Abstract
The hepatic peptide hormone hepcidin controls the duodenal absorption of iron, its storage and its systemic distribution. Hepcidin production is often insufficient in chronic hepatitis C and alcoholic liver disease, leading to hyperabsorption of iron and its accumulation in the liver. Hepatocyte growth factor (HGF) and epidermal growth factor (EGF) mediate the hepatic regeneration after liver injury. We examined the effect of the growth factors on hepcidin synthesis by hepatocytes. Results: HGF and EGF treatment of primary mouse hepatocytes, as well as EGF administration in mice, suppressed hepcidin mRNA synthesis. The suppression of hepcidin by these growth factors was transcriptional, and was mediated by a direct effect of HGF and EGF on the BMP pathway regulating hepcidin synthesis. We further showed that growth factors interfered with nuclear localization of activated Smads and increased the nuclear pool of the BMP transcriptional co-repressor TG-interacting factor (TGIF). In a kinase screen with small-molecule kinase inhibitors, inhibitors in the PI3 kinase pathway and in the MEK/ERK pathway prevented HGF suppression of hepcidin in primary mouse hepatocytes. Conclusion: HGF and EGF suppress hepatic hepcidin synthesis, in part through PI3 kinase MEK/ERK kinase pathways which may be modulating the nuclear localization of BMP pathway transcriptional regulators including activated Smads1/5/8 and the co-repressor TGIF. EGF, HGF and possibly other growth factors that activate similar pathways may contribute to hepcidin suppression in chronic liver diseases, promote iron accumulation in the liver and exacerbate the destructive disease processes.
Keywords: Chronic liver disease, iron overload, hepatocyte growth factor, epidermal growth factor, bone morphogenetic protein pathway
The hepatic hormone hepcidin controls the flows of iron from dietary absorption, storage and recycling into blood plasma, and thereby regulates plasma iron concentrations and stores (1). In turn, plasma iron concentrations and hepatic iron stores transcriptionally modulate hepcidin synthesis (2, 3), completing a homeostatic feedback loop. The BMP pathway is essential for iron and hepcidin regulation (4). The BMP receptor complex and a range of BMP ligands, including BMP6, induce hepcidin expression by activating Smad4 and Smad1/5/8 (5–7). Human mutations that cause hereditary hemochromatosis either ablate the hepcidin gene (rare) or affect iron-specific hepcidin regulatory proteins that are thought to interact with the BMP pathway (1).
Hepcidin insufficiency and hepatic iron loading are seen in chronic hepatitis of multiple etiologies, including alcoholic hepatitis and viral hepatitis (8–10) and the resulting chronic iron loading in the liver worsens disease prognosis (11). The mechanism of hepcidin suppression in chronic hepatitis is not known.
Chronic hepatitis is characterized by repeated liver injury and repair. Growth factors mitogenic for hepatocytes are important mediators of the liver repair and regeneration. Hepatocyte growth factor (HGF) and epidermal growth factor (EGF) are well-characterized mediators of hepatic regeneration following experimental injury (12–14). We explored the modulation of hepcidin synthesis by these growth factors.
METHODS*
* Detailed methods are provided in the Supplement
Reagents
Murine EGF, HGF, IGF-1 and IGF-2, rat PDGF-BB, and human BMP6 were from R&D Systems, Minneapolis, MN. Recombinant mouse IL-6 and recombinant human EGF were from Millipore, Billerica, MA. Kinase inhibitors EHT1864, PHA665752, NSC23766, 10-DEBC hydrochloride were from Tocris Bioscience, St. Louis, MO, and U73112, LY294002, Calphostin, JNK Inhibitor II, STAT3 inhibitor VII, U0126, ERK inhibitor peptide II FR180204, Akt inhibitor II and Akt inhibitor X from Millipore.
Cell culture and transient transfections
Hepatocytes were isolated from six to eight-week old wild-type C57BL/6 mice by a two-step portal vein collagenase perfusion method, and used within hours or after 18h incubation with serum-free William’s E Medium (serum-starved). Transfection of hepatocytes and HepG2 cells was done with Nucleofector (Lonza Group Ltd, Basel, Switzerland) according to the manufacturer’s instructions. The hepcidin-luciferase reporter included human hepcidin promoter spanning −1 to −2997 (15), and the BRE-luciferase reporter was obtained from H.Y. Lin (16). Luciferase activity was measured by a Veritas Microplate Luminometer (Turner Biosystems, now Promega, Sunnyvale CA).
RNA isolation and real-time quantitative PCR
Quantitative real-time RT-PCR data are presented as either fold-change relative to control or using the ddCt method which naturally yields a logarithmic scale. Fold-change was calculated by the method of Pfaffl (17), where the target gene (Hepc1 or ID1) was referenced to a housekeeping gene (β-actin) and the data presented as a ratio to the control treatment within each experiment. The average relative expression value of the triplicate control treatments was assigned as 1 in each experiment. Relative quantification was performed using the comparative Ct, or ddCt method (18). The target gene (Hepc1 or ID1) was first normalized to a reference housekeeping gene (β-actin) and then presented as the difference from the control treatment within each experiment. The average ddCt value of the triplicate control treatments was zero in each experiment.
Cell Fractionation and Western Blots
Cells for whole-cell lysates were plated in 60 mm collagen-coated dishes (BD Biocoat, Franklin Lakes, NJ) and cells for fractionated lysates (nuclear and cytosolic) were plated in 10-cm collagen-coated dishes. After treatment, whole-cell lysates were collected in ice-cold NETT buffer (150 mM NaCl, 10 mM EDTA, 10 mM Tris, 1% Triton X-100) containing HALT protease/phosphatase inhibitor cocktail (ThermoScientific/Pierce, Rockford, IL). Fractioned lysates were separated into nuclear and cytosolic fractions using the NE-PER kit (Pierce) according to the manufacturer’s instructions. Lysates (30 μg of fractionated lysates, 50 μg of whole-cell lysates) were electrophoresed on 4–20% LongLife iGels (NuSep Inc, Lawrenceville, GA) and transferred to Immobilon-P PVDF membrane (EMD Millipore) for Western blotting, visualization by chemiluminescence, and quantification with ChemiDoc XRS+ System with Image Lab software (Biorad, Hercules, CA).
Kinase inhibitor screen
Twenty-four hours after isolation, serum-starved hepatocytes were pre-treated with kinase inhibitors in a 0.5 μl volume of DMSO. Each kinase inhibitor was added to duplicate or triplicate wells and after 1h, 20 ng/ml HGF was added, then 1h later 10 ng/ml BMP6 was added. Depending on the kinase inhibitor, cultures were incubated for a minimum of 8 hours or overnight prior to sample collection.
Injection of mice with recombinant human EGF
WT 6-week-old C57BL/6 mice were mildly iron depleted by being placed on a diet with less than 4 ppm iron (Harlan Teklad) for seven days prior to the experiment. On the day of injection, mice received a series of three intraperitoneal (IP) injections (spaced six hours apart) of 50 μg human EGF (Millipore Corporation, Billerica, MA) or saline. Some mice were injected with 5 mg holotransferrin (Sanquin, Amsterdam, The Netherlands) with the third dose of EGF or saline. Liver samples were collected 24 hours after the first dose of EGF was administered.
RESULTS
HGF and EGF suppress hepcidin mRNA in vitro and in vivo
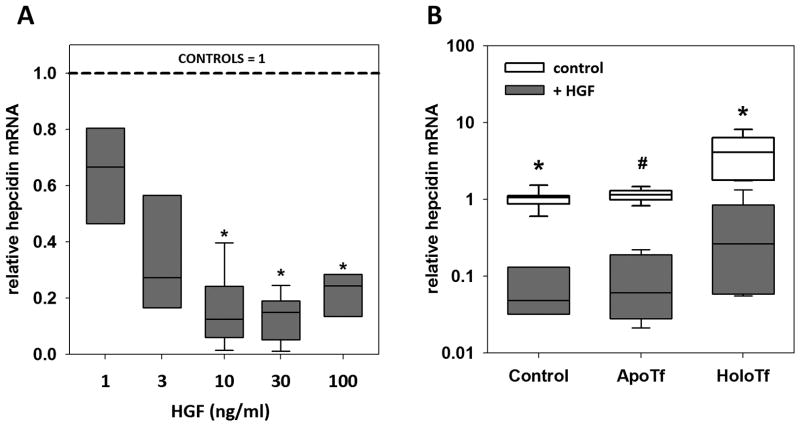
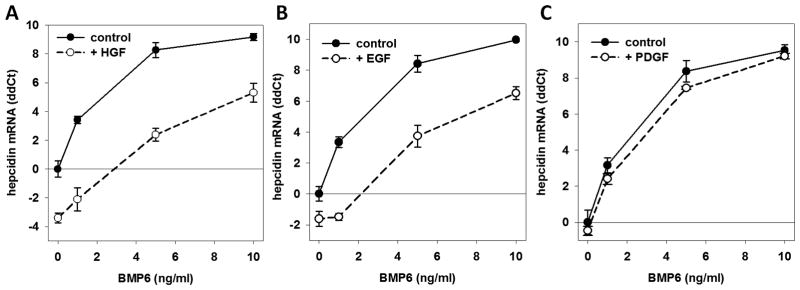
HGF dose-dependently suppressed hepcidin mRNA in hepatocytes (Figure 1, A). When hepcidin was induced by its physiological stimuli, holotransferrin or BMP, HGF significantly lowered both baseline hepcidin expression and the maximal induction of hepcidin by holotransferrin (Figure 1, B) or by a range of BMP6 concentrations (Figure 2, A). At each concentration of BMP6, HGF addition caused 10- to 20-fold suppression of hepcidin mRNA. In experiments where IL-6 was used as the inducing cytokine, HGF suppression of hepcidin mRNA was overcome by increasing concentrations of IL-6, even though it is a less potent hepcidin inducer than BMP6. (Supplementary Figure S1). Among other growth factors tested, only EGF suppressed hepcidin mRNA similarly to HGF (Figure 2, B). PDGF (Figure 2, C) and IGFs −1 and −2 (Supplementary Figure S2) had no effect on BMP induction of hepcidin mRNA.
Figure 1. HGF suppresses hepcidin mRNA expression.
Fresh primary mouse hepatocytes were treated with HGF for 18h in William’s E Medium with 5% FBS. The box plot represents the 25th and 75th percentile, the band the 50th percentile, and whiskers the minimum and maximum of the data. (A) Hepcidin mRNA was dose-dependently suppressed by HGF. * p<0.05 by one-way ANOVA on ranks as compared to untreated control. (B) Hepatocytes were treated with HGF (20 ng/ml) and apo- or holo-transferrin (30 μM). * p < 0.001 by Mann-Whitney rank sum test, # p < 0.001 by t-test.
Figure 2. HGF and EGF suppress hepcidin mRNA induction by BMP-6.
Hepatocytes were serum-starved for 18h prior to treatment with increasing concentrations of BMP6 and (A) 20 ng/ml HGF, (B) 20 ng/ml EGF and (C) 20 ng/ml PDGF-BB. The plots represent hepcidin mRNA concentration relative to untreated controls. (P< 0.001, paired t-test comparing controls vs. HGF or EGF treatment).
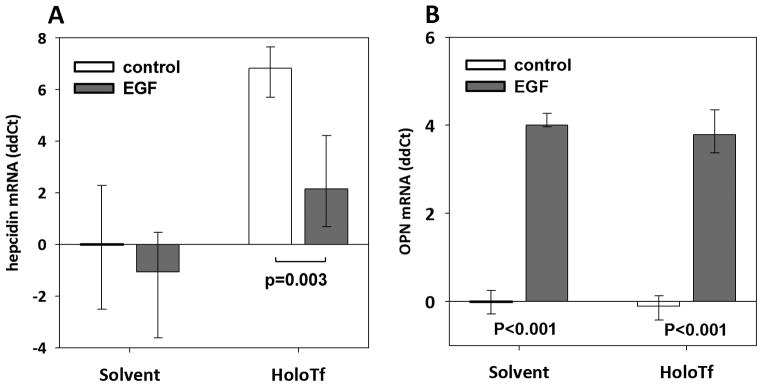
To test whether the suppressive effect of growth factors could be relevant in vivo, we injected mice with EGF, holotransferrin, or their combination (Figure 3), using recombinant human EGF because of much lower cost. Increased expression of the known EGF target transcript osteopontin confirmed that EGF had a detectable effect in the liver (Figure 3, B). EGF significantly suppressed hepcidin responses to holotransferrin (Figure 3, A), with hepcidin mRNA approximately 20-fold lower than in mice that received holotransferrin alone.
Figure 3. In vivo, EGF suppresses hepatic hepcidin mRNA induction by holotransferrin.
6-week-old C57BL/6 male mice received three EGF or saline intraperitoneal injections over 12h, with and without 5 mg holotransferrin co-administration at the last injection. The upper and lower limits of the box plots represent the 25th and 75th percentile, respectively; the band represents the median, n=8 per treatment group. Hepatic hepcidin (A) or osteopontin (B) mRNA expression was analyzed 24 h after the first EGF injection.
HGF represses transcription from the hepcidin promoter and other BMP-responsive promoters
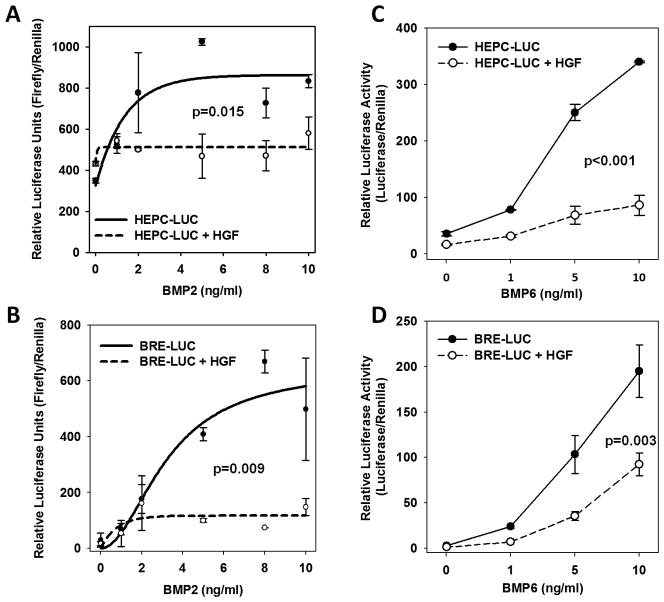
In primary mouse hepatocytes transfected with hepcidin promoter-luciferase reporter, HGF strongly suppressed the induction of the hepcidin promoter by BMP2 (Figure 4, A). We tested a broader range of BMPs in HepG2 cells transfected with hepcidin-luciferase reporter and found that HGF suppressed the induction of the hepcidin reporter by BMP-2, 4 6 and 9) (Figure 4, C, Supplementary Figure S3A). Thus HGF is a broadly active transcriptional suppressor of the BMP response of the hepcidin promoter.
Figure 4. HGF suppresses hepcidin transcription.
(A)* and (B)* Hepatocytes were transiently transfected with a hepcidin-luciferase reporter (HEPC-LUC) or BMP-responsive luciferase reporter (BRE_LUC) and thymidine-kinase Renilla transfection control reporter. Cells were treated with human BMP2, with and without HGF and incubated for 24h. (C)# and (D)* HepG2 cells were transiently transfected with the hepcidin-luciferase reporter (HEPC-LUC) or BMP-responsive luciferase reporter (BRE_LUC) and Renilla transfection control. Cells were treated with human BMP6, with and without HGF and incubated for 24h. Statistical significance was tested with paired t-test* or Wilcoxon signed rank test#.
We also tested the effect of HGF on another BMP-sensitive luciferase reporter containing the BMP-responsive element (BRE) from the promoter of the gene for ID1 (inhibitor of DNA binding 1), a known direct target gene for BMP (16). In transfected mouse hepatocytes and HepG2s, HGF suppressed the induction of the BRE-luciferase reporter by BMP-2, −4, −6 and −9 (Figure 4, B and D, and Supplementary Figure S3B). Further, in primary mouse hepatocytes HGF and EGF similarly modulated the BMP-dependent induction of ID1 mRNA (Supplementary Figure S4). Taken together, these data indicate that HGF and EGF inhibit transcription of BMP-sensitive genes including hepcidin, likely by modulating BMP pathway signaling or BMP-dependent assembly of transcriptional machinery.
The effect of HGF and EGF on signaling in the BMP pathway
When BMPs bind to their receptor (BMP-R), the receptor phosphorylates and activates cytosolic signaling proteins R-Smads 1, 5 and/or 8, which form complexes with the common mediator Smad4. These complexes translocate into the nucleus where they transactivate BMP-dependent transcription (19). The induction of hepcidin mRNA by BMP6 occurred within 4h, and co-stimulation with HGF or EGF suppressed the maximal induction of hepcidin mRNA within the same time-frame (Supplementary Figure S5). The short timeframe favored a mechanism based on rapid, covalent modifications of signaling mediators rather than the synthesis of new transcriptional regulators. We hypothesized that HGF and EGF were initiating kinase signaling that resulted in decreased activation of Smad1/5/8, or in inhibitory modification of Smad1/5/8, through prevention or removal of the activating C-terminal phosphorylation (20) or by targeting Smads 1/5/8 for degradation.
HGF and EGF do not decrease Smad1/5/8 activation
BMP-dependent activating phosphorylation of Smad 1/5/8 was equal in the growth factor-treated and BMP-only series (Figure 5, A, B), indicating that despite the presence of HGF or EGF the BMP signal was still fully transduced to the R-Smads. This makes it unlikely that BMP ligand-trap proteins, modification or degradation of BMP-Rs, or R-Smad de-activation play an important role in the modulation of the BMP effect by HGF or EGF. Further, total nuclear Smad1/5/8 is not decreased by HGF treatment (Figure 5A)..
Figure 5. HGF and EGF do not inhibit BMP activation of signaling mediators Smad1/5/8.
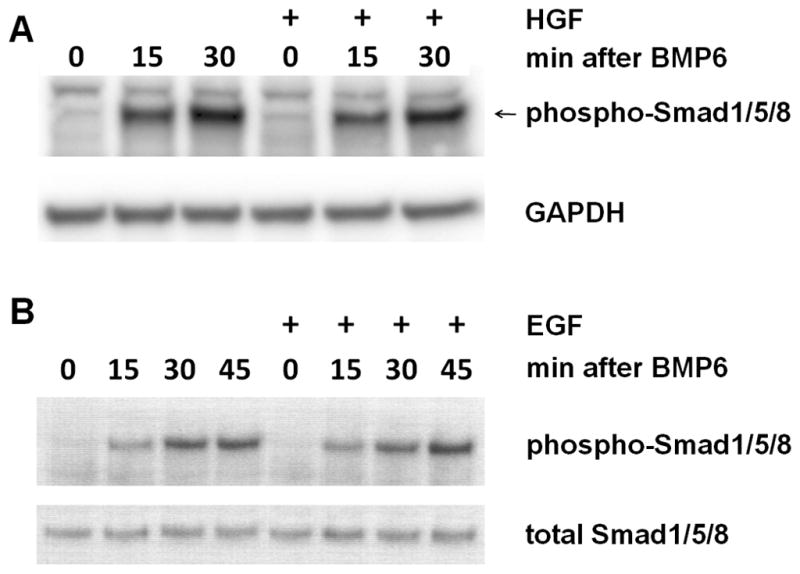
Serum-starved hepatocytes were treated with 20 ng/ml murine HGF (A) or EGF (B) 1h before addition of human BMP6 (10 ng/ml). Western blots of whole-cell lysate were probed with anti-pSmad1/5/8. As loading controls, blots were probed with anti-GAPDH or total Smad1/5/8.
HGF and EGF do not induce inhibitory Smads
Additional modulation at the ligand-receptor level is provided by BMP pathway inhibitors including BAMBI (21), Smad 6 (22, 23) and Smad7, the latter already known to play a role in hepcidin regulation (7, 24). The concentrations of BAMBI and inhibitory Smads determine their effect on signaling (23). We used whole-cell lysates of primary mouse hepatocyte cultures treated with BMP and HGF or EGF to examine the protein levels of the three inhibitors. After overnight incubation, neither Smads 6 and 7 (Supplementary Figure S6A, B) nor BAMBI (data not shown) were induced by growth factor treatments.
HGF or EGF does not decrease protein levels of R-Smads1 and 5 or common mediator Smad4
Growth factors have been reported to decrease the total Smad1 pool by proteasomal degradation (25). Treatment with HGF had no effect on total Smad1 or Smad5 (Supplementary Figure S6C, D) in the 2h following HGF treatment or overnight (data not shown). Treatment with EGF also did not cause a change in total Smad 1/5/8 in whole cell lysates (Figure 5, B).
The common mediator Smad4 is also a target for regulatory input and its ubiquitination leads to its degradation in the proteasome (26). Decreased Smad4 in the context of hepcidin reporter suppression by hypoxia was recently described (27). From hepatocytes treated with BMP6 with and without HGF, we blotted nuclear lysates for Smad4. Smad4 levels in the nucleus were unaffected by HGF, indicating that the BMP signal had adequate access to co-Smad for formation of transcription complexes (Supplementary Figure S6E). Thus, the mechanism for growth factor suppression of hepcidin does not include overall degradation of the receptor-activated Smad pool or Smad4.
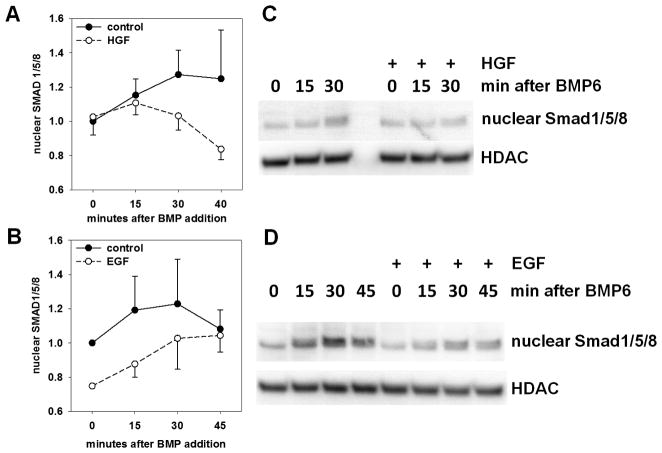
BMP-dependent nuclear localization of Smad1/5/8 is modestly suppressed by HGF and EGF
The linker region between the two globular domains of Smad1 can be phosphorylated by several kinases, including the mitogen-activated protein kinase (MAPK) ERK2, cyclin-dependent kinases (CDK) and glycogen synthase kinase-3β (GSK3β) (25) and the modification inhibits nuclear translocation of Smads. Growth factors including HGF and EGF induce linker phosphorylation (28) acting to limit BMP signaling during development.
After growth factor treatment of BMP6-stimulated hepatocytes, immunoblots for phospho-Smad1/5/8 showed moderately decreased nuclear localization of phospho-Smad/1/5/8 (Figure 6). The difference between the growth-factor treated nuclear lysates and the control lysates was statistically significant for both growth factors by pairwise t-test when four repeated experiments were analyzed together for each growth factor.
Figure 6. HGF and EGF modestly decrease BMP-stimulated nuclear import of SMADs.
Serum-starved hepatocytes were pre-treated with 20 ng/ml murine HGF (A and C) or 20 ng/ml murine EGF (B and D) 1h before treatment with human BMP6 (40 ng/ml). Nuclear lysates were analyzed by Western blotting and digital imaging. Histone deacetylase was used as a loading control. Four independent experiments were performed for A and B. Pairwise t-test: p = 0.04 for HGF; p = 0.004 for EGF.
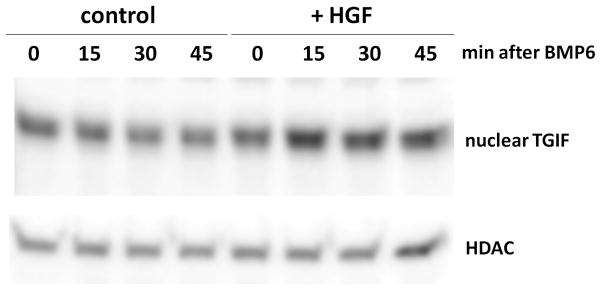
Nuclear levels of the transcriptional co-repressor TGIF are increased after treatment with HGF
We next considered modes of BMP pathway suppression that target the Smad transcriptional complex. The Smad transcriptional complex is nucleated by R-Smad/Smad4, but the binding affinity is regulated by DNA-binding co-activators or co-repressors such as TGIF (29). Figure 7 demonstrates increased protein levels of TGIF in nuclear lysates of cells treated with HGF, suggesting a mechanism for HGF interference with Smad transcriptional complexes.
Figure 7. HGF increases the nuclear protein levels of BMP-corepressor TGIF.
Serum-starved hepatocytes were treated with murine HGF 40 ng/ml 1h prior to the addition of 25 ng/ml BMP6. The nuclear fraction of the cell lysate was blotted and probed for the BMP co-repressor TGIF, with histone deacetylase as a loading control.
Taken together, these data indicate that the mechanism for HGF suppression is downstream of the multiple levels of Smad regulation and may involve a combination of decreased nuclear localization of activated Smad1/5/8 as well as induction of transcriptional co-repressors such as TGIF.
Which of the kinase pathways downstream of HGF/Met are suppressing hepcidin?
HGF activates signaling pathways through its receptor, tyrosine kinase Met. Met signaling is complex, branching into multiple distinct but interacting signaling modules, so that HGF suppression of BMP signaling to hepcidin may be the product of more than one downstream signal from HGF/Met (Supplementary Figure S7). Using primary hepatocytes treated with BMP6, we performed a limited screen with small-molecule kinase inhibitors against individual kinase pathways known to be activated by HGF.
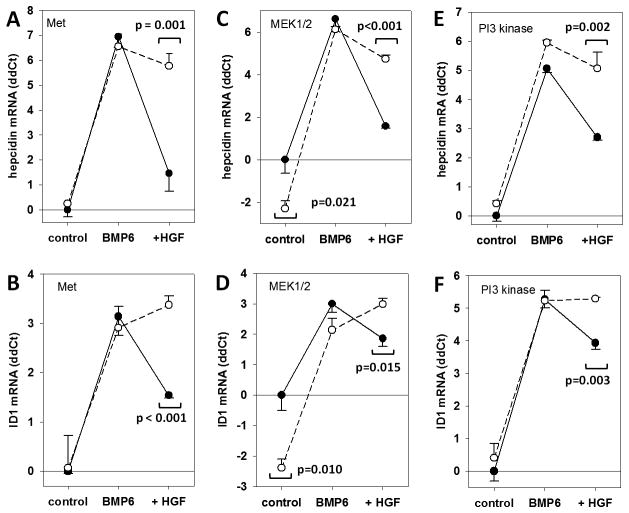
The proof of principle experiment tested for inhibition of HGF signaling by a kinase inhibitor for the Met receptor itself (PHA665752). Inhibition of the Met receptor abrogated HGF suppression of both hepcidin mRNA and ID1 mRNA (Figure 8, A, B). Interestingly, the dose required to inhibit HGF (1 μM) was 20-times the IC50 (25–50 nM) for inhibition of receptor activation in epithelial cell lines (kidney, lung and mammary cells). The requirement for high doses of inhibitor may be due to the hepatocyte cell membrane resistance to permeation of small molecule kinase inhibitors, akin to difficulties with the transfection of primary hepatocyte using liposomal methods. Alternatively, the known catabolic activity of hepatocytes towards small organic molecules may cause rapid degradation of many of our inhibitors. Bearing this in mind, we examined a higher range of inhibitor concentrations.
Figure 8. Reversal of HGF effect by PI3 kinase or MEK1/2 inhibition.
Serum-starved hepatocytes were pre-treated for 1h with one of the following kinase inhibitors: 1 μM Met inhibitor PHA665752, 8 μM PI3 kinase inhibitor LY294002, or 25 μM U0126 MEK1/2 inhibitor, then treated with BMP6 with or without HGF for 18h. Inhibition of the HGF receptor Met blocked the suppressive effect of HGF on hepcidin (A) (p=0.001, t-test), and on the unrelated BMP-responsive gene ID1 (B) (p<0.001, t-test). (C) Inhibition of MEK1/2 also appears to reverse hepcidin suppression by HGF, however, the inhibitor also suppressed baseline hepcidin (p= 0.021). (D) The MEK1/2 inhibitor also reversed the HGF suppression of ID1 (p=0.015, t-test) and suppressed ID1 at baseline (p=0.010, t-test). Inhibition of PI3 kinase fully reversed the suppression of (E) hepcidin (p=0.002, t-test) as well as (F) ID1 (p=0.003, t-test).
MAPK/ERK signaling plays only a partial role in HGF crosstalk with hepcidin
Two MAPK pathways are known to be activated by HGF: Rac1/JNK and Ras/MEK/ERK. Two Rac1 inhibitors (5 μM EHT1864, 94 μM NSC23766) did not inhibit HGF suppression of hepcidin mRNA, nor did JNK inhibition (1 μM, JNK Inhibitor II). With MEK1/2 inhibitor U0126, we observed partial reversal of HGF suppression of hepcidin mRNA (Figure 8, C) as well as ID1 mRNA (Figure 8, D). The ERK inhibitor peptide II (5 μM) recapitulated these data (data not shown). The high dose of U0126 (25 μM) reversed HGF suppression of hepcidin and ID1, but it also affected the baseline hepcidin and ID1 mRNA, indicating that the activity of the inhibitor at 25 μM may have effects not specific to HGF. These data indicate at most a partial role for HGF/MEK signaling to hepcidin; alternatively, inhibition of MEK1/2 may result in mild hepcidin increase by mechanisms independent of HGF.
Major pathways not involved: PKC, PLC and STAT3
Broadening our focus, we sought to rule out other major pathways downstream of the Met receptor (Supplementary Figure S7). Small-molecule inhibitors of protein kinase C (1.25 μM Calphostin), phospholipase C (5 μM U73112) or STAT3 (1 μM inhibitor VII) neither affected BMP induction of hepcidin, nor did they reverse suppression of hepcidin by HGF. We conclude that none of these major pathways plays a role in the regulation of hepcidin by HGF.
PI3 kinase inhibition reverses hepcidin suppression by HGF
Treatment of primary mouse hepatocytes with PI3K inhibitor LY294002 at a moderate concentration (8 μM, 5X IC50) significantly reversed HGF suppression of hepcidin (p = 0.04, t-test compared to controls) (Figure 8, E) without affecting baseline hepcidin mRNA in the controls or maximal hepcidin induction by BMP6. ID1 suppression was similarly reversed (Figure 8, F). Increased phosphorylation of AKT confirmed activation of PI3K by HGF, and loss of AKT phosphorylation confirmed the effectiveness of the PI3K inhibitor (Supplementary Figure 8). Pre-treatment with the Met inhibitor also prevented AKT activation (Supplementary Figure S9A). In agreement with hepcidin mRNA suppression in primary hepatocytes, only HGF and EGF, but not PDGF, IGF-1 and IGF-2, caused activation of AKT (Supplementary Figure S9B).
DISCUSSION
We report the growth factors HGF and EGF as a new category of hepcidin suppressors that robustly block hepcidin transcriptional regulation by known physiologic inducers, iron and BMPs. The ability of EGF to suppress iron-induced hepcidin mRNA was also confirmed in mice. Our data also indicate that HGF and EGF regulate hepcidin by suppressing BMP signaling upstream of the hepcidin promoter, a suppressive effect that extends to the unrelated BMP-sensitive promoter and mRNA transcript of ID1 The rapid onset of suppression suggests direct molecular crosstalk between BMP and growth factor signaling mediators. The crosstalk does not extend to the IL-6 pathway as HGF does not significantly suppress hepcidin mRNA at higher IL-6 concentrations.
Growth factor regulation of BMP signaling through MAPK-mediated nuclear exclusion of R-Smads has been extensively reported in culture systems using transfected, highly overexpressed tagged Smad constructs. The data from such studies suggest that Smad linker phosphorylation by growth factor-activated MAPK/ERK nearly entirely abrogates BMP-dependent nuclear localization of activated Smads (25). Our findings indicate that in hepatocytes the endogenous R-Smad pool is less strictly regulated. The trend we observed for nuclear exclusion of activated Smads and the increased regulatory phosphorylation at MAPK motifs on the Smad linker is modest at best and seems unlikely to account for the dramatic inhibition of hepcidin induction by HGF and EGF. Furthermore, the activation of the R-Smads was not suppressed by the growth factors, nor was the cellular pool of Smads1 and 5 and co-Smad4 degraded. We also detected no evidence of transcriptional induction of BMP negative regulators such as inhibitory Smads.
R-Smad by itself interacts weakly with its cognate promoter element and its association with other transcription factors is thought to be required for optimal activity. The increased protein levels of a transcriptional co-repressor, TGIF, in the nuclei of hepatocytes treated with HGF suggests a likely mechanism for HGF crosstalk with BMP signaling. Phosphorylation of the co-repressor TGIF by EGF-activated Ras/MEK signaling has been reported; TGIF phosphorylation resulted in stabilization of the repressor and formation of R-Smad/TGIF transcriptionally suppressive complexes (30). We surmise that HGF may suppress hepcidin induction by BMP through MAPK stabilization of TGIF.
HGF is a pleiotropic growth factor that activates a multitude of downstream signaling pathways; many of the mitogenic, morphogenic and motogenic effects of Met are regulated by more than one of these downstream signals. Our kinase inhibitor screen in primary hepatocytes identified at least two signaling pathways (MEK and PI3K) that appear to regulate hepcidin. The activity of the MEK1/2 inhibitor U0126 in our studies suggested a role for MEK in HGF suppression. It was previously reported that Ras/MEK activation by EGF results in phosphorylation and stabilization of the Smad transcriptional co-repressor TGIF (30). HGF may cause a similar stabilization of TGIF via MEK activation. A more detailed exploration of the similarities and differences between HGF and EGF pathways will be undertaken in a future study.
In view of the role of growth factors HGF, EGF, and TGF-α, which also binds to the EGF receptor, as mediators of the hepatic regenerative response (14), the suppression of hepcidin by growth factors may be relevant to hepcidin deficiency and hepatic iron loading in chronic liver diseases. Elevated liver tissue concentrations of growth factors in chronic viral and alcoholic hepatitis could be repressing maximal hepcidin response to iron thereby increasing dietary iron absorption and worsening the liver injury. As in hereditary hemochromatosis, the relative lack of hepcidin induction by iron in chronic hepatitis results in chronic hyperabsorption of dietary iron. Excess iron accumulates particularly in the liver due to the avid uptake of non-transferrin-bound iron (NTBI) by hepatocytes, as well as the first-pass effect of portal circulation from the gut. The iron deposition is often parenchymal and compounds preexisting liver injury from hepatitis, worsening disease prognosis. In CHC, iron correlates with development of cirrhosis and hepatocellular carcinoma (HCC) (11). The role of iron in disease progression has been supported by studies in which phlebotomy improved disease indices in nonalcoholic steatohepatitis and chronic hepatitis C (31, 32). However, the effects of iron on hepatitis C may be complex; excess iron promotes tissue damage but it also suppresses viral replication, perhaps accounting for the divergent outcomes of phlebotomy interventions (33).
Regulation of hepcidin by growth factors may be important for normal iron homeostasis as well. Hepcidin must be physiologically suppressed during early years of life, when continuing growth and development require greater iron absorption than in the mature adult (34). Though few studies exist of hepcidin levels in children and adolescents, a recent study in children and adult patients undergoing hemodialysis found that the pediatric control group had serum hepcidin concentrations that were only a third as high as the adult control group (35). Interestingly, hepatic hepcidin mRNA is not detectable by Northern blot in mice from E15.5 to postnatal day 56 apart from a transient induction at birth extending to postnatal day 2 (36). Hepcidin expression, therefore, only reaches a high level in the adult mouse liver, concordant with the human studies suggesting that hepcidin is repressed during early growth and maturation.
Finally, better understanding of pathways that mediate hepcidin suppression may help identify useful targets for new treatments for iron restrictive disorders (anemia of inflammation, anemia of chronic kidney disease) in which hepcidin excess contributes to the pathogenesis of anemia and to erythropoietin resistance.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical assistance of Victoria Gabayan with all the mouse studies in this manuscript.
Financial Support:
This work was supported by Roche Foundation for Anemia Research (RoFAR) to T.G., National Institutes of Health grants R01 DK065029 (to T.G.), R01 DK082717 (to E.N.), the Will Rogers Fund (to T.G.), Ruth L. Kirschstein National Research Service Award (NRSA) F30 DK082151 (to J.G.) and Ruth L. Kirschstein NRSA GM007185 (to E.R.).
List of Abbreviations
- HGF
hepatocyte growth factor
- EGF
epidermal growth factor
- BMP
bone morphogenetic protein
- Smad
sons of mothers against decapentaplegic
- TGIF
TG-interacting factor
- PDGF
platelet-derived growth factor
- MEK/ERK, pathway
mitogen-activated kinase pathway, (MEK = mitogen-activated ERK kinase, ERK= extracellular signal-regulated kinase)
- MAPK
mitogen activated protein kinase
- Holo-Tf
holo-transferrin
- Apo-Tf
. Apotransferrin
- Met
HGF receptor (Met protooncogene)
- PI3-kinase
phosphoinositide 3-kinase
Contributor Information
Julia B. Goodnough, Email: JBGoodnough@mednet.ucla.edu.
Emilio Ramos, Email: emramos@ucla.edu.
Elizabeta Nemeth, Email: enemeth@mednet.ucla.edu.
Tomas Ganz, Email: tganz@mednet.ucla.edu.
Reference List
- 1.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Annu Rev Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 2.Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, et al. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology. 2011;53:1333–1341. doi: 10.1002/hep.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corradini E, Meynard D, Wu Q, Chen S, Ventura P, Pietrangelo A, et al. Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)-SMAD signaling pathway in mice. Hepatology. 2011;54:273–284. doi: 10.1002/hep.24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 6.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 8.Girelli D, Pasino M, Goodnough JB, Nemeth E, Guido M, Castagna A, et al. Reduced serum hepcidin levels in patients with chronic hepatitis C. J Hepatol. 2009;51:845–852. doi: 10.1016/j.jhep.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita N, Sugimoto R, Takeo M, Urawa N, Mifuji R, Tanaka H, et al. Hepcidin expression in the liver: relatively low level in patients with chronic hepatitis C. Mol Med. 2007;13:97–104. doi: 10.2119/2006-00057.Fujita. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishina S, Hino K, Korenaga M, Vecchi C, Pietrangelo A, Mizukami Y, et al. Hepatitis C virus-induced reactive oxygen species raise hepatic iron level in mice by reducing hepcidin transcription. Gastroenterology. 2008;134:226–238. doi: 10.1053/j.gastro.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Lambrecht RW, Sterling RK, Naishadham D, Stoddard AM, Rogers T, Morishima C, et al. Iron levels in hepatocytes and portal tract cells predict progression and outcomes of patients with advanced chronic hepatitis C. Gastroenterology. 2011;140:1490–1500. doi: 10.1053/j.gastro.2011.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhave VS, Paranjpe S, Bowen WC, Donthamsetty S, Bell AW, Khillan JS, et al. Genes inducing iPS phenotype play a role in hepatocyte survival and proliferation in vitro and liver regeneration in vivo. Hepatology. 2011:n/a. doi: 10.1002/hep.24507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Factor VM, Seo D, Ishikawa T, Kaposi-Novak P, Marquardt JU, Andersen JB, et al. Loss of c-Met disrupts gene expression program required for G2/M progression during liver regeneration in mice. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0012739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 15.Maes K, Nemeth E, Roodman GD, Huston A, Esteve F, Freytes C, et al. In anemia of multiple myeloma, hepcidin is induced by increased bone morphogenetic protein 2. Blood. 2010;116:3635–3644. doi: 10.1182/blood-2010-03-274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korchynskyi O, ten Dijke P. Identification and Functional Characterization of Distinct Critically Important Bone Morphogenetic Protein-specific Response Elements in the Id1 Promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Kawabata M, Miyazono K. Signal transduction of the TGF-beta superfamily by Smad proteins. J Biochem. 1999;125:9–16. doi: 10.1093/oxfordjournals.jbchem.a022273. [DOI] [PubMed] [Google Scholar]
- 20.Knockaert M, Sapkota G, Alarcon C, Massague J, Brivanlou AH. Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc Natl Acad Sci U S A. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, et al. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 22.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 24.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, M++ller A, Boutros M, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 25.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 27.Chaston TB, Matak P, Pourvali K, Srai SK, McKie AT, Sharp PA. Hypoxia inhibits hepcidin expression in HuH7 hepatoma cells via decreased SMAD4 signaling. Am J Physiol Cell Physiol. 2011;300:C888–C895. doi: 10.1152/ajpcell.00121.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 29.Wotton D, Massague J. Smad transcriptional corepressors in TGF beta family signaling. Curr Top Microbiol Immunol. 2001;254:145–164. [PubMed] [Google Scholar]
- 30.Lo RS, Wotton D, Massague J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 2001;20:128–136. doi: 10.1093/emboj/20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato J, Miyanishi K, Kobune M, Nakamura T, Takada K, Takimoto R, et al. Long-term phlebotomy with low-iron diet therapy lowers risk of development of hepatocellular carcinoma from chronic hepatitis C. J Gastroenterol. 2007;42:830–836. doi: 10.1007/s00535-007-2095-z. [DOI] [PubMed] [Google Scholar]
- 32.Fargion S, Valenti L, Fracanzani AL. Beyond hereditary hemochromatosis: new insights into the relationship between iron overload and chronic liver diseases. Dig Liver Dis. 2011;43:89–95. doi: 10.1016/j.dld.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Mueller S. Increased iron in HCV infection: collateral damage or antiviral defense? J Hepatol. 2010;53:990–992. doi: 10.1016/j.jhep.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Lonnerdal B, Kelleher SL. Iron metabolism in infants and children. Food Nutr Bull. 2007;28:S491–S499. doi: 10.1177/15648265070284S402. [DOI] [PubMed] [Google Scholar]
- 35.Zaritsky J, Young B, Gales B, Wang HJ, Rastogi A, Westerman M, et al. Reduction of serum hepcidin by hemodialysis in pediatric and adult patients. Clin J Am Soc Nephrol. 2010;5:1010–1014. doi: 10.2215/CJN.08161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci U S A. 2002;99:4596–4601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.