending into BA10; p = 0.00006, uncorrected; p = 0.05, SVC for FWE), left parahippocampal gyrus (p = 0.0003, uncorrected; p = 0.05, SVC for FWE), and right hippocampus (p = 0.0007, uncorrected). Met carriers also showed greater rCBF in the left insula and right lateral temporal regions not hypothesized. Details of these findings are reported in Table 1.
Figure 1.
BDNF allelic variation affects rCBF. Areas of significant between-genotype group differences: Met carriers showed greater rCBF values than Val homozygotes. Sagittal views (top) and rCBF values (bottom) for BA25 (p = 0.00006, p < 0.05, SVC for FWE) (left), right hippocampus (p = 0.001, uncorrected) (middle), and left parahippocampal gyrus (p = 0.0003, p < 0.05, SVC for FWE) (right). Error bars are ±1 SEM. There were no regions in which rCBF in Val homozygotes was greater.
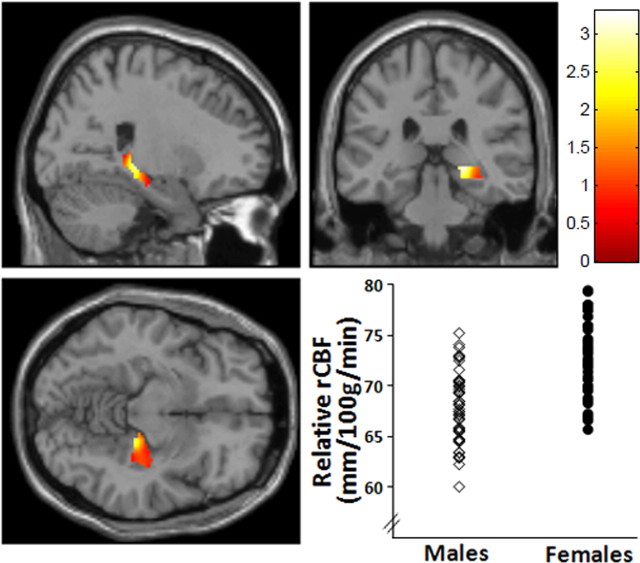
Sex and rCBF
Whole-brain voxelwise analysis examining the main effect of sex in this cohort revealed an extensive group of regions in which resting rCBF was higher in women than in men. These included regions hypothesized a priori [medial PFC (mPFC)/BA10, p ≤ 0.002, FDR corrected; right hippocampus, p = 0.0001, FDR corrected; and bilateral parahippocampal gyri, p ≤ 0.002, FDR corrected]. There were no regions in which rCBF for men were greater than women in either genotype group. Details of these findings are reported in Table 2, and representative data are shown in Figure 2.
Table 2.
Locales within regions hypothesized a priori (PFC and medial temporal cortex) in which resting rCBF was higher in women than in men
| Brain region | MNI coordinates |
Cluster size | Z values | p values | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| R hippocampus | 22 | −30 | −2 | 16822 | 5.66 | 0.000000007* |
| L parahippocampal gyrus (BA36) | −38 | 4 | −32 | 209 | 4.56 | 0.000003* |
| R parahippocampal gyrus (BA36) | 32 | 8 | −38 | 254 | 4.08 | 0.00002 |
| L superior frontal gyrus (BA10) | −16 | 52 | −4 | 453 | 4.47 | 0.000004* |
| L superior frontal gyrus (BA8) | −38 | 26 | 46 | 123 | 4.24 | 0.00001 |
| R superior frontal gyrus (BA10) | 16 | 58 | −6 | 2724 | 4.06 | 0.00002 |
| R middle frontal gyrus (BA10) | 50 | 12 | 50 | 216 | 4.33 | 0.000008 |
| R middle frontal gyrus (BA10) | 40 | 52 | 22 | 2724 | 3.98 | 0.00003 |
| L middle frontal gyrus (BA10) | −34 | 60 | 6 | 453 | 3.89 | 0.00005 |
| L middle frontal gyrus (BA6) | −36 | 4 | 58 | 74 | 3.53 | 0.0002 |
| R inferior prefrontal gyrus (BA47) | 34 | 46 | −2 | 2724 | 4.20 | 0.00001 |
| L inferior prefrontal gyrus (BA47) | −52 | 24 | −8 | 10 | 3.33 | 0.0004 |
| R orbital frontal gyrus (BA11) | 40 | 58 | −10 | 2724 | 4.05 | 0.00003 |
| R DLPFC (BA46) | 52 | 30 | 30 | 77 | 4.04 | 0.00003 |
| L DLPFC (BA9) | −4 | 58 | 34 | 2724 | 3.77 | 0.00008 |
| Dorsal anterior cingulate (BA32) | 0 | 26 | 32 | 2724 | 3.90 | 0.00005 |
No regions showed the opposite effect.
* FWE corrected at p < 0.05. Cluster size reported at p < 0.001. Additional regions, including thalamus, right putamen, right postcentral gyrus, left cerebellum, bilateral superior temporal gyrus, angular gyrus, supramarginal gyrus, and visual association cortex, also showed higher rCBF for women than men at p ≤ 0.0006, uncorrected. L, Left; R, right.
Figure 2.
Sex differences in rCBF. Statistical parametric map and graph (MNI coordinates x, y, z = 22, −30, −2) showing sex differences in right hippocampus in which rCBF was higher in females than males. p < 0.05, FWE corrected. Additional regions in which this effect was seen are given in Table 2.
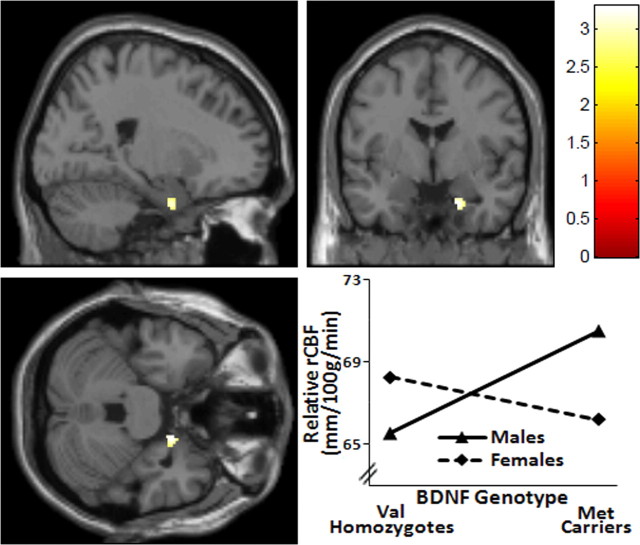
Sex × genotype interactions on rCBF
Whole-brain voxelwise analysis evaluating the relationship between BDNF genotype and sex revealed interactions in several brain regions as detailed in Table 3. Specifically, genotype effects differed in men versus women in the right parahippocampal gyrus (p = 0.0006, uncorrected) and the right dorsolateral PFC (DLPFC, BA9, p = 0.0009, uncorrected), as well as in the right caudate (p = 0.0003, uncorrected), left fusiform gyrus (BA37, p = 0.0005, uncorrected), and left inferior temporal gyrus (BA20, p = 0.0009, uncorrected). For all of these regions, rCBF was higher in females than males for the Val homozygotes, whereas the opposite relationship was seen for Met carriers. This consistent pattern is demonstrated in Figure 3 for right parahippocampal gyrus. Conversely, this pattern was reversed in only a single locus in the left precuneus, in which rCBF was higher in males compared with females for Val homozygotes, and the opposite was seen for Met carriers (BA7, p = 0.0001, uncorrected).
Table 3.
Regions showing significant interaction between BDNF allelic variations and sex differences on rCBF
| Brain region | MNI coordinates |
Cluster size | t values | p values | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Val > Met (female > male) | ||||||
| R caudate | 18 | 4 | 18 | 29 | 3.52 | 0.0003 |
| L fusiform gyrus | −28 | −48 | −14 | 63 | 3.39 | 0.0005 |
| R parahippocampal gyrus | 18 | −2 | −30 | 69 | 3.33 | 0.0006 |
| L inferior temporal gyrus (BA20) | −30 | −8 | −42 | 227 | 3.23 | 0.0009 |
| R DLPFC (BA9) | 16 | 60 | 32 | 21 | 3.16 | 0.0009 |
| Val > Met (male > female) | ||||||
| L precuneus (BA7) | −10 | −54 | 62 | 358 | 3.83 | 0.0001 |
Cluster size reported at p < 0.001. L, Left; R, right.
Figure 3.
BDNF genotype × sex interaction affects rCBF. Statistical parametric map and graph (MNI coordinates x, y, z = 18, −2, −30) showing sex × genotype interaction in right parahippocampus in which rCBF was higher in females than males for the Val homozygotes, whereas the opposite relationship was seen for Met carriers. p = 0.0006, uncorrected.
BDNF genotype and functional connectivity
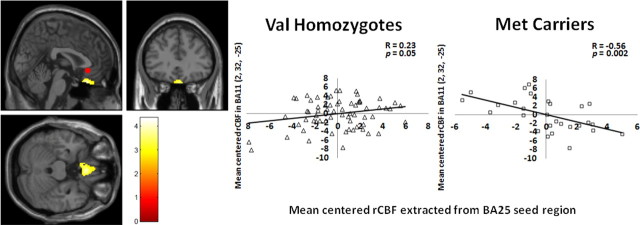
Three separate between-genotype analyses of functional connectivity were performed using seed regions from left BA25 in the PFC, right hippocampus, and left parahippocampus as defined by the between-genotypes analysis shown in Table 1. Detailed results of these analyses are given in Table 4. Between-genotype differences in inter-regional correlations of these three regions showed a predominant pattern such that the correlations were positive in Val homozygotes but robustly negative in Met carriers. This predominant pattern is demonstrated in Figure 4 for the relationship between the BA25 seed region and inferomedial BA11. Findings in the opposite direction (inter-regional functional connectivity positive in Met carriers but negative in Val homozygotes) were sparse (Table 4).
Table 4.
Regions showing differential functional connectivity of rCBF for BDNF Val homozygotes and Met carriers
| Seed/brain region | MNI coordinates |
Cluster size | t values | p values | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Connectivity with BA25 | ||||||
| Val > Met | ||||||
| R fusiform gyrus (BA37) | 48 | −64 | −20 | 119 | 4.33 | 0.00002 |
| R precuneus (BA7) | 10 | −84 | 44 | 122 | 4.11 | 0.00005 |
| L fusiform gyrus (BA37) | −46 | −68 | −18 | 248 | 4.00 | 0.00007 |
| Orbital frontal gyrus (BA11) | 2 | 26 | −24 | 70 | 3.88 | 0.0001 |
| R inferior temporal gyrus (BA20) | 52 | −30 | −22 | 46 | 3.80 | 0.0001 |
| R inferior temporal gyrus (BA20) | 50 | 14 | −30 | 23 | 3.67 | 0.0002 |
| R precuneus (BA7) | 24 | −64 | 60 | 44 | 3.59 | 0.0002 |
| L inferior temporal gyrus (BA20) | −50 | −32 | −22 | 16 | 3.47 | 0.0004 |
| Met > Val | ||||||
| R PFC (BA10) | 14 | 56 | 22 | 12 | 3.56 | 0.0003 |
| L precentral gyrus (BA4) | −30 | −26 | 52 | 19 | 3.48 | 0.0004 |
| Connectivity with right hippocampus | ||||||
| Val > Met | ||||||
| R middle temporal gyrus (BA21) | 50 | −36 | −12 | 251 | 4.11 | 0.00004 |
| R inferior frontal gyrus (BA45) | 46 | 28 | 6 | 70 | 3.98 | 0.00007 |
| R parahippocampal gyrus (BA28) | 22 | −12 | −24 | 47 | 3.79 | 0.0001 |
| R inferior frontal gyrus (BA47) | 24 | 26 | −20 | 19 | 3.45 | 0.0004 |
| Connectivity with left parahippocampus | ||||||
| Val > Met | ||||||
| L inferior frontal gyrus (BA44) | −52 | 8 | 8 | 88 | 3.78 | 0.0001 |
| L fusiform gyrus (BA37) | −44 | −48 | −20 | 25 | 3.61 | 0.0003 |
| L middle frontal gyrus (BA8) | −22 | 26 | 48 | 13 | 3.49 | 0.0004 |
| R inferior frontal gyrus (BA45) | 48 | 22 | 8 | 12 | 3.45 | 0.0004 |
| Met > Val | ||||||
| L precentral gyrus (BA4) | 14 | −30 | 58 | 23 | 3.63 | 0.0002 |
Cluster size reported at p < 0.001. L, Left; R, right.
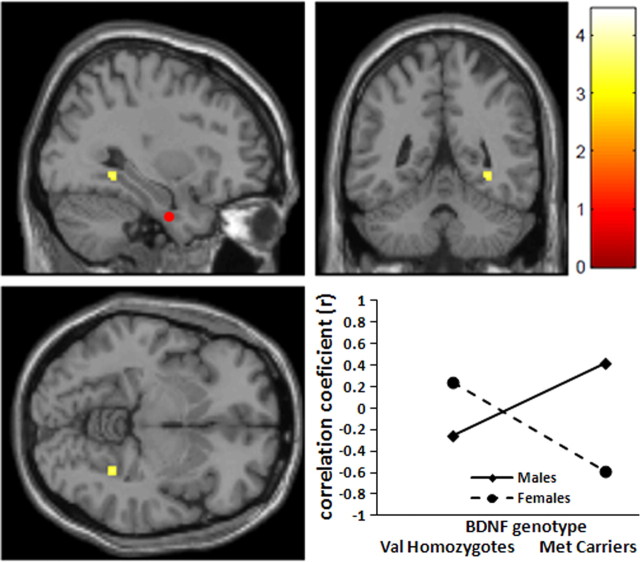
Figure 4.
BDNF allelic variation affects rCBF functional connectivity. Statistical parametric map and graphs showing differential functional connectivity between genotype groups. Between-group difference in functional connectivity between the BA25 seed region (red circle) and BA11; between-group difference in the slopes of the correlations was significant at the p = 0.0001 level. Post hoc analysis of extracted rCBF values (at MNI coordinates x, y, z = 2, 26, −24) demonstrated a positive correlation for the Val homozygotes (r = 0.23, p = 0.05) and a negative correlation for the Met carriers (r = −0.56, p = 0.002).
Sex × genotype interactions on functional connectivity
To investigate sex × genotype interactions on functional connectivity, we used the same approach as in the genotype analysis of functional connectivity mentioned above: connectivity maps for three seed regions from left BA25, right hippocampus, and left parahippocampus as defined in Table 1 were derived for the entire group regardless of genotype or sex, and these three connectivity maps were used as search areas for our analysis of sex × genotype effects on functional connectivity. A consistent pattern of interactions was seen for functional connectivity with all three seed regions such that, for Val homozygotes, inter-regional correlations were more robustly positive for females than males, whereas for Met carriers, this pattern was reversed in that correlations for males were more robustly positive than for females. An example of this pattern is shown in Figure 5 for the relationship between the left parahippocampal seed region and the right parahippocampal gyrus. No interactions in the opposite directions were observed. Complete details are given in Table 5.
Figure 5.
BDNF genotype × sex interaction affects rCBF functional connectivity. Statistical parametric map and graph (MNI coordinates x, y, z = 32, −48, −6) showing sex × genotype interaction (p = 0.0004, uncorrected) in functional connectivity between the left parahippocampal seed region (red circle indicated on the right rather than left sagittal view for illustration) and the right parahippocampal gyrus. The correlation was robustly positive for female Val homozygotes, whereas in male Val homozygotes, the correlation was negative; conversely, for Met carriers, the inter-regional correlation was robustly positive for males but negative for females.
Table 5.
Regions showing interaction between sex and BDNF Val66Met polymorphism on rCBF functional connectivity
| Seed/brain region | MNI coordinates |
Cluster size | t values | P values | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Connectivity with BA25 | ||||||
| Val > Met (female > male) | ||||||
| R inferior frontal gyrus (BA45) | 36 | 22 | 8 | 66 | 3.80 | 0.0001 |
| R hippocampus | 34 | −42 | −2 | 21 | 3.70 | 0.0002 |
| L middle frontal gyrus (BA8) | −40 | 28 | 46 | 44 | 3.64 | 0.0002 |
| Ventral anterior cingulated (BA24) | 4 | −4 | 46 | 27 | 3.47 | 0.0004 |
| Connectivity with right hippocampus | ||||||
| Val > Met (female > male) | ||||||
| L DLPFC (BA9) | −54 | 14 | 34 | 470 | 4.69 | 0.000005* |
| L middle frontal gyrus (BA6) | −38 | 2 | 50 | 166 | 4.61 | 0.000007* |
| L middle frontal gyrus (BA10) | −22 | 56 | 22 | 212 | 4.59 | 0.000008* |
| L middle temporal gyrus (BA39) | −50 | −72 | 28 | 471 | 4.52 | 0.00001* |
| R DLPFC (BA9) | 56 | 12 | 38 | 184 | 4.29 | 0.00002 |
| R cingulate gyrus (BA29) | 8 | −48 | 8 | 98 | 4.23 | 0.00003 |
| R hippocampus | 30 | −16 | −20 | 29 | 4.19 | 0.00003 |
| R postcentral gyrus (BA4) | 18 | −36 | 74 | 300 | 4.18 | 0.00003 |
| R DLPFC (BA9) | 28 | 44 | 38 | 148 | 3.97 | 0.00007 |
| R precentral gyrus (BA4) | 24 | 20 | 58 | 161 | 3.97 | 0.00007 |
| R superior frontal gyrus (BA8) | 10 | 38 | 54 | 42 | 3.80 | 0.0001 |
| R cerebellum | 44 | −68 | −26 | 250 | 3.77 | 0.0002 |
| R postcentral gyrus (BA4) | 28 | −44 | 64 | 45 | 3.76 | 0.0002 |
| L middle frontal gyrus (BA10) | −26 | 64 | −4 | 87 | 3.72 | 0.0002 |
| R precentral gyrus (BA4) | 30 | −16 | 64 | 47 | 3.59 | 0.0003 |
| R parahippocampal gyrus (BA35) | 20 | −34 | −12 | 61 | 3.59 | 0.0003 |
| Dorsal anterior cingulate (BA32) | −4 | 32 | −10 | 45 | 3.58 | 0.0003 |
| L medial frontal gyrus (BA6) | −18 | −2 | 64 | 28 | 3.52 | 0.0003 |
| L postcentral gyrus (BA4) | −22 | −32 | 72 | 36 | 3.42 | 0.0005 |
| Connectivity with left parahippocampus | ||||||
| Val > Met (female > male) | ||||||
| L precentral gyrus (BA6) | −60 | −18 | 44 | 120 | 4.44 | 0.00001 |
| R fusiform gyrus (BA18) | 26 | −78 | −20 | 159 | 4.17 | 0.00004 |
| R precentral gyrus (BA8) | 8 | −26 | 74 | 96 | 3.99 | 0.00007 |
| L fusiform gyrus (BA18) | −28 | −86 | −22 | 111 | 3.77 | 0.0002 |
| R postcentral gyrus (BA4) | 58 | −20 | 54 | 17 | 3.72 | 0.0002 |
| R middle frontal gyrus (BA8) | 34 | 32 | 52 | 16 | 3.61 | 0.0003 |
| R ventral anterior cingulated (BA24) | 14 | −6 | 46 | 33 | 3.44 | 0.0003 |
| R middle temporal gyrus | 38 | 6 | −24 | 24 | 3.54 | 0.0003 |
| R parahippocampal gyrus | 32 | −48 | −6 | 18 | 3.51 | 0.0004 |
| L parahippocampal gyrus (BA28) | −24 | 4 | −22 | 13 | 3.36 | 0.0004 |
* FWE corrected at p < 0.05. Cluster size reported at p < 0.001. L, Left; R, right.
Discussion
BDNF is a key regulator of long-term potentiation and promotes synaptic plasticity and efficacy via excitatory glutamatergic neurotransmission in the hippocampus (Messaoudi et al., 1998; Drake et al., 1999; Tartaglia et al., 2001; Bramham and Messaoudi, 2005). The BDNF Val66Met polymorphism attenuates activity-dependent but not constitutive BDNF signaling in cultured hippocampal neurons (Egan et al., 2003; Lu, 2003) and has been linked to both hippocampal and prefrontal abnormalities by functional neuroimaging. Previous fMRI experiments, whose measurements inherently depend on computing differences between two brain states and thus provide only relative measures of activation over baseline (DeYoe et al., 1994), have shown abnormal hippocampal recruitment during memory-related tasks compared with control tasks (Egan et al., 2003; Hariri et al., 2003; Hashimoto et al., 2008). Other fMRI studies have also found activation differences in mPFC dependent on BDNF genotype in which Met carriers showed reduced ventral mPFC activity during memory extinction (Soliman et al., 2010). However, it has remained unclear until the present work whether this polymorphism has implications for the cerebral metabolic landscape during the so-called basal or resting state. We used a gold-standard PET method that allows task-independent brain function, specifically rCBF, a parameter tightly coupled to cerebral glucose metabolic rate, to be measured and mapped without reference to any other brain state.
Our data demonstrate that, even at rest, rCBF in bilateral hippocampal/parahippocampal and medial frontal regions of healthy individuals is affected by the BDNF Val66Met polymorphism. In addition to providing an important perspective on fMRI activation studies, these data suggest that there exists a potent, genetically mediated bias in the basal activity of frontotemporal circuitry. Because BDNF generally facilitates neural activity in hippocampus and cortex (Henderson, 1996; Benraiss et al., 2001), this bias in Met carriers—increased regional activity at baseline—could reflect a neural-systems-level accommodation for the relatively inefficient, activity-dependent BDNF signaling associated with Met alleles at the cellular level (Egan et al., 2003). Whether this is additionally related to previously reported compensatory increases in peripheral serum concentration of BDNF in healthy Met carriers (Lang et al., 2009) is unclear, and, given the broad impact of BDNF on neuronal development, activity, and plasticity, delineating the precise roots of such speculated accommodation necessarily requires additional research. Nonetheless, it is remarkable that even subtle differences in this molecule result in measurable neurofunctional alterations, particularly in key components of the default resting state network (Raichle et al., 2001), in a cognitively and affectively unchallenged state.
Our finding of increased resting rCBF in women compared with men is consistent with several previous studies (Devous et al., 1986; Yoshii et al., 1988; Slosman et al., 2001) and has been hypothesized to reflect physiologic compensation for the smaller brain size found in women (Ho et al., 1980a,b; Ankney, 1992). The observation that sex effects in several regions within frontotemporal structures are interactively modulated by BDNF allelic variation is in line with a substantial body of animal literature demonstrating parallel and interacting actions of estradiol and BDNF in the brain. Not only do estrogen receptors, BDNF, and trkB show regional coexpression in the hippocampus and frontal cortex (Miranda et al., 1993; Sohrabji and Lewis, 2006), both BDNF and estradiol increase adult neurogenesis (Ormerod et al., 2004; Sairanen et al., 2005), provide neuroprotection (Kiprianova et al., 1999; Shughrue and Merchenthaler, 2003), and facilitate memory formation (Mizuno et al., 2000; Frick et al., 2002). Additionally, BDNF mRNA levels are reduced in ovariectomized female rats and are rescued by estradiol replacement in the hippocampus and cortex (Singh et al., 1995; Sohrabji et al., 1995; Liu et al., 2001). Despite this ample preclinical evidence for sex × genotype interaction, there is a paucity of human data demonstrating true statistical interactions between genotype and sex, although several authors have reported genotype effects occurring only in one sex and not the other (Henningsson et al., 2009; van Wingen et al., 2010). The generally greater magnitude of BDNF genotype effects in men than women in these select regions agrees with studies finding genotype effects on frontotemporal measures selectively in men, including those examining serotonin transporter availability (Henningsson et al., 2009) and activation during memory tasks (van Wingen et al., 2010). However, our findings of reversed sex effects on resting rCBF in Val homozygotes versus Met carriers indicate that the combined effects of sex and BDNF signaling on neurophysiological function are likely to be complex.
Because both BDNF and estradiol play important roles in axonal guidance and dendritic arborization (Dominguez et al., 2004) as well as in synaptogenesis (Tanapat et al., 1999; Scharfman and MacLusky, 2005), we postulated that BDNF function, as predicted by genotype, both alone and in conjunction with sex, may modulate the functional connectivity of areas impacted by BDNF genotype. In keeping with this hypothesis, significantly greater positive correlations between rCBF in the BA25 and that in mPFC and between hippocampus and parahippocampus were seen in Val homozygotes compared with Met carriers during rest, consistent with previous documentation of greater resting network connectivity in these areas in Val homozygotic children (Thomason et al., 2009). These findings are complimented by the sex × genotype interactions, which demonstrated consistent genotype-determined differences in sexually dimorphic patterns of inter-regional cooperativity within BA25 and hippocampal/parahippocampal regions. The correlations of rCBF between these regions were modulated differentially by sex, with correlations more robustly positive for Val homozygotic females than males, whereas for Met carriers, this pattern was reversed. These results together suggest that, when investigating genotypic modulation on neurophysiology, sex difference is an important factor that should be considered in explaining the findings.
Our results are also relevant in considering the neural substrate of neuropsychiatric disorders. For example, the “neurotrophin hypothesis ” of depression speculates that incompetent BDNF function is directly related to the pathophysiology of depression. Although previous genetic association studies of BDNF Val66Met polymorphism in depression have generated inconsistent results (Surtees et al., 2007; Chen et al., 2008; Verhagen et al., 2010), our data suggest that several brain regions known to display depression-related abnormalities could also be modulated by the BDNF Val66Met polymorphism. The hippocampus, BA25, and mPFC have been shown to exhibit abnormal depression-related variations, including hyperperfusion in BA25 in chronic/treatment-resistant patients (Mayberg, 1994; Drevets et al., 1997; Mayberg et al., 2005; Duhameau et al., 2010), hyperactivation of hippocampus to emotional stimuli (Lau et al., 2010), and hyperrecruitment of PFC when downregulating amygdala responses to negative stimuli (Johnstone et al., 2007). Our finding of increased resting rCBF in these same regions in healthy Met carriers without clinical depression mirrors this systems-level pathological phenotype and suggests that inefficient BDNF protein processing may translate to limbic functional changes that could have importance in illness states. Conversely, abnormally increased subgenual cingulate and default network functional connectivity observed in depressed patients (Greicius et al., 2007) was manifested differently in Met carriers in our study, suggesting that the confluence of BDNF genotype effects and depression pathophysiology is intricate and requires additional investigation.
There are several caveats that warrant mention regarding the present work. First, we were unable to document menstrual cycle phase on all women in the study, which prevents the investigation of the effect of this important variable in our data. Future research, including such hormonally related parameters, will provide crucial information. Second, although our study controlled for age, race, sex, and past psychiatric illness, genes interact with many factors, including early life stress, hormones, and the environment to influence the underlying neurobiology of the intermediate phenotype. Although our sample size is adequate, these factors could contribute variance to findings in the present study. In addition, resting state by definition implies unrestricted thought process, and variation in the content of the thought process or in the response to the scanning session may be affected by genotype, and these effects also may interact with sex and other personality-related features. Such experiential variability could be reflected in our rCBF results.
In conclusion, our data identify in healthy individuals BDNF genotype-determined differences in basal resting activity and inter-regional activity relationships within medial frontotemporal nodes important in neuropsychiatric illnesses, such as depression. The fact that sex differences in these measures in these regions are modulated by BDNF genotype serves as an important impetus to further examine BDNF–gonadal hormone interactions on resting network dynamics in both healthy individuals and patients with major depression.
Footnotes
This research was supported by the Intramural Research Program, National Institute of Mental Health, National Institutes of Health.
The authors declare no competing financial interests.
References
- Ankney CD. Sex differences in relative brain size: the mismeasure of woman, too? Intelligence. 1992;16:329–336. [Google Scholar]
- Benraiss A, Chmielnicki E, Lerner K, Roh D, Goldman SA. Adenoviral brain-derived neurotrophic factor induces both neostriatal and olfactory neuronal recruitment from endogenous progenitor cells in the adult forebrain. J Neurosci. 2001;21:6718–6731. doi: 10.1523/JNEUROSCI.21-17-06718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, Monteggia LM, Self DW, Nestler EJ. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lawlor DA, Lewis SJ, Yuan W, Abdollahi MR, Timpson NJ, Day IN, Ebrahim S, Smith GD, Shugart YY. Genetic association study of BDNF in depression: finding from two cohort studies and a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:814–821. doi: 10.1002/ajmg.b.30686. [DOI] [PubMed] [Google Scholar]
- Conner JM, Lauterborn JC, Yan Q, Gall CM, Varon S. Distribution of brain-derived neurotrophic factor (BDNF) protein and mRNA in the normal adult rat CNS: evidence for anterograde axonal transport. J Neurosci. 1997;17:2295–2313. doi: 10.1523/JNEUROSCI.17-07-02295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devous MD, Sr, Stokely EM, Chehabi HH, Bonte FJ. Normal distribution of regional cerebral blood flow measured by dynamic single-photon emission tomography. J Cereb Blood Flow Metab. 1986;6:95–104. doi: 10.1038/jcbfm.1986.12. [DOI] [PubMed] [Google Scholar]
- DeYoe EA, Bandettini P, Neitz J, Miller D, Winans P. Functional magnetic-resonance-imaging (fMRI) of the human brain. J Neurosci Methods. 1994;54:171–187. doi: 10.1016/0165-0270(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Jalali C, de Lacalle S. Morphological effects of estrogen on cholinergic neurons in vitro involves activation of extracellular signal-regulated kinases. J Neurosci. 2004;24:982–990. doi: 10.1523/JNEUROSCI.2586-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake CT, Milner TA, Patterson SL. Ultrastructural localization of full-length trkB immunoreactivity in rat hippocampus suggests multiple roles in modulating activity-dependent synaptic plasticity. J Neurosci. 1999;19:8009–8026. doi: 10.1523/JNEUROSCI.19-18-08009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Simpson JR, Jr, Todd RD, Reich T, Vannier M, Raichle ME. Subgenual prefrontal cortex abnormalities in mood disorders. Nature. 1997;386:824–827. doi: 10.1038/386824a0. [DOI] [PubMed] [Google Scholar]
- Duhameau B, Ferré JC, Jannin P, Gauvrit JY, Vérin M, Millet B, Drapier D. Chronic and treatment-resistant depression: a study using arterial spin labeling perfusion MRI at 3 Tesla. Psychiatry Res. 2010;182:111–116. doi: 10.1016/j.pscychresns.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured clinical interview for DSM-IV axis I disorders research version (SCID-I) New York: New York State Psychiatric Institute, Biometrics Research; 1996. [Google Scholar]
- Frick KM, Fernandez SM, Bulinski SC. Estrogen replacement improves spatial reference memory and increases hippocampal synaptophysin in aged female mice. Neuroscience. 2002;115:547–558. doi: 10.1016/s0306-4522(02)00377-9. [DOI] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RA, Schofield PR, Gordon E, Kemp AH, Williams LM. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Mol Psychiatry. 2009;14:681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, Reiss AL, Schatzberg AF. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry. 2007;62:429–437. doi: 10.1016/j.biopsych.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val(66)met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Moriguchi Y, Yamashita F, Mori T, Nemoto K, Okada T, Hori H, Noguchi H, Kunugi H, Ohnishi T. Dose-dependent effect of the Val66Met polymorphism of the brain-derived neurotrophic factor gene on memory-related hippocampal activity. Neurosci Res. 2008;61:360–367. doi: 10.1016/j.neures.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Henderson CE. Role of neurotrophic factors in neuronal development. Curr Opin Neurobiol. 1996;6:64–70. doi: 10.1016/s0959-4388(96)80010-9. [DOI] [PubMed] [Google Scholar]
- Henningsson S, Borg J, Lundberg J, Bah J, Lindström M, Ryding E, Jovanovic H, Saijo T, Inoue M, Rosén I, Träskman-Bendz L, Farde L, Eriksson E. Genetic variation in brain-derived neurotrophic factor is associated with serotonin transporter but not serotonin-1A receptor availability in men. Biol Psychiatry. 2009;66:477–485. doi: 10.1016/j.biopsych.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA. Reginal distribution of brain-derived neurotrophic factor messenger-RNA in the adult-mouse brain. EMBO J. 1990;9:2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KC, Roessmann U, Straumfjord JV, Monroe G. Analysis of brain weight. I. Adult brain weight in relation to sex, race, and age. Arch Pathol Lab Med. 1980a;104:635–639. [PubMed] [Google Scholar]
- Ho KC, Roessmann U, Straumfjord JV, Monroe G. Analysis of brain weight. II. Adult brain weight in relation to body height, weight, and surface area. Arch Pathol Lab Med. 1980b;104:640–645. [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley GW, Benson DL, Jones EG, Isackson PJ. Developmentalexpression of brain derived neurotrophic factor messenger-RNA by neurons of fetal and adult monkey prefrontal cortex. Brain Res Dev Brain Res. 1992;70:53–63. doi: 10.1016/0165-3806(92)90103-4. [DOI] [PubMed] [Google Scholar]
- Ji Y, Lu Y, Yang F, Shen W, Tang TT, Feng L, Duan S, Lu B. Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons. Nat Neurosci. 2010;13:302–309. doi: 10.1038/nn.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone T, van Reekum CM, Urry HL, Kalin NH, Davidson RJ. Failure to regulate: counterproductive recruitment of top-down prefrontal-subcortical circuitry in major depression. J Neurosci. 2007;27:8877–8884. doi: 10.1523/JNEUROSCI.2063-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprianova I, Freiman TM, Desiderato S, Schwab S, Galmbacher R, Gillardon F, Spranger M. Brain-derived neurotrophic factor prevents neuronal death and glial activation after global ischemia in the rat. J Neurosci Res. 1999;56:21–27. doi: 10.1002/(SICI)1097-4547(19990401)56:1<21::AID-JNR3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Lang UE, Hellweg R, Sander T, Gallinat J. The Met allele of the BDNF Val66Met polymorphism is associated with increased BDNF serum concentrations. Mol Psychiatry. 2009;14:120–122. doi: 10.1038/mp.2008.80. [DOI] [PubMed] [Google Scholar]
- Lau JY, Goldman D, Buzas B, Hodgkinson C, Leibenluft E, Nelson E, Sankin L, Pine DS, Ernst M. BDNF gene polymorphism (Val66Met) predicts amygdala and anterior hippocampus responses to emotional faces in anxious and depressed adolescents. Neuroimage. 2010;53:952–961. doi: 10.1016/j.neuroimage.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnarsson S, Björklund A, Ernfors P. Learning deficit in BDNF mutant mice. Eur J Neurosci. 1997;9:2581–2587. doi: 10.1111/j.1460-9568.1997.tb01687.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fowler CD, Young LJ, Yan Q, Insel TR, Wang Z. Expression and estrogen regulation of brain-derived neurotrophic factor gene and protein in the forebrain of female prairie voles. J Comp Neurol. 2001;433:499–514. doi: 10.1002/cne.1156. [DOI] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS. Frontal-lobe dysfunction in secondary depression. J Neuropsychiatry Clin Neurosci. 1994;6:428–442. doi: 10.1176/jnp.6.4.428. [DOI] [PubMed] [Google Scholar]
- Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, Schwalb JM, Kennedy SH. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–660. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Messaoudi E, Bârdsen K, Srebro B, Bramham CR. Acute intrahippocampal infusion of BDNF induces lasting potentiation of synaptic transmission in the rat dentate gyrus. J Neurophysiol. 1998;79:496–499. doi: 10.1152/jn.1998.79.1.496. [DOI] [PubMed] [Google Scholar]
- Miranda RC, Sohrabji F, Toran-Allerand CD. Neuronal colocalization of messenger-RNAs for neurotrophins and their receptors in the developing central-nervous-system suggests a potential for autocrine interactions. Proc Natl Acad Sci U S A. 1993;90:6439–6443. doi: 10.1073/pnas.90.14.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Yamada K, Olariu A, Nawa H, Nabeshima T. Involvement of brain-derived neurotrophic factor in spatial memory formation and maintenance in a radial arm maze test in rats. J Neurosci. 2000;20:7116–7121. doi: 10.1523/JNEUROSCI.20-18-07116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DD, Cole NB, Segal M. Brain-derived neurotrophic factor mediates estradiol-induced dendritic spine formation in hippocampal neurons. Proc Natl Acad Sci U S A. 1998;95:11412–11417. doi: 10.1073/pnas.95.19.11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P, Sinclair M, Crombie C, Walker N, St Clair DM. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005;10:208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- Ninan I, Bath KG, Dagar K, Perez-Castro R, Plummer MR, Lee FS, Chao MV. The BDNF Val66Met polymorphism impairs NMDA receptor-dependent synaptic plasticity in the hippocampus. J Neurosci. 2010;30:8866–8870. doi: 10.1523/JNEUROSCI.1405-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod BK, Lee TT, Galea LA. Estradiol enhances neurogenesis in the dentate gyri of adult male meadow voles by increasing the survival of young granule neurons. Neuroscience. 2004;128:645–654. doi: 10.1016/j.neuroscience.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen M, Lucas G, Ernfors P, Castrén M, Castrén E. Brain-derived neurotrophic factor and antidepressant drugs have different but coordinated effects on neuronal turnover, proliferation, and survival in the adult dentate gyrus. J Neurosci. 2005;25:1089–1094. doi: 10.1523/JNEUROSCI.3741-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman HE, MacLusky NJ. Estrogen and brain-derived neurotrophic factor (BDNF) in hippocampus: complexity of steroid hormone-growth factor interactions in the adult CNS. Front Neuroendocrinol. 2006;27:415–435. doi: 10.1016/j.yfrne.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman H, Goodman J, Macleod A, Phani S, Antonelli C, Croll S. Increased neurogenesis and the ectopic granule cells after intrahippocampal BDNF infusion in adult rats. Exp Neurol. 2005;192:348–356. doi: 10.1016/j.expneurol.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Iyo M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. Am J Med Genet B Neuropsychiatr Genet. 2004;126B:122–123. doi: 10.1002/ajmg.b.20118. [DOI] [PubMed] [Google Scholar]
- Shughrue PJ, Merchenthaler I. Estrogen prevents the loss of CA1 hippocampal neurons in gerbils after ischemic injury. Neuroscience. 2003;116:851–861. doi: 10.1016/s0306-4522(02)00790-x. [DOI] [PubMed] [Google Scholar]
- Singh M, Meyer EM, Simpkins JW. The effect of ovariectomy and estradiol replacement on brain-derived neurotrophic factor messenger-ribonucleic-acid expression in cortical and hippocampal brain-regions of female Sprague-Dawley rats. Endocrinology. 1995;136:2320–2324. doi: 10.1210/endo.136.5.7720680. [DOI] [PubMed] [Google Scholar]
- Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Mol Psychiatry. 2002;7:579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- Slosman DO, Chicherio C, Ludwig C, Genton L, de Ribaupierre S, Hans D, Pichard C, Mayer E, Annoni JM, de Ribaupierre A. 133Xe SPECT cerebral blood flow study in a healthy population: determination of T-scores. J Nucl Med. 2001;42:864–870. [PubMed] [Google Scholar]
- Sohrabji F, Lewis DK. Estrogen-BDNF interactions: implications for neurodegenerative diseases. Front Neuroendocrinol. 2006;27:404–414. doi: 10.1016/j.yfrne.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabji F, Miranda RC, Toran-Allerand CD. Identification of a putative estrogen response element in the gene encoding brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:11110–11114. doi: 10.1073/pnas.92.24.11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, Jing D, Tottenham N, Amso D, Somerville LH, Voss HU, Glover G, Ballon DJ, Liston C, Teslovich T, Van Kempen T, Lee FS, Casey BJ. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees PG, Wainwright NW, Willis-Owen SA, Sandhu MS, Luben R, Day NE, Flint J. No association between the BDNF Val66Met polymorphism and mood status in a non-clinical community sample of 7389 older adults. J Psychiatr Res. 2007;41:404–409. doi: 10.1016/j.jpsychires.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Tanapat P, Hastings NB, Reeves AJ, Gould E. Estrogen stimulates a transient increase in the number of new neurons in the dentate gyrus of the adult female rat. J Neurosci. 1999;19:5792–5801. doi: 10.1523/JNEUROSCI.19-14-05792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia N, Du J, Tyler WJ, Neale E, Pozzo-Miller L, Lu B. Protein synthesis-dependent and -independent regulation of hippocampal synapses by brain-derived neurotrophic factor. J Biol Chem. 2001;276:37585–37593. doi: 10.1074/jbc.M101683200. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Yoo DJ, Glover GH, Gotlib IH. BDNF genotype modulates resting functional connectivity in children. Front Hum Neurosci. 2009;3:55. doi: 10.3389/neuro.09.055.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WJ, Pozzo-Miller L. Miniature synaptic transmission and BDNF modulate dendritic spine growth and form in rat CA1 neurones. J Physiol. 2003;553:497–509. doi: 10.1113/jphysiol.2003.052639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wingen G, Rijpkema M, Franke B, van Eijndhoven P, Tendolkar I, Verkes RJ, Buitelaar J, Fernández G. The brain-derived neurotrophic factor Val66Met polymorphism affects memory formation and retrieval of biologically salient stimuli. Neuroimage. 2010;50:1212–1218. doi: 10.1016/j.neuroimage.2010.01.058. [DOI] [PubMed] [Google Scholar]
- Ventriglia M, Bocchio Chiavetto L, Benussi L, Binetti G, Zanetti O, Riva MA, Gennarelli M. Association between the BDNF 196 A/G polymorphism and sporadic Alzheimer's disease. Mol Psychiatry. 2002;7:136–137. doi: 10.1038/sj.mp.4000952. [DOI] [PubMed] [Google Scholar]
- Verhagen M, van der Meij A, van Deurzen PA, Janzing JG, Arias-Vásquez A, Buitelaar JK, Franke B. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry. 2010;15:260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Ligons DL, Romanczyk T, Ungaro G, Hyde TM, Herman MM, Weinberger DR, Kleinman JE. Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2005;10:637–650. doi: 10.1038/sj.mp.4001678. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Geneva: World Health Organization; 2008. The global burden of disease: 2004 update, Table A2, Burden of disease in DALYs by cause, sex and income group in WHO regions, estimates for 2004. http://www.who.int/healthinfo/global_burden_disease/GBD_report_2004update_full.pdf. [Google Scholar]
- Yang F, Je HS, Ji Y, Nagappan G, Hempstead B, Lu B. Pro-BDNF-induced synaptic depression and retraction at developing neuromuscular synapses. J Cell Biol. 2009;185:727–741. doi: 10.1083/jcb.200811147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii F, Barker WW, Chang JY, Loewenstein D, Apicella A, Smith D, Boothe T, Ginsberg MD, Pascal S, Duara R. Sensitivity of cerebral glucose metabolism to age, gender, brain volume, brain atrophy, and cerebrovascular risk factors. J Cereb Blood Flow Metab. 1988;8:654–661. doi: 10.1038/jcbfm.1988.112. [DOI] [PubMed] [Google Scholar]