Abstract
Higher order chromatin structure in eukaryotes can lead to differential gene expression in response to the same transcription factor; however, how transcription factor inputs integrate with quantitative features of the chromatin environment to regulate gene expression is not clear. In vitro models of HIV gene regulation, in which repressive mechanisms acting locally at an integration site keep proviruses transcriptionally silent until appropriately stimulated, provide a powerful system to study gene expression regulation in different chromatin environments. Here we quantified HIV expression as a function of activating transcription factor nuclear factor-κB RelA/p65 (RelA) levels and chromatin features at a panel of viral integration sites. Variable RelA overexpression demonstrated that the viral genomic location sets a threshold RelA level necessary to induce gene expression. However, once the induction threshold is reached, gene expression increases similarly for all integration sites. Furthermore, we found that higher induction thresholds are associated with repressive histone marks and a decreased sensitivity to nuclease digestion at the LTR promoter. Increasing chromatin accessibility via inhibition of histone deacetylation or DNA methylation lowered the induction threshold, demonstrating that chromatin accessibility sets the level of RelA required to activate gene expression. Finally, a functional relationship between gene expression, RelA level, and chromatin accessibility accurately predicted synergistic HIV activation in response to combinatorial pharmacological perturbations. Different genomic environments thus set a threshold for transcription factor activation of a key viral promoter, which may point toward biological principles that underlie selective gene expression and inform strategies for combinatorial therapies to combat latent HIV.
Introduction
A central question in eukaryotic gene expression is how the activation of gene expression depends simultaneously on transcription factor availability and quantitative features of the chromatin environment at different genomic locations1 (Fig. 1A). Eukaryotic transcription factors commonly regulate multiple genes, yet extracellular stimuli that activate transcription factors result in selective expression of only a subset of these genes. The sequence and arrangement of transcription factor binding sites in different promoters cannot fully explain differential responses to the same transcription factor2. Another important input, chromatin features of the genomic locus, can also provide regulatory selectivity in response to transcription factor activation, including in complex processes such as inflammation3, 4 and development5. It would therefore be informative to quantify how the placement of a particular gene in the genome impacts its responsiveness to an input transcription factor signal and features of the local chromatin environment. Such a quantitative understanding of how chromatin environment impacts gene regulation may also improve rational design of therapies to reverse gene expression dysregulation induced by chromatin changes6.
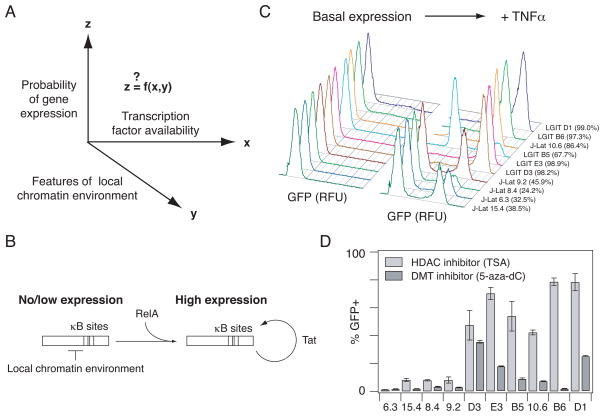
Fig. 1.
In vitro models of HIV gene expression provide an experimental system to study RelA-mediated gene expression in a range of chromatin environments. (A) There is general interest in how gene expression probability varies as a function of transcription factor availability and quantitative features of the local chromatin environment. (B) Schematic describing RelA-mediated gene expression in the HIV vectors before and after the Tat-mediated positive feedback loop is activated. (C) Representative flow cytometry histograms of GFP expression for the panel of clones each infected with a single integration of an inactive HIV provirus under basal conditions (left) and after stimulation with TNFα (20 ng/ml) for 48 hours (right). Percentage of TNFα-activated cells is indicated in parentheses. Clones are ordered according to increasing basal gene expression. (D) Infected clonal populations were stimulated with 400 nM TSA for 24 hours (light gray bars) or 5 μM 5-aza-dC for 48 hours (dark gray bars). Experiments were performed in biological triplicate. Data are presented as the mean ± standard deviation.
Studies in S. cerevisiae recently demonstrated that chromatin provides a mechanism for tuning gene expression in response to transcription factors by setting a gene induction threshold that is decoupled from gene expression range7, 8. However, it is unclear if a similar relationship holds for genes in multicellular organisms, in which gene expression attenuation and silencing are mediated by more complex repressive chromatin modifications9. To address this question, we studied activation of the human retrovirus human immunodeficiency virus-1 (HIV). Because HIV integrates into the genome of its host cell in a semi-random fashion and responds to host transcription factors, it provides a unique opportunity to study activation of the same gene by the same transcription factor in different chromatin environments without altering promoter architecture10, 11.
Following infection and integration into the host chromosome, initial expression from the HIV long terminal repeat (LTR) promoter is inefficient and subject to the availability of the host cell transcriptional machinery and to local factors operating at the integration site (Fig. 1B)12–14. In some cases, chromatin-mediated repression of HIV gene expression – including histone deacetylation, histone methylation, and DNA methylation – results in inactive viral gene expression that may be related to viral latency, in which the virus adopts a quiescent phenotype but can be reactivated when stimulated with the appropriate transcriptional cues15–19. Within inactive HIV-1 promoters, a nucleosome is precisely positioned immediately downstream of the transcription start site (TSS), and transcriptional activation of silent proviruses is strongly correlated with its removal via chromatin remodeling complexes20, 21. Upon such LTR activation, a virally-encoded transcriptional activator (Tat) feeds back on the LTR to amplify gene expression nearly 100-fold (Fig. 1B)22, 23, and stochastic effects in this process may also contribute to viral latency24–26. Thus, inactive HIV integrated at different genomic locations offers a biomedically relevant system to study the probability of gene activation from the same mammalian promoter in a spectrum of repressive chromatin environments.
Like most cellular promoters, the HIV LTR is also strongly regulated by global host factors, prominently including the transcription factor nuclear factor-κB (NF-κB) p65/RelA. Transcription factors of the NF-κB/Rel family control complex transcriptional patterns in both the innate and adaptive immune responses, and these diverse patterns in part result from differences in the chromatin structure of target genes3, 27. Upon stimulation with a NF-κB pathway activator, such as the inflammatory cytokine tumor necrosis factor-α (TNFα), RelA translocates to the nucleus and binds to the HIV LTR to stimulate gene expression20, 28. Specifically, NF-κB RelA promotes elongation by RNA polymerase II (RNAPII) in the absence of Tat29 and is thought to be important in mediating the activation of silent proviruses30. Thus, as a model system, the HIV LTR provides a common promoter architecture to quantitatively explore how NF-κB RelA mediates gene expression in different chromatin environments.
Here we quantified viral gene expression as a function of NF-κB RelA level and quantitative features of the chromatin environment at the viral integration site. In cell populations containing different clonal integrations of the LTR promoter, we found that the threshold level of RelA necessary to initiate gene expression in the cell population varied monotonically with the degree of chromatin accessibility at the LTR promoter. Furthermore, upon onset, gene expression increased as a function of additional RelA increases in a non-linear manner similar for all clones. Moreover, increasing chromatin accessibility via small molecule inhibition of either histone deacetylation or DNA methylation reduced the RelA threshold without otherwise changing this gene activation function. Finally, an empirical gene activation function describing the dependence of HIV gene expression on RelA level and chromatin accessibility accurately predicted synergistic activation in response to combinatorial treatment with chromatin- or DNA- modifying enzyme inhibitors and TNFα. Thus, our results demonstrate that chromatin accessibility at LTR promoters, mediated by complex epigenetic modifications acting at the integration site, sets a threshold level of RelA required for promoter activation, after which the activation profile is conserved across genomic locations. These findings point to a general mechanism by which genomic location may establish differential gene expression in response to the same transcription factor. These results may also aid efforts to develop combinatorial therapies to reverse chromatin repression and purge latent HIV reservoirs31, 32.
Results
Inactive HIV infections in Jurkat T cells show varying degrees of repression and differential response to NF-κB pathway activation
Inactive HIV infections of Jurkat leukemic T cells provide an opportunity to study gene expression in response to the same transcription factor from a single promoter located in different genomic environments. Here, we studied two in vitro models previously used to study HIV latency, in which clonal populations of Jurkat cells harbor a single viral integration at different genomic locations15, 24. LGIT-infected clones contain a minimal, replication-incompetent HIV- based lentiviral vector with Tat and GFP under the control of the LTR promoter (Fig. S1A)24, whereas J-Lat clones contain a full-length, replication-incompetent HIV virus with GFP in place of the Nef gene (Fig. S1B)15. In the early stages of viral gene expression, Tat and GFP are the primary proteins expressed from the full-length virus, and the mechanism of transcriptional activation is thus similar for both models18, 25. Also, both J-Lat and LGIT exhibit bimodal gene expression, where the virus can exist in a non- expressing state, or where basal expression is amplified by the Tat positive feedback loop to yield transactivated expression (Fig. 1B).
To explore a range of behaviors, we selected complementary sets of LGIT clones – in which a small fraction of the cells exhibit active transcription and the rest remain inactive – or J-Lat clones – which are generally more silent since they were originally selected to have no basal gene expression unless stimulated with TNFα (Fig. S1B)15. We compared five LGIT clones (B5, B6, D1, D3, and E3) and five J-Lat clones (6.3, 8.4, 9.2, 10.6, and 15.4) that showed low or no GFP expression from the LTR promoter in the absence of stimulation, as measured by flow cytometry (Fig. 1C). All clones were activated to some extent by NF-κB RelA via TNFα stimulation, indicating that all integrated promoters could support viral transcription (Fig. 1C); however, activation occurred to varying degrees. In general, TNFα stimulation activated a smaller fraction of J-Lat clonal populations compared to LGIT clonal populations, except for J-Lat 10.6, which was activated to a greater extent than LGIT B5. Moreover, the TNFα dose required to activate gene expression across clonal populations varied more than 10-fold (Fig. S2). Thus, the J-Lat and LGIT in vitro latency models display a range of gene expression activation in response to the transcription factor RelA in different genomic environments.
We hypothesized that differences in TNFα-mediated activation may be due to epigenetic modifications at the LTR promoter that result in higher order chromatin structure, as suggested in previous studies17, 18. Local genomic features of the integration site did not reveal any systematic differences among the clones (Table S1). To chemically probe the nature of repression at the site of integration in each clone, we added trichostatin A (TSA), an inhibitor of class I and II mammalian HDACs, or 5-aza-2′-deoxycytidine (5-aza-dC), which inhibits DNA methyltransferase (DMT) activity. Similar to TNFα treatment, TSA or 5-aza-dC stimulation activated gene expression to varying extents across the clonal populations (Fig. 1D). Therefore, the panel of inactive integrated proviruses is subject to varying degrees of chromatin repression across integration sites and exhibit differential responses to TNFα-mediated RelA activation.
LTR activation by tunable overexpression of the transcription factor NF-κB RelA revealed an activation threshold that varied significantly across clones
To more quantitatively and directly analyze how RelA activates HIV gene expression in different chromatin environments, we modified a tetracycline inducible expression system33 for variable expression of a mCherry-RelA fusion protein (iRelA, Fig. 2A). Treatment with increasing doses of doxycycline (DOX) induced a steady increase in total RelA expression relative to endogenous levels, ranging from approximately a 0.2-fold increase in RelA fusion protein relative to endogenous RelA in the absence of DOX (due to basal expression from the Tet promoter) to a 5-fold increase at high DOX concentrations (Fig. 2B and Fig. S3A). Total mCherry fluorescence varied with DOX dosage in a similar manner as protein level (Fig. S3B), confirming that the two measurements are monotonically related. Deletion of the κB sites from the HIV LTR promoter abolished activation by the inducible (iRelA) vector and TNFα, but retained activation by TSA (Fig. S3C), indicating that RelA overexpression activated the LTR via specifically binding to the LTR κB sites.
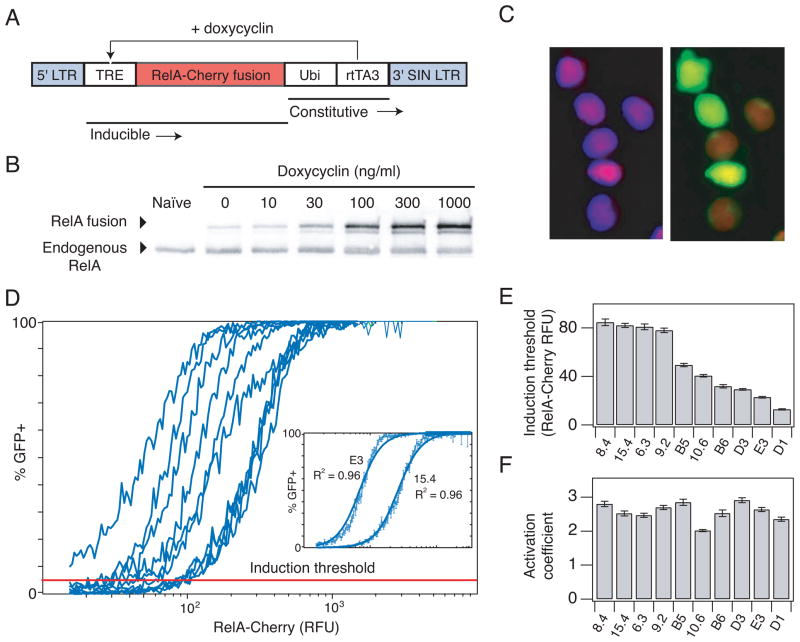
Fig. 2.
Inducing HIV gene expression by overexpression of RelA reveals an induction threshold of gene activation. (A) Schematic of the inducible RelA (iRelA) vector. (B) Immunoblot of total RelA-Cherry fusion protein and endogenous protein levels in clone 6.3 infected with iRelA 4 days after DOX induction. (C) Microscopy picture of clone 6.3 infected with iRelA 4 days after induction with 30 ng/ml DOX. (Left) DAPI and mCherry overlay. (Right) GFP and mCherry overlay. (D) Combined flow cytometry data for HIV-infected clones expressing iRelA in response to a range of DOX concentrations. More than 50,000 single cell events were divided into 256 bins of mCherry fluorescence, and the fraction of GFP+ cells was calculated and plotted for each bin. (Inset) Least squares fit line for clone 15.4 and E3. (E) Induction threshold (defined as the mCherry- RelA level at which 5% of the population expressed GFP) and (F) activation coefficient (defined as the Hill coefficient calculated from fitting Hill functions to the curves in (D)) for each clone. Error bars in (D–F) represent standard deviations and were calculated by bootstrapping.
We introduced iRelA into the panel of clones and stimulated them across the full range of RelA expression until GFP expression and RelA levels reached steady-state 4 days post DOX addition (Fig. S3D). Within individual cells, mCherry-RelA predominantly localized to the nucleus for all but the lowest RelA levels (Fig. 2C and S3E), suggesting that RelA expression had largely overcome cytoplasmic sequestration by I-κB. Stimulation of an i-RelA-infected population of cells at a particular DOX concentration produced a wide distribution of mCherry-RelA expression (Fig. S3F). Therefore, to quantify gene activation in the population directly as a function of mCherry-RelA across this full range, we pooled flow cytometry measurements across all DOX levels and subdivided the single cell data into 256 mCherry-RelA bins (Fig. S3G). Gene expression for each clone varied from minimal activation with low mCherry-RelA to fully activated (i.e. 100% of the population expressing GFP) at maximal RelA levels (Fig. 2D). The resulting gene activation curves were fit to the Hill equation after log transforming it into a linear equation:
The experimental gene activation curves were well described using the fit parameters, K and n (Fig. 2D, inset and Fig. S4) and the quality of the fits was independent of the total number of subdivisions (bins). Strikingly, we observed that gene expression in each clonal population is induced at a different level of RelA (mCherry), but after induction the increase in the GFP+ fraction as a function of RelA is similar (Fig. 2D). The mCherry-RelA level at which 5% of the population expressed GFP was defined as the induction threshold (Fig. 2D, red line), calculated using the Hill “gene activation” functions. Note that the relative difference in threshold of activation among clones was independent of the GFP level chosen for computing this metric (Fig. S5A).
The induction threshold exhibited a considerable 6-fold range of variation in mCherry fluorescence units (Fig. 2E), which was also reflected by variation in the fit parameter K, i.e. the mCherry-RelA level at half maximal GFP induction (Fig. S5B–C). In contrast, the apparent Hill coefficient n, which describes the steepness in the rise of the gene activation function, did not vary more than 1.5-fold among clones (Fig. 2F). The Hill coefficients, which we will refer to as the activation coefficients, were greater than 2, suggesting possible cooperativity in RelA- and Tat-mediated LTR activation (Fig. 1B). Notably, clones that responded more strongly to drug treatments (Fig. 1C–D) also exhibited lower induction thresholds. Tat transcripts were undetectable below the induction threshold for both LGIT- and J-Lat-infected clones, and Tat did not increase significantly until after the induction threshold was reached, indicating that any difference in transcription and Tat production between the two vectors did not affect the threshold (Fig. S6). Taken together, these data suggest that the genomic environment at the integration site affected the induction threshold of the LTR in response to RelA, but did not significantly affect progressive RelA-mediated increases in gene expression within the population once the gene had been induced.
Chromatin accessibility at the LTR across clones is strongly correlated with the RelA induction threshold
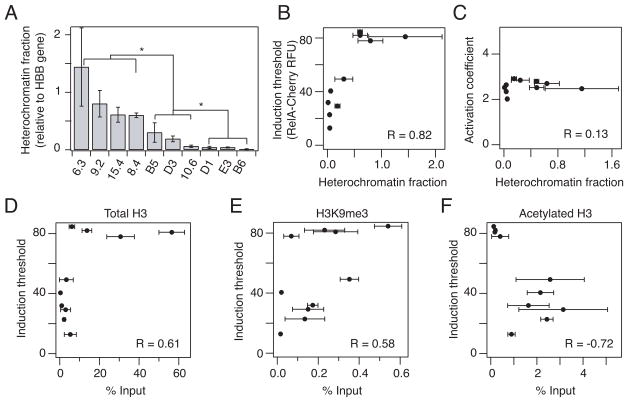
We reasoned that the local chromatin environment may affect the induction threshold by modulating chromatin accessibility at the promoter7, 34. To quantitatively compare general chromatin accessibility, we measured the extent to which chromatin limited the sensitivity to DNAse I digestion near the transcription start site (TSS) of the LTR in each clonal population35, 36. Nuclease sensitivity assay measurements of the LTR were normalized to the same measurement made on the highly repressed hemoglobin-β (HBB) reference gene37 for each clone, and we refer to this normalized metric as the heterochromatin fraction (see Materials and Methods).
The panel of inactive clones harbored proviruses in a wide range of chromatin environments, with heterochromatin fractions varying 100-fold (from clone 6.3 down to clone B6) (Fig. 3A). The differences in heterochromatin fraction could be resolved into three groups (p < 0.05 by one-way ANOVA): strong repression (> 0.5), intermediate repression (0.05–0.5), and weak repression (< 0.05). Importantly, the induction threshold (Fig. 2E) showed a strong positive correlation with heterochromatin fraction (Fig. 3B; Pearson R = 0.82, p = 0.01), suggesting that chromatin accessibility at the promoter may be a determinant of RelA levels required to initiate gene expression. In contrast, the activation coefficient n did not show a significant correlation with nuclease sensitivity (Fig. 3C), consistent with the observation that this coefficient does not vary across clones (Fig. 2F). These results suggest that activation following initial gene expression may be an intrinsic property of the promoter, whereas initiation of gene expression is strongly correlated to the local chromatin environment at the site of integration.
Fig. 3.
Chromatin accessibility is correlated with RelA induction threshold. (A) Heterochromatin fraction was quantified with a DNAse I sensitivity assay. Quantitative PCR was performed in triplicate and normalized to a hemoglobin-β (HBB) reference gene. (B–C) Correlation of heterochromatin fraction with (B) induction threshold and (C) activation coefficient extracted from the fits in Fig. 2D–F. (D–F) Chromatin immunoprecipitation for (D) total H3, (E) H3K9me3 and (F) acetylated H3 bound to the HIV promoter in unstimulated clones was correlated to the induction threshold. Quantitative PCR was performed in triplicate and normalized to an input control. Data are presented as the mean ± standard deviation. Differences are labeled as significant (*) if p < 0.05. Pearson correlation coefficient R is indicated on plot.
We next measured if the nuclease sensitivity assay was consistent with known molecular determinants of heterochromatin, and how these determinants correlated with the induction threshold and activation coefficient induced by RelA overexpression. Using chromatin immunoprecipitation, we measured the total amount of histone 3 (H3), presumably higher with increased nucleosome occupancy near the promoter; the level of H3 tri-methylation at lysine 9 (H3K9me3), associated with repressed promoters; and the level of H3 acetylation (AcH3), associated with active promoters. As anticipated, total H3 increased with increasing heterochromatin fraction and was positively correlated with the induction threshold (Fig. 3D; R = 0.61, p = 0.06). H3K9me3 levels were also generally higher for clones with higher heterochromatin fractions and also showed a positive correlation with the induction threshold (Fig. 3E; R = 0.58, p = 0.08). In contrast, total AcH3 was generally lower for increased heterochromatin fraction and negatively correlated with the induction threshold (Fig. 3F; Pearson R = −0.72, p = 0.02). The activation coefficient was not significantly correlated with total histone levels or histone modifications (Fig. S7). Therefore, the threshold level of RelA necessary to activate gene expression is significantly correlated with chromatin accessibility and molecular determinants of heterochromatin across loci.
Activation of gene expression is more strongly associated with a decrease in heterochromatin rather than an increase in RNAPII binding or phosphorylation
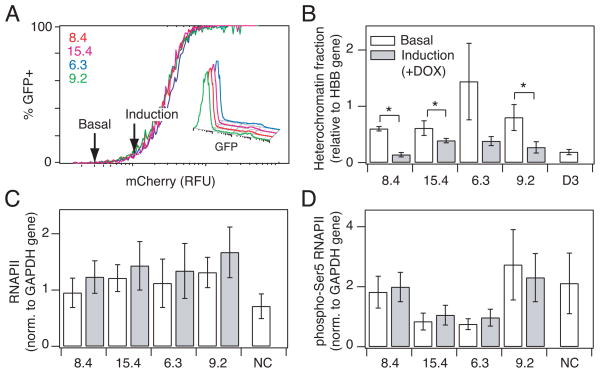
For strongly repressed clones (6.3, 9.2, 15.4, and 8.4), significant increases in RelA levels are necessary to reach an induction threshold (Fig. 2D). Therefore, we used these clones to test what quantitative features at the promoter change between the basal state and the point at which gene expression has just been initiated. Based on the measured correlations between the induction threshold and chromatin structure (Fig. 3), we hypothesized that at the point of gene expression onset, the heterochromatin fraction at the promoter may be reduced to that of clones that have induction thresholds close to the basal RelA level. We thus measured the heterochromatin fraction for each clone at a low DOX concentration (20 ng/ml) that approximately increased RelA to the induction threshold, at which point a small fraction of cells expressed GFP (5–8%; Fig. 4A, inset). The heterochromatin fraction at the induction threshold was compared to heterochromatin at the basal level for each clone (Fig. 3A), at which point <1% of cells express GFP. The level of heterochromatin at the induction threshold was reduced for all four clones, and three exhibited statistically significant decreases relative to the basal state (Fig. 4B, p < 0.05). Moreover, at the induction threshold, the measured heterochromatin fraction was not significantly different from that of clones displaying intermediate levels of repression (clones B5, 10.6, and D3; p = 0.09 by ANOVA), consistent with the hypothesis that chromatin accessibility becomes equalized at the induction threshold.
Fig. 4.
Induction of gene expression is associated with a decrease in heterochromatin fraction. (A) Selected clones were treated with 20 ng/ml DOX to hold the clonal populations at the point at which gene expression in the population is just induced (arrow). (Inset) Flow histograms showing a low fraction of cells expressing GFP for each clone at the point of induction. (B) Heterochromatin fraction as quantified by nuclease sensitivity for clones at basal (white) and induction (gray) level of RelA. Quantitative PCR was performed in triplicate and normalized to a HBB reference gene. (C–D) Chromatin immunoprecipitation comparing (C) RNA polymerase II and (D) phospho- Ser5 RNAPII bound to the LTR promoter at basal (white) and induction (gray) level of RelA. Quantitative PCR was performed in triplicate and normalized to a GAPDH control gene. Data are presented as the mean ± standard deviation. Changes are labeled as significant (*) if p < 0.05.
An alternative to alleviating promoter repression at the induction threshold would be increased recruitment of positive regulators of transcription, including RNAPII and the associated factors required for transcription initiation. We used chromatin immunoprecipitation to measure the level of total RNAPII and RNAPII phosphorylated at serine 5 (pSer5-RNAPII associated with transcription initiation) at the promoter. No significant differences in LTR-bound RNAPII or pSer5-RNAPII were measured in the basal state across the entire panel of clones (Fig. S8A–B) and both were low relative to an actively expressing GFP+ HIV-infected population (Fig. S8C–D). Moreover, no significant changes were measured between basal conditions and threshold conditions at induction for either RNAPII (Fig. 4C) or pSer5-RNAPII bound to the promoter (Fig. 4D), consistent with our measurements of Tat transcription (Fig. S6). Overall, we conclude that the heterochromatin fractions in different clones begin to converge as they reach a gene expression threshold, prior to significant increases in Tat, RNAPII binding and phosphorylation at the promoter.
Increasing chromatin accessibility via small molecule inhibitors lowers the RelA induction threshold
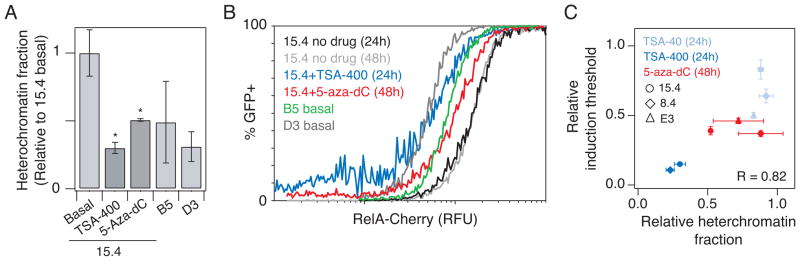
If chromatin accessibility at the integration site is a determinant of the RelA induction threshold, then increasing chromatin accessibility at the LTR promoter of strongly repressed clones, which have relatively high induction thresholds, may shift the gene activation response curves to resemble more weakly repressed clones. While TSA or 5-aza-dC did not highly activate gene expression in clone 15.4 (approximately 1–2% for both drugs), these compounds may still modulate chromatin accessibility. We thus treated 15.4 with TSA (40 or 400 nM) or 5-aza-dC (5 μM) and analyzed nuclease sensitivity following incubation times previously demonstrated to be sufficient for producing measurable changes in H3 acetylation (4 hours for TSA)16 or DNA methylation (48 hours for 5-aza-dC)38. Nuclease sensitivity depended on TSA dosage (Fig. S9A). In addition, the higher 400 nM TSA dosage induced an approximately 3-fold decrease in the heterochromatin fraction, and 5 μM 5-aza-dC decreased the heterochromatin fraction by 2-fold, bringing these fractions into intermediate levels of basal heterochromatin (clones B5 and D3; Fig. 5A).
Fig. 5.
Increasing chromatin accessibility via drug treatment lowers the RelA induction threshold. (A) Heterochromatin fraction for clone 15.4 was quantified with a DNAse I sensitivity assay following stimulation with 400 nM TSA for 4 hours or with 5 μM 5-aza- dC for 48 hours. Quantitative PCR was performed in triplicate and normalized to the hemoglobin reference gene. Relative heterochromatin fraction was calculated by normalizing clone 15.4 with drugs, B5 and D3 to the unstimulated 15.4 control. (B) Combined flow cytometry data for 15.4 expressing iRelA in response to a range of DOX concentrations and simultaneous stimulation with 400 nM TSA for 24 hours (dark blue), 5 μM 5-aza-dC for 48 hours (red), and no drug treatment controls at 24 and 48 hours (black and light gray, respectively). iRelA dose response curves for clone B5 (green) and D3 (dark gray) without TSA or 5-aza-dC are included for comparison. (C) Relative change in induction threshold versus relative change in heterochromatin fraction for clones 15.4 (circles), 8.4 (diamonds) and E3 (triangles). Data for 15.4 are calculated from results presented in (A) and (B), and data for 8.4 and E3 are calculated from experiments presented in Fig. S10. All points are calculated by normalizing the value of heterochromatin fraction or threshold for the clone in the presence of drugs to the corresponding value for the unstimulated control clone. Data are presented as the mean ± standard deviation. Changes are labeled as significant (*) if p < 0.05. Pearson correlation coefficient R is indicated on plot.
To determine whether these shifts in chromatin accessibility lower the induction threshold for clone 15.4, we repeated the DOX induction of RelA-mediated gene activation in the presence of inhibitors at time points before these compounds affected cell viability (24 hours for TSA and 48 hours for 5-aza-dC). We then fit the resulting curves to the Hill equation (as in Fig. 2D) and extracted new values for the threshold and activation coefficient that define a new gene activation function. As anticipated, the induction threshold in the presence of either TSA or 5-aza-dC was significantly decreased compared to the control (7-fold and 2.5-fold, respectively; Fig. S9B) and importantly resulted in gene activation curves that resembled those of clones that had intermediate heterochromatin fractions (Fig. 5B and Fig. 3A). By comparison, the activation coefficient was modestly lower following drug treatment (approximately 25% and 40%, respectively; Fig. S9C). Furthermore, TSA and 5-aza-dC had similar effects on another repressed clone, 8.4, again inducing increased nuclease sensitivity and a lower induction threshold (Fig. S10A–D). Finally, we investigated whether increasing the chromatin accessibility could further reduce the RelA induction threshold of even a weakly repressed clone. Consistent with results for the two highly repressed clones, reducing the heterochromatin fraction for the weakly repressed clone E3 with TSA or 5-aza-dC caused a decrease in induction threshold (Fig. S10E–H).
When TSA and 5-aza-dC results were combined for all clones tested (15.4, 8.4, and E3), we observed that the change in heterochromatin fraction induced by inhibitor treatment showed a strong positive correlation with the resulting change in RelA induction threshold (Fig. 5C; R = 0.82, p = 0.03). This observation further supports the correlative relationship between chromatin accessibility and the RelA level required for induction observed for clones across different integration positions (Fig. 3B). Taken together, these data demonstrate that chromatin accessibility at the HIV promoter sets a threshold for transcription factor-induced activation and that altering chromatin accessibility via multiple epigenetic pathways shifts this induction threshold.
Gene activation functions account for synergistic increases in HIV gene activation following treatment with epigenetic modifiers and TNFα
Our small molecule perturbation data demonstrated that when the heterochromatin fraction for a repressed clone (e.g. clone 15.4) is decreased via small molecule inhibitors, chromatin accessibility is increased and the gene activation curve (or function) shifts such that it responds at lower RelA levels, similar to more weakly repressed clones. Since more weakly repressed clones also respond more robustly to TNFα stimulation (Fig. 1C), we considered whether the empirically measured gene activation function, i.e. gene expression as a function of RelA, could accurately predict gene expression in response to combined HDAC inhibition and NF-κB activation. Such predictions may be relevant to HIV latency therapy, as combinatorial treatment with a HDAC inhibitor and an activator of the TNF pathway has recently been observed to result in synergistic activation for in vitro HIV latency models (including J-Lat and LGIT)18, 31, 32.
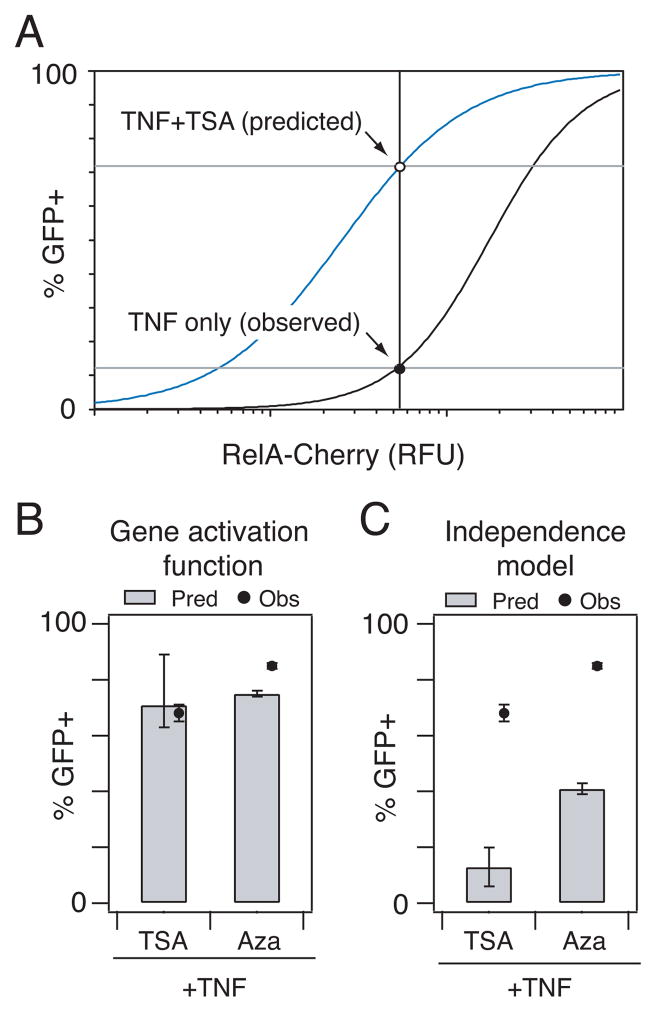
To predict potential synergistic effects, we first inferred the approximate mCherry-RelA level associated with TNFα stimulation of clone 15.4 from earlier data (Fig. 1C). We then used this mCherry level and the 15.4+TSA activation function (Fig. 5B) to predict the population fraction activated in response to both TSA and TNFα (Fig. 6A). The 15.4+TSA gene activation function predicted a combined response of 71%, very close to the measured response of 68% (Fig. 6B). In contrast, when these data were used to predict combined responses under the assumption of Bliss independence 39, the expected activation in response to TNFα+TSA was 13% (i.e., 12% in response to TNFα only and 1% in response to TSA only) (Fig. 6C).
Fig. 6.
Gene activation function accurately predicts synergistic activation of HIV gene expression by simultaneous treatment with TNFα and HDAC or DMT inhibitors. (A) The empirically-derived gene activation function for clone 15.4+TSA was used to predict its response to combinatorial perturbation with TSA and TNFα. Approximate mCherry- RelA increases associated with TNFα treatment alone were estimated by locating the point on the gene activation curve for basal clone 15.4 that corresponded to the percentage of GFP+ cells that responded to TNFα treatment (~12%) (black line). This estimated TNFα-induced value of mCherry-RelA was used to predict the fraction of GFP+ cells expected for a combination of TNFα and TSA by solving the gene activation function for 15.4 treated with TSA (blue line). (B–C) Predicted (bars) and observed (dots) percentage of GFP+ cells following stimulation with TSA+TNFα or 5-aza-dC+TNFα based on (B) gene activation functions or (C) a Bliss independence model of drug response. Experiments were performed in biological triplicate and are presented as the mean ± standard deviation. Error bars for prediction were calculated as described in Materials and Methods.
Also, the gene activation curve for 15.4+5-aza-dC accurately predicted synergistic gene activation in response to combined TNFα and 5-aza-dC stimulation (75% predicted activation versus 84% measured activation), while the Bliss independence model predicted only 41% activation (Fig. 6B–C). Gene activation functions derived for clone 8.4 treated with TSA or 5-aza-dC also predicted gene expression in response to a combination of TSA+TNFα or 5-aza-dC+TNFα more accurately than a Bliss independence model of drug response (Fig. S11). Our analysis collectively suggests that the predicted synergy occurred because treatment with TSA or 5-aza-dC lowered the RelA induction threshold significantly via increasing chromatin accessibility (Fig. 5C), such that TNFα-induced RelA activation resulted in a non-linear increase in population gene expression. Our prediction and observation that TSA and 5-aza-dC combine non- linearly with TNFα to stimulate gene expression is similar to the experimental synergy observed in vitro between activators of RelA and HDAC or DMT inhibitors in combinatorial anti-latency therapy strategies18, 31, 32, 38.
Discussion
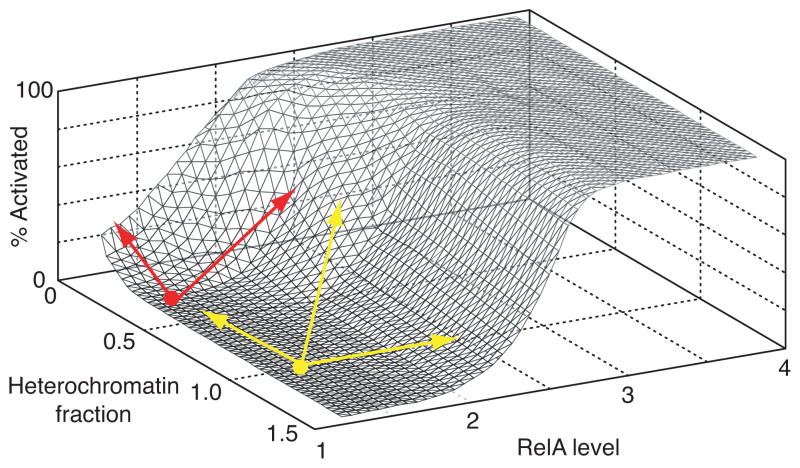
We have investigated how RelA level and features of the local chromatin environment quantitatively regulate the activation of HIV gene expression in a cell population. We demonstrated that gene expression is only induced when the cellular RelA level is brought above an induction threshold set by chromatin accessibility at the integration site, or conversely if chromatin accessibility is increased such that the induction threshold dips below the basal RelA levels (Fig. 2D and Fig. 5). A 3-D surface was constructed to incorporate and summarize data from Figs. 2 and 5 and to thereby show gene activation as a function of RelA for different genomic locations (Fig. 7). This functional surface – which offers the information discussed in Fig. 1A – indicates that the semi-random integration of HIV into the human genome causes it to sample a wide spectrum of chromatin environments that would lead integrated virus to respond differentially to global cellular activation, or to small molecule interventions designed to therapeutically activate gene expression.
Fig. 7.
3-D surface plot demonstrates gene activation as a function of RelA for different genomic locations. The plot was empirically derived by combining the gene activation functions for a subset of clones ranging from high to low repression. Surface plot provides a quantitative depiction of the function hypothesized in Fig. 1A. Yellow and red points and arrows describe behavior in different regimes of promoter repression. See text for discussion.
To qualitatively understand how the genomic environment of latent HIV infections may alter the response to small molecule activation, three regimes of gene expression “potential” may be considered (Fig. 7). In regime 1, proviruses are close to the induction threshold such that increasing either chromatin accessibility or RelA level will result in almost full activation of the population (Fig. 7, red). In regime 2, the level of RelA required to reach the induction threshold is sufficiently far from basal RelA such that increasing chromatin accessibility or raising RelA level alone will not be enough to activate the population, but moving along both axes will lead to activation (Fig. 7, yellow). Finally, it may be possible to have a promoter with sufficiently low chromatin accessibility (i.e. near the lower left corner of the functional surface) such that no combination of epigenetic modifiers and RelA activators will overcome the induction threshold and activate gene expression, though this scenario is outside the range of our experimental data. If other transcription factors that activate HIV display gene activation functions that are similar to RelA, then these infections may be difficult to activate therapeutically, but also may never result in a productive infection in activated T cells in vivo.
The vectors compared in our study contain differences in sequence, Tat expression and splicing, and viral accessory proteins that could affect the threshold behavior. However, we demonstrated that Tat transcription is extremely low prior to reaching the induction threshold (Fig. S6). Furthermore, when measurements of chromatin accessibility and induction threshold under different conditions are separated by vector type, the strong correlation between heterochromatin fraction and induction threshold is maintained (Fig. S12). Although we think it is likely that each vector and selection strategy may optimally select for a particular range of chromatin environments, our data strongly support chromatin accessibility as the primary determinant of the induction threshold.
Induction thresholds set by chromatin have previously been shown in S. cerevisiae to be a mechanism for fine-tuning gene expression in response to transcription factors. Specifically, the affinity of the transcription factor Pho4 for its binding site in the PHO5 promoter sets a threshold for PHO5 activation by determining the level of Pho4 necessary to remodel a nucleosome positioned over the TSS7, 8. Interestingly, other transcription factor binding sites in the PHO5 promoter serve to scale expression after chromatin remodeling, suggesting that the two steps are independent. This is similar to our finding that the induction threshold for gene expression in the population is set by the local chromatin environment, but the increase in RelA-mediated gene activation and maximum fraction of activation achievable in the population is not. In the PHO5 study, the affinity of the Pho4 binding site was directly modified by introducing promoter mutations7, 8. In our study, the affinity of RelA for the κB sites on the LTR promoters is the same, and it is instead the chromatin accessibility at the site of integration that tunes the level of cellular RelA required for sufficient chromatin remodeling to activate gene expression. Further measurements are needed to determine if RelA binding to the promoter is directly or indirectly affected by changes in the affinity of nucleosomes for the LTR promoter.
The more general idea that chromosomal location modulates gene expression has been increasingly investigated since the study of position effect variegation40. Our results explore how chromatin context quantitatively impacts activation by a single transcription factor input, and suggest that chromatin environment within the mammalian genome can threshold the activation of different genes to the same transcription factor, without significantly affecting the transcription factor-mediated expression after gene expression is induced in the population. Such a mechanism potentially contributes to observed differential activation of genes in response to proinflammatory stimuli27, where stimulation by proinflammatory cytokines resulted in two waves of NF-κB recruitment to target genes – early and late – that are primarily differentiated by the chromatin configuration at the promoter and not the affinity of the binding site3. Our analysis was performed at steady-state but could be extended to examine the role of a chromatin threshold in the dynamics of NF-κB recruitment and gene activation.
A recent genome-wide study of glucocorticoid receptor (GR) binding demonstrated that for a large majority of GR binding motifs, cell-specific differences in pre-existing patterns of chromatin accessibility at GR binding sites were a primary determinant of cell-selective GR occupancy, leading to cell-specific gene expression patterns34. Our results also show that chromatin accessibility prior to stimulation plays a major role in determining NF-κB-mediated gene expression from the LTR, and thus appear to support an emerging general mechanism of how chromatin modulates transcription factor–gene interaction specificity in diverse biological systems. Because transcription factor binding in response to exogenous stimuli underlies all biological processes, a quantitative understanding of how these interactions are regulated by the local chromatin environment are important to decipher input-output responses of a cell.
Materials and Methods
Plasmids
LGIT has been previously described24. The inducible RelA (iRelA) vector was based on a single lentiviral vector platform for tetracycline-regulated expression of the product 33. The mCherry fluorescent protein was fused to the N-terminus of RelA by splice overlap PCR 41 and then cloned into the pEN-Tmcs (ATCC). The pEN-Cherry- RelA fusion plasmid was cloned into the pSLIK-Venus plasmid (ATCC) by LR recombination reaction (Invitrogen) as previously described33, and the IRES-Venus sequence was removed. Cloning details and the final plasmid map is available upon request.
Cell culture
Jurkat cells and HEK 293T cells (used for lentiviral packaging) were cultured as previously described26. LGIT clones were sorted and cultured as previously described26. J-Lat full length clones15 were obtained from the laboratory of Dr. Eric Verdin via the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Viral harvesting and infection of iRelA cell lines
Lentiviral vectors were packaged as previously described42. For infection with the iRelA vector, 3×105 LGIT and J-Lat clones were grown in 12-well plates and infected at a multiplicity of infection of 0.6. Four days later, infected cells were stimulated with 1 μg/ml doxycyclin for 48 hours and the top quartile of the mCherry-expressing population was sorted on a Cytopeia InFlux cell sorter (BD Biosciences). The sorted iRelA cell lines populations were expanded and frozen stocks were stored in liquid nitrogen.
Drug Stimulation
The LGIT and J-Lat cell lines and the corresponding iRelA cell lines were treated with the following pharmacological agents for the indicated times and analyzed by flow cytometry: TNF-α at 20 ng/mL (24 or 48 hours post-stimulation), TSA at 40 nM or 400 nM (24 hours), and 5-aza-dC at 5 μM (48 hours). For the iRelA cell line stimulations, cells were treated with DOX at 0, 10, 30, 100 or 300 ng/mL or as indicated in the text.
Fitting the gene activation functions
For each iRelA cell line, flow cytometry data collected from DOX stimulation at 0, 10, 30, 100 and 300 ng/mL were combined. The data were binned into 256 GFP and mCherry channels. For each mCherry channel, the percentage of GFP+ cells was computed and plotted, as in Fig. 2D. The Hill equation was log transformed into a linear equation and the curves in Fig. 2D were fit by least squares as shown in Fig. S4. The quality of the fits did not improve by changing the number of bins. The slope and intercept obtained from the least squares regression was used to compute the threshold and activation (Hill) coefficient (Fig. 2E–F). iRelA cell lines stimulated with chromatin modifying enzymes and DOX were analyzed similarly. Standard deviations for the threshold and activation coefficient were bootstrapped using 1000 bootstrapped data samples.
Nuclease sensitivity assay
The nuclease sensitivity assay was performed using the EpiQ™ Chromatin Analysis Kit (Bio-Rad) with minor modifications of the manufacturer’s protocol. Briefly, 250,000 cells were incubated with DNAse I for 1 hour. Enzyme concentrations were adjusted to account for the range of nuclease sensitivities being compared (1X for measuring drug response in LGIT E3 and 3X for measuring basal chromatin across clones and drug response in J-Lat clones). Following extraction and purification of the genomic DNA, the level of HBB and LTR were quantified by qPCR. The primers used were: 5′-GGACTTTCCGCTGGGGACTTTCCAGGG-3′ (forward) and 5′-GCGCGCTTCAGCAAGCCGAGTCCTGCGTCGAG-3′ (reverse). Primers were designed to prime within the DNase hypersensitive site located inside the core promoter and cover the binding site of nucleosome-1, a nucleosome whose remodeling is associated with activation of the latent promoter20, 21.
Chromatin immunoprecipitation
Upstate EZ ChIP Kit Reagents (Upstate) and protocols were used with minor modifications as previously described 26. DNA isolated from ChIP was quantified by quantitative PCR (BioRad iCycler, iQ5) using the EpiQ Chromatin SYBR Supermix. qPCR was performed in triplicate and melt curves were run to ensure product specificity. See Supplementary Methods for additional protocol details including a list of antibodies and primers.
Combinatorial drug predictions
The GFP+ fraction activated by TNFα+TSA or TNFα+5-aza-dC according to the model of Bliss independence (μTNF+inh,BLISS) was calculated as follows: μTNF+inh,BLISS = 1 – (1-μTNF)*(1-μinh) where μTNF and σTNF and μinh and σinh are the mean and standard deviation of the GFP+ fraction activated by TNFα and by TSA or 5-aza-dC, respectively. For predictions using the gene activation functions, for each clone of interest we located the point on the basal gene activation curve that corresponded to μTNF ± σTNF and used this to estimate the approximate mCherry-RelA increase, nTNF-RelA ± eTNF-RelA associated with TNFα treatment alone (where eTNF-RelA is the uncertainty in mCherry-RelA associated with σTNF). Finally, μTNF+inh,GA ± σTNF,GA was calculated by solving the empirical gene activation function for the clone of interest in the presence of drug treatment (clone+TSA or clone+5-aza-dC) at the point nTNF-RelA ± eTNF- RelA.
Statistical analysis
We used Student’s t-test to compare two means, and two-factor ANOVA to compare heterochromatin fraction across different clonal groups. Significance of Pearson correlation coefficients was calculated according to the following formula for the t statistic: t = r*[(1−r2)/(n−2)]−1/2 where r is the Pearson correlation coefficient and n is the sample size.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM73058 (to D.V.S. and A.P.A.) and National Institutes of Health National Research Service Award 1F32AI072996-01A2 (to K.M.-J.).
Footnotes
Author contributions: K.M.-J., S.S.D. and D.V.S. designed research; K.M.-J., S.S.D., and N.P. performed research; J.E.F. contributed analytic tools; K.M.-J., S.S.D. and D.V.S. analyzed the data; K.M.-J., S.S.D., A.P.A., and D.V.S. wrote the paper.
The authors declare no conflict of interest.
References
- 1.Segal E, Widom J. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Trends Genet. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saccani S, Pantano S, Natoli G. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smale ST. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambeyron S, Bickmore WA. Genes & Development. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portela A, Esteller M. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 7.Lam FH, Steger DJ, O’shea EK. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HD, O’shea EK. Nat Struct Mol Biol. 2008;15:1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kundu S, Peterson CL. Biochim Biophys Acta. 2009;1790:445–455. doi: 10.1016/j.bbagen.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skupsky R, Burnett JC, Foley JE, Schaffer DV, Arkin AP. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller-Jensen K, Dey SS, Schaffer DV, Arkin AP. Trends in Biotechnology. 2011 doi: 10.1016/j.tibtech.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. Trends Mol Med. 2004;10:525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu S-Y, Chiang C-M, Karn J. EMBO J. 2006;25:1–9. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barboric M, Yik JHN, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. Nucleic Acids Research. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan A, Bisgrove D, Verdin E. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams SA, Chen L-F, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Journal of Virology. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I, Hope TJ. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakre S, Chavez L, Shirakawa K, Verdin E. Current Opinion in HIV and AIDS. 2011;6:19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- 20.Verdin E, Paras P, Van Lint C. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.el Kharroubi A, Verdin E. J Biol Chem. 1994;269:19916–19924. [PubMed] [Google Scholar]
- 22.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 23.Feinberg MB, Baltimore D, Frankel AD. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Weinberger LS, Dar RD, Simpson ML. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 26.Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. PLoS Pathog. 2009;5:e1000260. doi: 10.1371/journal.ppat.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natoli G. Cold Spring Harb Perspect Biol. 2009:1. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Proc Natl Acad Sci U S A. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 30.Chan JK, Greene WC. Curr Opin HIV AIDS. 2011;6:12–18. doi: 10.1097/COH.0b013e32834124fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuse S, Calao M, Kabeya K, Guiguen A, Gatot J-S, Quivy V, Vanhulle C, Lamine A, Vaira D, Demonte D, Martinelli V, Veithen E, Cherrier T, Avettand V, Poutrel S, Piette J, de Launoit Y, Moutschen M, Burny A, Rouzioux C, De Wit S, Herbein G, Rohr O, Collette Y, Lambotte O, Clumeck N, Van Lint C. PLoS ONE. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnett JC, Lim K-I, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. Journal of Virology. 2010;84:5958–5974. doi: 10.1128/JVI.00161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin K-J, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang J-I, Rebres R, Roach T, Seaman W, Simon MI, Fraser IDC. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- 36.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Nat Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- 38.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. PLoS Pathog. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.BLISS CI. Bacteriol Rev. 1956;20:243–258. doi: 10.1128/br.20.4.243-258.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henikoff S. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 41.Heckman KL, Pease LR. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 42.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.