Abstract
The ontogeny of the first four families of cytochromes P450 (P450s) (i.e., Cyp1–Cyp4) can affect the biotransformation of drugs and dietary chemicals in liver, resulting in unique pharmacological reactions in children. Because genome-scale investigations have identified many novel P450 isoforms, it is critical to perform a systematic characterization of these P450s during liver development. In this study, livers were collected from C57BL/6 mice 2 days before birth and at various postnatal ages (0–45 days of age). The mRNA levels for 75 P450 isoforms (Cyp1–Cyp4) were quantified with branched DNA assays and reverse transcription-polymerase chain reaction assays. More than half of the mouse P450s are conserved in humans, but there are more isoforms in mice. The P450 mRNA levels increased after birth in mouse liver, forming four distinct ontogenic patterns. The majority of P450s form a total of eight genomic clusters, namely, Cyp1a1 and Cyp1a2 genes on chromosome 9 (cluster 1), Cyp2a, Cyp2b, Cyp2f, Cyp2g, and Cyp2t genes on chromosome 7 (cluster 2), Cyp2c genes on chromosome 19 (cluster 3), Cyp2d genes on chromosome 15 (cluster 4), Cyp2j genes on chromosome 4 (cluster 5), Cyp3a genes on chromosome 5 (cluster 6), Cyp4a, Cyp4b, and Cyp4x genes on chromosome 4 (cluster 7), and Cyp4f genes on chromosome 17 (cluster 8). Some P450 isoforms within the same genomic cluster showed similar ontogenic patterns. In conclusion, the present study revealed four patterns of ontogeny for P450s in liver and showed that many P450s within a genomic cluster exhibited similar ontogenic patterns, which suggests that some P450s within a cluster are likely regulated by a common pathway during liver development.
Introduction
The cytochromes P450 (P450s) are membrane-bound enzymes with widespread diverse functions. According to the most-recently updated genomic database, 57 functional P450s have been identified in humans and 102 P450 isoforms in mice (Nelson et al., 2004). The P450 proteins are categorized into families and subfamilies on the basis of their sequence similarities. Humans have 18 families of P450s, which include 44 subfamily members (CYP1–CYP5, CYP7, CYP8, CYP11, CYP17, CYP19, CYP20, CYP21, CYP24, CYP26, CYP27, CYP39, CYP46, and CYP57) (Nelson et al., 2004).
It is generally considered that P450s in families 1 to 4 are critical and inducible components of phase I xenobiotic metabolism in various species (Wei et al., 2000; Estabrook, 2003; Kang et al., 2007; Li et al., 2007), and many are also important for metabolizing lipids, including steroids (CYP2), bile acids (CYP3A), fatty acids (CYP4), and many other endogenous compounds (Nebert and Russell, 2002). In contrast, other P450 families specialize in endobiotic metabolism (Nebert and Russell, 2002). Whereas genetic mutations in P450s are responsible for various types of inborn errors of metabolism and human diseases (Nebert and Russell, 2002; Caldwell, 2004), induction of some P450s is a risk factor for adverse drug interactions. Previous studies revealed that levels of the mRNAs for Cyp1 to Cyp4 are increased by ligands for four classes of xenoreceptors, namely, the aryl hydrocarbon receptor, the constitutive androstane receptor (CAR), the pregnane X receptor, and peroxisome proliferator-activated receptor α (Petrick and Klaassen, 2007). Although these receptors have overlapping targets, it is generally considered that the aryl hydrocarbon receptor is responsible for mRNA induction of CYP1, CAR for CYP2, pregnane X receptor for CYP3, and peroxisome proliferator-activated receptor α for CYP4.
The liver is an essential organ for drug metabolism and nutrient homeostasis. Many well characterized P450s have been found to be enriched in adult liver (Nebert and Russell, 2002). Before birth, the liver is mainly a hematopoietic organ, with very limited capacity for drug metabolism, in both humans and mice. After birth, however, the liver becomes the major organ for processing drugs and other chemicals. In 1959, it was reported that a number of drugs that were metabolized by P450s in liver microsomes from adult rabbits were not metabolized in livers of newborn rabbits (Fouts and Adamson, 1959). At that time, only one or two P450 enzymes were thought to exist. Later, when several P450 genes had been cloned, the ontogeny of a few P450s was characterized in human livers (de Wildt et al., 1999; Blake et al., 2005; Leeder et al., 2005; Gaedigk et al., 2006; Hines, 2007). Those data were rather fragmentary, however, in that at most 10 P450 isoforms were studied in each investigation, and no investigators aimed to characterize all of the drug-metabolizing P450s systematically. Two exciting findings then were reported. First, the Human Genome Project was declared complete in 2003, which indicated that there were 57 human P450s. Second, the National Center for Biotechnology Information database was released, which indicated that there were 102 mouse P450s (Nelson et al., 2004). Many of the newly identified P450s are still considered “orphans,” that is, they have no known functions. Very little is known about the ontogeny of novel P450s during postnatal liver development.
Mice have been a popular research tool for studies of the functions and ontogeny of P450s (Choudhary et al., 2003, 2005; Hart et al., 2009). For mechanistic studies, moral, ethical, and technical limitations in studies with human fetal and neonatal livers have precluded in-depth investigations of the mechanisms underlying regulation of the ontogeny of human P450s (Rowell and Zlotkin, 1997). To elucidate the regulatory mechanisms governing the maturation of P450s, the mouse model is used in the present study to minimize the influences of genetics, environment, diet, and tissue scarcity. Previously, we identified three patterns of P450 gene expression during postnatal liver maturation in mice (Hart et al., 2009). However, only 19 P450s were included. Whether the novel P450 genes in the first four families are expressed in liver and their ontogenic patterns during postnatal liver maturation remain to be determined.
The purpose of the present study was to determine systematically the ontogeny of 75 P450s in the Cyp1 to Cyp4 families in mouse liver (Renaud et al., 2011). We also compared their sequence homology with human forms and determined the correlations between their chromosomal locations and coordinate gene expression. Successful completion of the blueprint for the ontogeny of novel P450s should assist in predictions of the functions of the orphan P450s during liver development.
Materials and Methods
Animals.
Eight-week-old C57BL/6 breeding pairs were purchased from Charles River Laboratories (Wilmington, MA). Mice were housed according to Association for Assessment and Accreditation of Laboratory Animal Care guidelines and were bred under standard conditions at the University of Kansas Medical Center. Breeding pairs were established at 4:00 PM and separated the following day at 8:00 AM. The body weights of female mice were recorded each day, to monitor pregnancy status. Livers from offspring were collected at the following ages: 2 days before birth and 0 (immediately after birth and before the start of lactation), 1, 3, 5, 10, 15, 20, 30, and 45 days of age. The time points used in the present study were selected on the basis of previous publications on the ontogeny of drug-processing genes in developing mouse liver and other critical drug-metabolizing organs (Cheng et al., 2005; Maher et al., 2005; Alnouti and Klaassen, 2006, 2008; Alnouti et al., 2006; Cheng and Klaassen, 2009; Cui et al., 2009). Because of the small size of the livers in young mice (from 2 days before birth to 5 days of age), samples from male and female offspring (same litter) were pooled at each age to achieve the minimal amount of liver needed for subsequent analyses. After 10 days of age, male and female mice were separated. Livers were frozen immediately in liquid nitrogen and stored at −80°C. All animal procedures were reviewed and approved by the institutional animal care and use committee at the University of Kansas Medical Center.
RNA Isolation.
Total RNA was isolated by using RNAzol Bee reagent (Tel-Test Inc., Friendswood, TX) according to the manufacturer's protocol. RNA concentrations were quantified by using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE), at a wavelength of 260 nm. The integrity of the total RNA samples was evaluated through formaldehyde-agarose gel electrophoresis and confirmed with observation of 18S and 28S rRNA bands.
Selection of P450 Genes.
Because the first four P450 gene families (Cyp1, Cyp2, Cyp3, and Cyp4) are known to be important in drug metabolism, 75 known or novel P450 isoforms in these gene families were included in the present study. These 75 P450 genes were selected on the basis of searches of PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), the cytochrome P450 database established by Dr. David Nelson (http://drnelson.uthsc.edu/cytochromeP450.html), HighWire (http://highwire.stanford.edu/), and BioGPS (http://biogps.gnf.org). Pseudogenes were excluded from the present study. The human homologs for the mouse P450s (if present), as well as their DNA and protein identities, are shown in Supplemental Table 1.
mRNA Quantification.
The mRNA levels for 75 P450s from the Cyp1, Cyp2, Cyp3, and Cyp4 gene families were determined from total RNA samples (n = 4 or 5 per group). The majority of these P450 mRNAs (55 mRNAs) were quantified with the QuantiGene Plex 2.0 branched DNA assay (panels 21211, 21212, and 21213; Panomics/Affymetrix, Fremont, CA). In brief, individual bead-based oligonucleotide probe sets specific for each gene examined were developed by Panomics/Affymetrix (the probe sequences are available at www.panomics.com). Samples (400 ng of total RNA per sample) were analyzed by using a Bio-Plex 200 system array reader with Luminex 100 X-MAP technology, and data were acquired by using Bio-Plex Data Manager 5.0 software (Bio-Rad Laboratories, Hercules, CA). Assays were performed according to the manufacturer's protocol. Data are expressed as relative light units specific to mRNA expression, with normalization to levels of the internal control Gapdh. The gene names, accession numbers, and panel information are shown in Supplemental Table 2. Because of high levels of sequence homology, the other 19 P450 mRNAs were quantified with quantitative RT-PCR, to avoid cross-reaction. Reverse transcription of total RNA to cDNA was performed by using high-capacity reverse transcription kits (Applied Biosystems, Foster City, CA). In brief, equal volumes of 50 ng/μl RNA and 2× reverse transcription reaction mixture were combined and placed in an Eppendorf Mastercycler (Eppendorf North America, New York, NY), under the following conditions: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. Quantitative PCR was performed with the resulting cDNA. Primers for quantitative PCR were designed by using Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast), and the bit scores and e-values were determined through a BLAST search (www.blast.ncbi.nlm.nih.gov/Blast.cgi) of the mouse genomic and transcript database. Primer target specificity was determined to investigate whether off-target sequence matches had a bit score difference of >6 bits, compared with the target sequence. In this regard, it should be noted that the primers for Cyp3a59 (bit score = 40.1; e-value = 0.013) matched the off-target sequence of Cyp3a25 (bit score = 36.1; e-value = 0.21). All primers were synthesized by Integrated DNA Technologies (Coralville, IA). For each reaction, the PCR mixture contained 12.5 μl of Applied Biosystems SYBR Green PCR Master Mix, 2.5 μl of a 3 μM forward and reverse primer mixture, 5 μl of RNase-free H2O, and 5 μl of a 2 ng/μl cDNA solution. Reactions were seeded in a 96-well optical reaction plate (Applied Biosystems), and fluorescence was quantified in real time by using an Applied Biosystems 7300 real-time PCR system under the following conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles of amplification (95°C for 15 s and 60°C for 1 min), and finally dissociation (95°C for 15 s, 60°C for 30 s, and 95°C for 15 s). Standard curves were generated for each target to determine the relative amount of transcript present in the sample. Relative amounts of P450 transcripts were then normalized to amounts of the housekeeping transcript Gapdh. The gene names, accession numbers, and forward and reverse primer sequences are listed in Supplemental Table 3.
Statistical Analysis.
Statistical differences in P450 mRNA expression between male and female mice were determined by using two-tailed Student's t tests, with significance set at p < 0.05. The mRNA ontogeny of the 73 detected P450 isoforms was analyzed with a two-way hierarchical clustering method (JMP 8.0 software; SAS Institute, Cary, NC) using Ward's minimal variance, and results were visualized with a dendrogram. Distances between genes reflect the significance of associations. Red represents relatively higher and blue lower expression.
Results
Sequence Homology of P450s between Mice and Humans.
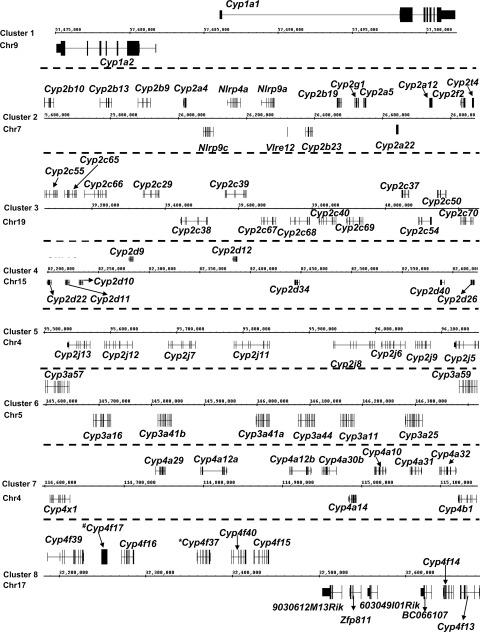
In this study, the National Center for Biotechnology Information HomoloGene Database (http://www.ncbi.nlm.nih.gov/) was used to determine the homologous P450 genes in mice that are conserved for human P450s, as well as P450 isoforms that are unique in mice. Supplemental Table 1 lists the mouse P450 gene names, their human homologs (if present), mouse gene identification numbers, mouse mRNA and protein accession numbers, identities of DNA and protein sequences for mice and humans, and chromosomal locations of the mouse P450 genes. Among these 75 P450s in mice, 39 genes are conserved in humans, whereas 36 genes are mouse P450 isoforms with no apparent human homologs. For the conserved P450s, some mouse genes display more variations in their isoforms than do human genes, because many human P450s have multiple mouse homologs. For example, human CYP2C19 is homologous to three mouse P450 isoforms, namely, Cyp2c37, Cyp2c50, and Cyp2c54; human CYP3A4 is homologous to five mouse P450 isoforms, namely, Cyp3a11, Cyp3a16, Cyp3a41a, Cyp3a41b, and Cyp3a44; human CYP3A43 is homologous to mouse Cyp3a57 and Cyp3a59; and human CYP4A22 is homologous to mouse Cyp4a12a and Cyp4a12b (Supplemental Table 1). The other human P450s have only one homologous gene in mice. The majority of P450s form a total of eight genomic clusters on various mouse chromosomes (Fig. 1). The only mouse P450 genes found in isolation (i.e., they do not form clusters with other P450s) include Cyp1b1, Cyp2c44, Cyp2e1, Cyp2u1, Cyp2w1, Cyp3a13, Cyp4f18, and Cyp4v3. The Cyp1a and Cyp3a gene families have human homologs for all of the genes within these two genomic clusters (chromosome 9 for Cyp1a and chromosome 5 for Cyp3a) (Fig. 1, clusters 1 and 6), which highlights their functional significance during evolution. In contrast, in cluster 2, only 4 of 11 mouse Cyp2a to Cyp2af genes have human homologs; in cluster 3, only 5 of 14 mouse Cyp2c genes have human homologs. Cyp2d22 is the only gene within cluster 4 that has a human homolog (CYP2D6), and Cyp2j6 is the only gene within cluster 5 that has a human homolog (CYP2J2). In addition, 6 of 10 Cyp4 genes within cluster 7 and five of seven Cyp4 genes within cluster 8 have human homologs (Supplemental Table 1; Fig. 1).
Fig. 1.
Genomic locations of P450 gene clusters from Cyp1 (cluster 1), Cyp2 (clusters 2–5), Cyp3 (cluster 6), and Cyp4 (clusters 7 and 8) gene families. The image was obtained with the Affymetrix Integrated Genome Browser. Genes above the chromatin coordinates are transcribed from the plus strand, and genes below are transcribed from the minus strand. *, pseudogene; #, gene annotated in the National Center for Biotechnology Information database but not in the Integrated Genome Browser. Arrows connect gene names to the loci. Dashed lines separate panels from each other. Chr, chromosome.
Ontogeny of mRNA for Cyp1 Gene Family.
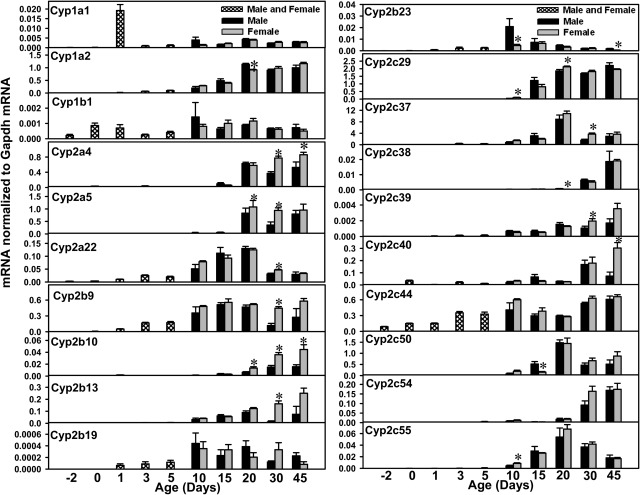
Within the Cyp1 gene family, Cyp1a1 and Cyp1a2 form a cluster on mouse chromosome 7 but are transcribed from opposite strands (Cyp1a1 from the plus strand and Cyp1a2 from the minus strand) (Fig. 1), whereas Cyp1b1 is not clustered with other genes (data not shown). More specifically, Cyp1a2 is embedded in the 5′-untranslated region of the Cyp1a1 gene locus. The mRNA levels for Cyp1a1, Cyp1a2, and Cyp1b1 were quantified during liver development in mice (Fig. 2). Cyp1a1 mRNA levels were lowest before birth but increased markedly after birth (1.7 times day −2 values) and peaked at 1 day of age (255 times day −2 levels). After 1 day of age, Cyp1a1 mRNA levels decreased and gradually reached adult levels after 30 days of age (∼35 times day −2 levels) (Fig. 2). More Cyp1a2 mRNA was detected, compared with Cyp1a1 and Cyp1b1, throughout postnatal liver maturation. Cyp1a2 mRNA expression was lowest before birth, increased 17-fold immediately after birth, and continued to increase until ∼20 days of age.
Fig. 2.
mRNA ontogenic expression of the Cyp1 family (Cyp1a1, Cyp1a2, and Cyp1b1) and part of the Cyp2 family (Cyp2a4, Cyp2a5, Cyp2a22, Cyp2b9, Cyp2b10, Cyp2b13, Cyp2b19, Cyp2b23, Cyp2c29, Cyp2c37, Cyp2c38, Cyp2c39, Cyp2c40, Cyp2c44, Cyp2c50, Cyp2c54, and Cyp2c55) in male and female livers from day −2 to day 45 of age. Total RNA was isolated from livers at each age (n = 5 per gender at each age). Gender was not differentiated from day −2 to 5 days of age (n = 5 at each age). The mRNA levels for Cyp2a5, Cyp2c37, Cyp2c40, Cyp2c50, and Cyp2c54 were determined with RT-PCR assays, whereas the mRNA levels for other P450s were quantified with the multiplex branched DNA assay, as described under Materials and Methods. Data are presented as the fluorescence intensity of mRNA (mean ± S.E.M.) normalized to Gapdh levels. *, statistically significant differences between male and female (p < 0.05).
Cyp1b1 mRNA levels also were lowest before birth and increased 3-fold immediately after birth. The Cyp1b1 mRNA levels remained relatively constant thereafter (Fig. 2).
Ontogeny of mRNA for Cyp2 Gene Family.
Compared with other P450 gene families, the Cyp2 family has the greatest number and most subfamilies of gene isoforms, including the Cyp2a subfamily, Cyp2b, Cyp2c, and Cyp2d subfamily, and Cyp2e, Cyp2f, Cyp2g, Cyp2j, Cyp2s, Cyp2t, Cyp2u, and Cyp2w. The mRNAs of these Cyp2 genes, except for Cyp2a12, Cyp2s1, Cyp2c65, Cyp2d13, and Cyp2r1 (because of potential cross-reaction among primers), were quantified during liver development in mice (Figs. 2–4). For the Cyp2a subfamily (Fig. 2), both Cyp2a4 and Cyp2a5 mRNAs were expressed at very low levels before birth, which gradually increased to adult values at 20 days of age and remained relatively stable thereafter. Cyp2a22 mRNA levels were also low before birth, followed by a gradual increase after birth until 20 days of age, when peak mRNA expression was observed. After 20 days of age, Cyp2a22 mRNA levels decreased ∼60% at both 30 and 45 days of age. Female-predominant expression of Cyp2a5 was observed at 20 and 30 days of age.
Fig. 3.
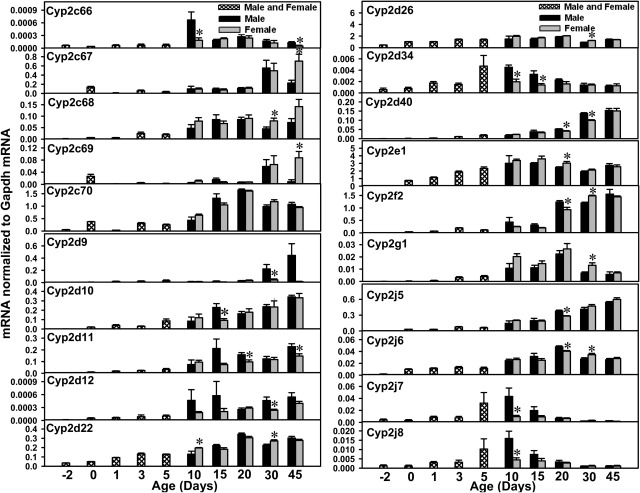
mRNA ontogenic expression of part of the Cyp2 family (Cyp2c66, Cyp2c67, Cyp2c68, Cyp2c69, Cyp2c70, Cyp2d9, Cyp2d10, Cyp2d11, Cyp2d12, Cyp2d22, Cyp2d26, Cyp2d34, Cyp2d40, Cyp2e1, Cyp2f2, Cyp2g1, Cyp2j5, Cyp2j6, Cyp2j7, and Cyp2j8) in male and female livers from day −2 to 45 days of age. Total RNA was isolated from livers at each age (n = 5 per gender at each age). Gender was not differentiated from day −2 to 5 days of age (n = 5 at each age). The mRNA expression levels for Cyp2c67, Cyp2c68, Cyp2c69, Cyp2d9, Cyp2d10, Cyp2d11, and Cyp2d12 were determined with RT-PCR assays, whereas the mRNA levels for other P450s were quantified with the multiplex branched DNA assay, as described under Materials and Methods. Data are presented as the fluorescence intensity of mRNA (mean ± S.E.M.) normalized to Gapdh levels. *, statistically significant differences between male and female (p < 0.05).
Fig. 4.
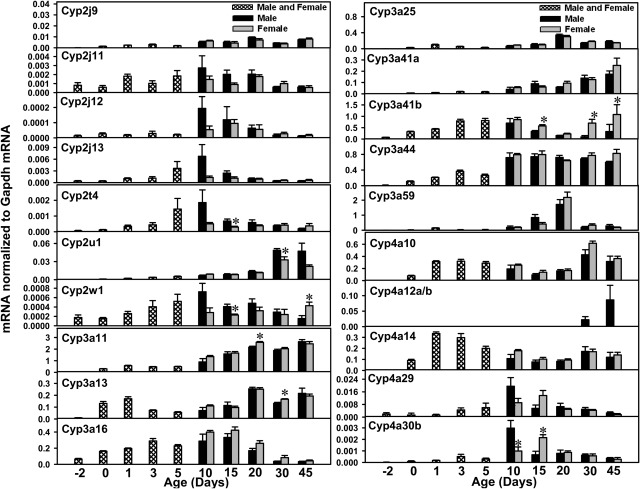
mRNA ontogenic expression of part of the Cyp2 family (Cyp2j9, Cyp2j11, Cyp2j13, Cyp2j13, Cyp2t4, Cyp2u1, and Cyp2w1), the Cyp3 family (Cyp3a11, Cyp2a13, Cyp2a16, Cyp3a25, Cyp3a59, Cyp3a41a, Cyp3a41b, and Cyp3a44), and part of the Cyp4 family (Cyp4a10, Cyp4a12a/b, Cyp4a14, Cyp4a29, and Cyp4a30b) in male and female livers from day −2 to 45 days of age. Total RNA was isolated from livers at each age (n = 5 per gender at each age). Gender was not differentiated from day −2 to day 5 of age (n = 5 at each age). The mRNA expression levels for Cyp2j12, Cyp3a59, Cyp3a41b, and Cyp4a12a/b were determined with RT-PCR assays, whereas the mRNA levels for other P450s were quantified with the multiplex branched DNA assay, as described under Materials and Methods. Data are presented as the fluorescence intensity of mRNA (mean ± S.E.M.) normalized to Gapdh levels. *, statistically significant differences between male and female (p < 0.05).
For the Cyp2b subfamily (Fig. 2), the mRNA levels for Cyp2b9, Cyp2b10, and Cyp2b13 seemed to be higher than those for Cyp2b19 and Cyp2b23 during liver development. The mRNA levels for these three liver-enriched Cyp2b genes were all low before birth, followed by gradual increases after birth. Peak mRNA levels for Cyp2b9 were observed at 15 days of age, for Cyp2b10 at 45 days of age, and for Cyp2b13 at 45 days of age. Female-predominant mRNA patterns were observed for Cyp2b9, Cyp2b10, and Cyp2b13. For Cyp2b19 and Cyp2b23, minimal mRNA expression was observed before birth, followed by gradual increases from 0 to 5 days of age; levels reached a peak at ∼10 days of age and gradually decreased thereafter. The mRNA levels for Cyp2b19 and Cyp2b23 in liver were relatively low throughout development.
For the Cyp2c subfamily (Fig. 2), Cyp2c29 mRNA levels gradually increased to adult levels by 20 days of age. Cyp2c37 mRNA levels were lowest before birth, increased after birth, and peaked at 20 days of age. After 20 days of age, Cyp2c37 mRNA levels decreased markedly to adult expression levels at 45 days of age. The mRNA levels for Cyp2c38, Cyp2c39, Cyp2c40, Cyp2c54, Cyp2c67, Cyp2c68, and Cyp2c69 were all low before birth but gradually increased to adult levels between 30 and 45 days of age. The mRNA levels for Cyp2c50, Cyp2c55, Cyp2c66, and Cyp2c70 were all low before birth but increased after birth, reaching peak values between 10 and 20 days of age. Cyp2c44 mRNA levels were low before birth and seemed to reach adult levels by 10 days of age. Most of the Cyp2b genes showed female-predominant expression patterns. Female-predominant mRNA expression was observed for Cyp2c39, Cyp2c40, Cyp2c67, Cyp2c68, and Cyp2c69. Male-predominant mRNA expression was observed at 20 days of age for Cyp2c38 (although gene expression in both genders was very low at this age) and at 10 and 45 days of age for Cyp2c66. Cyp2c65 mRNA levels were not determined in the present study because of high sequence homology with other P450s.
In the Cyp2d subfamily (Fig. 3), Cyp2d9 is an adult-enriched, male-predominant, P450 isoform; low Cyp2d9 mRNA expression was observed before 20 days of age but male Cyp2d9 mRNA levels increased markedly thereafter, whereas Cyp2d9 was expressed at low levels in female mice at all ages. The mRNA levels for Cyp2d10, Cyp2d11, Cyp2d12, Cyp2d22, Cyp2d26, and Cyp2d40 were low before birth and gradually increased to adult levels during development in both genders of mice. For Cyp2d22 and Cyp2d26, peak expression was observed at 20 days of age. Cyp2d34 mRNA levels were low before birth and at neonatal ages (days 0–3) but increased and reached a peak at 5 days of age, with a gradual decrease to prenatal expression.
Cyp2e1 mRNA levels were low before birth and increased markedly immediately after birth; levels continued to increase and reached adult levels at 5 days of age. Cyp2f2 mRNA levels were low before birth, increased gradually after birth, and reached adult expression at ∼20 days of age. Cyp2g1 mRNA levels were low before birth, gradually increased after birth until 20 days of age (when peak levels were observed), and decreased thereafter.
The mRNA levels for Cyp2j5, Cyp2j6, and Cyp2j9 were all low before birth and increased after birth, with peak levels being observed between 20 and 45 days of age. In contrast, the mRNA levels for Cyp2j7, Cyp2j8, Cyp2j11, Cyp2j12, and Cyp2j13 gradually increased after birth but peaked at ∼10 days of age. The mRNA expression of Cyp2j11 to Cyp2j13 seemed to be lower than that of the other Cyp2j isoforms throughout liver development.
Cyp2t4 mRNA levels were low before birth, increased after birth, appeared to reach maximal expression at 10 days of age, and reached adult levels at ∼30 to 45 days of age. Cyp2u1 mRNA levels were low from before birth to 20 days of age, followed by a marked increase at 30 days of age, when peak levels were observed. Cyp2u1 appeared to be a male-predominant P450, because Cyp2u1 mRNA levels tended to be lower in female mice after 30 days of age.
Cyp2w1 mRNA levels were low before birth, increased gradually after birth, and peaked at 10 days of age in male mice, whereas female Cyp2w1 levels remained stable from 10 to 30 days of age and increased to peak expression at 45 days of age. In general, the Cyp2w1 mRNA signals were very low throughout liver development (<0.08% of Gapdh mRNA levels).
Most of the Cyp2 genes are contained within four genomic clusters, including the Cyp2a to Cyp2f cluster on chromosome 7 (Fig. 1, cluster 2) (plus strand, Cyp2b10, Cyp2b13, Cyp2b9, Cyp2a4, Cyp2b19, Cyp2g1, Cyp2a5, Cyp2a12, Cyp2f2, and Cyp2t4; minus strand, Cyp2a22 and Cyp2b23), the Cyp2c cluster on chromosome 19 (Fig. 1, cluster 3) (plus strand, Cyp2c55, Cyp2c65, Cyp2c66, Cyp2c29, Cyp2c39, Cyp2c37, and Cyp2c50; minus strand, Cyp2c70, Cyp2c54, Cyp2c69, Cyp2c40, Cyp2c68, Cyp2c67, and Cyp2c38), the Cyp2d cluster on chromosome 15 (Fig. 1, cluster 4) (plus strand, Cyp2d9 and Cyp2d12; minus strand, Cyp2d26, Cyp2d40, Cyp2d34, Cyp2d10, Cyp2d11, and Cyp2d22), and the Cyp2j cluster on chromosome 4 (Fig. 1, cluster 5) (Cyp2j5, Cyp2j9, Cyp2j6, Cyp2j8, Cyp2j11, Cyp2j7, Cyp2j12, and Cyp2j13, all transcribed from the minus strand). The only Cyp2 genes that are not clustered with other P450s in the mouse genome include Cyp2c44, Cyp2e1, Cyp2u1, and Cyp2w1. Within the Cyp2a to Cyp2f cluster on chromosome 7 (Fig. 1, cluster 2), Cyp2b10, Cyp2b13, Cyp2b9, and Cyp2a4, which are clustered together on the left side (upstream), all displayed female-predominant mRNA expression patterns, with the highest expression levels being observed between 20 and 45 days of age (Fig. 2). Downstream of the Cyp2a4 gene, Cyp2b19, Cyp2g1, Cyp2a5, Cyp2a12, Cyp2f2, and Cyp2t4 form another cluster, separated from the Cyp2b10 to Cyp2a4 cluster by four non-P450 genes (Nlrp9c, Nlrp4a, Nlrp9a, and Vlre12) (Fig. 1, cluster 2). Within this cluster, the mRNA levels for Cyp2b19, Cyp2g1, and Cyp2t4 were highest at adolescent ages (10–20 days of age) (Figs. 2–4), whereas Cyp2a5 and Cyp2f2, in the middle of the cluster, were both enriched between 30 and 45 days of age (Figs. 2 and 3). Cyp2a12 mRNA levels were not determined in the present study because of high sequence homology.
Within the Cyp2c cluster on chromosome 19 (Fig. 1, cluster 3), on the left side of the cluster, the mRNA levels for Cyp2c55 and Cyp2c66 were both high at 10 to 20 days of age (Figs. 2 and 3). Cyp2c65 mRNA levels were not determined because of high sequence homology. Downstream of Cyp2c66, the mRNAs for Cyp2c29, Cyp2c39, Cyp2c69, Cyp2c40, Cyp2c68, Cyp2c67, and Cyp2c38 all displayed adult-enriched patterns, and many of them, such as Cyp2c39, Cyp2c40, Cyp2c67, Cyp2c68, and Cyp2c69, tended to exhibit higher levels in female mice (Figs. 2–4). On the right side of the Cyp2c cluster, the mRNA levels for Cyp2c37, Cyp2c50, and Cyp2c70 were all high at ∼20 days of age (Figs. 2 and 3). The only exception in this region was Cyp2c54, levels of which were high at ∼45 days of age (Fig. 2).
Within the Cyp2d cluster on chromosome 15 (Fig. 1, cluster 4), the mRNAs for most genes (Cyp2d9, Cyp2d12, Cyp2d40, Cyp2d10, Cyp2d11, and Cyp2d22) were most highly expressed at 20 to 45 days of age, except for Cyp2d26 and Cyp2d34, which were enriched at ∼10 days of age (Fig. 3). Many of these Cyp2d genes tended to exhibit male-predominant patterns between adolescent and adult ages (from 10 to 45 days of age), such as Cyp2d9 after 30 days of age, Cyp2d11 at 20 and 45 days of age, Cyp2d12 at 30 days of age, Cyp2d34 at 10 and 15 days of age, and Cyp2d40 at 20 and 30 days of age. In contrast, Cyp2d22 and Cyp2d26, which are located in the boundary region of the cluster, did not show male-predominant patterns.
Within the Cyp2j cluster on chromosome 4 (Fig. 1, cluster 5), on the left side of the cluster, the mRNA levels for Cyp2j13, Cyp2j12, Cyp2j7, Cyp2j11, and Cyp2j8 were all high at ∼10 days of age and tended to exhibit male-predominant patterns at that age. In the middle of the cluster, the mRNA levels for Cyp2j6 and Cyp2j9 were both high at 20 days of age. On the right side of the cluster, Cyp2j5 mRNA levels were high at 45 days of age.
In summary, levels for all of the 44 Cyp2 genes were high after birth. Gender differences were observed for many Cyp2 mRNA levels from adolescent to adult ages. In addition, some of the P450 isoforms within a genomic cluster tended to show similar ontogenic patterns.
Ontogeny of mRNA for Cyp3 Gene Family.
The Cyp3a subfamily includes eight members, which form a cluster on chromosome 5 (Fig. 1, cluster 6) that includes Cyp3a57 and Cyp3a59 (transcribed from the plus strand) and Cyp3a25, Cyp3a11, Cyp3a44, Cyp3a41a, Cyp3a41b, and Cyp3a16 (transcribed from the minus strand). Cyp3a13 is the only Cyp3a that is found in isolation (data not shown). The ontogeny of the Cyp3a mRNAs is most thoroughly characterized in the literature. Consistent with previous findings (Hart et al., 2009; Li et al., 2009), the mRNA levels for Cyp3a11, Cyp3a13, and Cyp3a25 increased after birth and peaked between 20 and 45 days of age (Fig. 4). Cyp3a16 mRNA levels were low before birth, increased markedly at neonatal ages, and decreased thereafter. Whereas Cyp3a41b mRNA levels in male livers increased at neonatal ages, they decreased thereafter. In female livers, however, Cyp3a41b mRNA levels increased again after 30 days of age and peaked at 45 days of age. The present study also determined the mRNA ontogeny of Cyp3a41a, Cyp3a44, and Cyp3a59 (Fig. 4). Cyp3a41a mRNA levels were low before birth and gradually increased to a peak at 45 days of age. Cyp3a44 mRNA levels were low before birth, increased to adult levels by 10 days of age, and remained relatively constant thereafter. Cyp3a59 mRNA levels were highest at 20 days of age in both genders and decreased markedly thereafter. Within the entire Cyp3a gene cluster on chromosome 5 (Fig. 1, cluster 6), only Cyp3a16 and Cyp3a59, which are located in the boundary regions of the cluster, displayed adolescence-enriched patterns (i.e., high levels at days 10–20), with low mRNA expression levels in adults. In contrast, the Cyp3a genes in the middle of the cluster, namely, Cyp3a25, Cyp3a11, Cyp3a44, Cyp3a41a, and Cyp3a41b, all showed highest expression levels at adult ages. Cyp3a57 mRNA levels were not determined in the present study because of high sequence homology.
Ontogeny of mRNA for Cyp4 Gene Family.
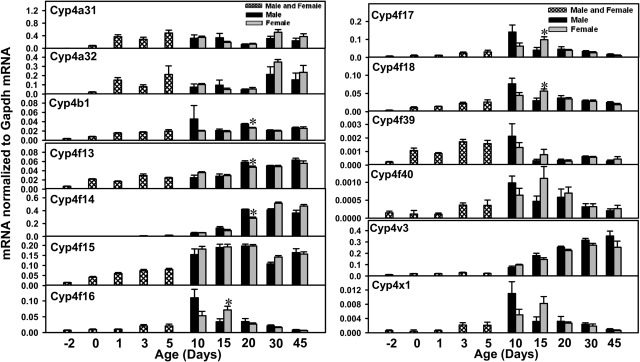
The Cyp4 gene family includes 19 members, i.e., the Cyp4a subfamily (Cyp4a10, Cyp4a12a, Cyp4a12b, Cyp4a14, Cyp4a29, Cyp4a30b, Cyp4a31, and Cyp4a32), Cyp4b1, the Cyp4f subfamily (Cyp4f13–Cyp4f18, Cyp4f39, and 4f40), Cyp4v3, and Cyp4x1. Cyp4a10 mRNA levels were low before birth but almost reached adult levels by 1 day of age. Because of high sequence homology, Cyp4a12a and Cyp4a12b were quantified together as Cyp4a12a/b (Fig. 4). Cyp4a12a/b mRNA was expressed only in male mice, and expression levels were relatively low before 30 days of age. Cyp4a14 mRNA levels were low before birth, with marked increases at neonatal ages. Cyp4a14 mRNA levels decreased moderately after 5 days of age and returned to adult levels after 30 days of age (∼1500 times day −2 levels) (Fig. 4). The mRNA levels for Cyp4a29 and Cyp4a30b were both low before birth and increased after birth, with peak levels at 10 days of age (Cyp4a29, 10 times day −2 levels in male mice and 4.7 times in female mice; Cyp4a30b, 213 times in male mice and 71 times in female mice). After 10 days of age, the mRNA levels for Cyp4a29 and Cyp4a30b in male mice decreased to prenatal levels, whereas the mRNA levels in female mice continued to increase, reached peaks at 15 days of age, and decreased thereafter. The mRNAs for Cyp4a31 and Cyp4a32 were expressed immediately after birth, with the first peak of expression being observed at 5 days of age; the mRNA levels decreased gradually but returned to high levels at 30 days of age (Fig. 5).
Fig. 5.
mRNA ontogenic expression of part of the Cyp4 family (Cyp4a31, Cyp4a32, Cyp4b1, Cyp4f13, Cyp4f14, Cyp4f15, Cyp4f16, Cyp4f17, Cyp4f18, Cyp4f39, Cyp4f40, Cyp4v3, and Cyp4x1) in male and female livers from day −2 to 45 days of age. Total RNA was isolated from livers at each age (n = 5 per gender at each age). Gender was not differentiated from day −2 to 5 days of age (n = 5 at each age). The mRNA expression levels for Cyp4a31 and Cyp4a32 were determined with RT-PCR assays, whereas the mRNA levels for other P450s were quantified with the multiplex branched DNA assay, as described under Materials and Methods. Data are presented as the fluorescence intensity of mRNA (mean ± S.E.M.) normalized to Gapdh levels. *, statistically significant differences between male and female (p < 0.05).
Cyp4b1 mRNA levels were low before birth and increased markedly after birth. Both male and female Cyp4b1 mRNA levels remained relatively stable thereafter, except that male Cyp4b1 mRNA levels increased further at 10 days of age (12.8 times day −2 levels) (Fig. 5).
Within the Cyp4f subfamily, the mRNAs for Cyp4f13 to Cyp4f15 were expressed at low levels before birth, with gradual increases after birth and adult expression levels being reached between 20 and 45 days of age in both genders (Fig. 5). In contrast, the mRNA levels for Cyp4a16 to Cyp4a18, Cyp4f39, and Cyp4f40 (Fig. 5) were all high at 10 to 15 days of age. Female-predominant expression was observed at 15 days of age for Cyp4f16, Cyp4f17, and Cyp4f18.
Cyp4v3 mRNA levels were low before birth and gradually increased to adult levels at ∼45 days of age. Cyp4x1 mRNA levels were high at ∼10 to 15 days of age (∼15 times day −2 levels) and decreased to prenatal levels thereafter.
The Cyp4 gene family forms two genomic clusters (Fig. 1, clusters 7 and 8). Cluster 7 contains Cyp4a29, Cyp4a12a, Cyp4a12b, Cyp4a30b, Cyp4a10, Cyp4a31, and Cyp4a32 from the plus strand and Cyp4b1, Cyp4a14, and Cyp4x1 from the minus strand. Cluster 8 is the Cyp4f cluster, which includes Cyp4f39, Cyp4f17, Cyp4f16, Cyp4f37, Cyp4f40, and Cyp4f15 from the plus strand and Cyp4f13 and Cyp4f14 from the minus strand, with four non-P450 genes (BC066107, 603049I01Rik, Zfp811, and 9030612M13Rik) between Cyp4f15 and Cyp4f13. Cyp4f18 and Cyp4v3 are the only Cyp4 genes found in isolation. Within cluster 7, Cyp4x1, Cyp4a29, and Cyp4a30b exhibited adolescence-enriched mRNA expression patterns, whereas Cyp4a31 and Cyp4a32 mRNA levels were high at neonatal and adult ages. Cyp4a12a/b mRNA levels were high only in male mice at adult ages.
In the Cyp4f cluster (Fig. 1, cluster 8), Cyp4f39, Cyp4f17, Cyp4f16, and Cyp4f40 are located on the left side of the cluster and all displayed an adolescence-enriched mRNA expression pattern, with highest expression levels being observed at ∼10 days of age. The mRNA levels for these genes decreased markedly thereafter. In contrast, mRNA levels for Cyp4f15, Cyp4f14, and Cyp4f13, which are located on the right side of the cluster, gradually increased after birth, reached adult levels after 20 days of age, and remained stable thereafter.
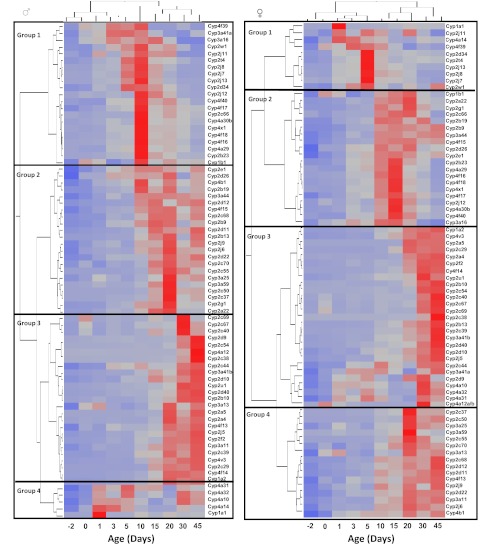
Hierarchical Clustering of P450s during Liver Development.
For unbiased classification of the expression patterns of the mouse P450 isoforms, the mRNA ontogenic expression of these P450s was analyzed in developing mouse liver (separated according to gender) with a two-way hierarchical clustering method (JMP 8.0) and results were observed as heat maps, as described under Materials and Methods. Four distinct classes of ontogenic expression patterns were identified, which were defined as groups 1, 2, 3, and 4 in the present study. In male mice, group 1, which showed low levels of prenatal and neonatal expression and highest expression levels at ∼10 days of age, consists of Cyp4f39, Cyp3a41a, Cyp3a16, Cyp2w1, Cyp2j11, Cyp2t1, Cyp2j8, Cyp2j7, Cyp2j13, Cyp2d34, Cyp2j12, Cyp4f40, Cyp4f17, Cyp2c66, Cyp4a30b, Cyp4x1, Cyp4f18, Cyp4f16, Cyp4a29, Cyp2b23, and Cyp1b1 (Fig. 6, left). Group 2, which exhibited prominent mRNA increases after 10 days of age, with the highest levels of expression being observed near 15 to 20 days of age, consists of Cyp2e1, Cyp2d26, Cyp4b1, Cyp2b19, Cyp3a44, Cyp2d12, Cyp4f15, Cyp2c68, Cyp2b9, Cyp2d11, Cyp2b13, Cyp2j9, Cyp2j6, Cyp2d22, Cyp2c70, Cyp2c55, Cyp3a25, Cyp3a59, Cyp2c50, Cyp2c37, Cyp2g1, and Cyp2a22. Group 3, for which levels were most enriched between 30 and 45 days of age, consists of Cyp2c69, Cyp2c67, Cyp2c40, Cyp2d9, Cyp2c54, Cyp4a12, Cyp2c38, Cyp2c44, Cyp3a41b, Cyp2d10, Cyp3a13, Cyp2a5, Cyp2a4, Cyp4f13, Cyp2j5, Cyp2f2, Cyp3a11, Cyp2c39, Cyp4v13, Cyp2c29, Cyp4f14, and Cyp1a2. Group 4, for which levels increased markedly at neonatal ages (especially from 1 to 5 days of age), decreased in the adolescent period (from 10 to 20 days of age), and increased moderately at adult ages (from 30 to 45 days of age), consists of Cyp4a31, Cyp4a32, Cyp4a10, Cyp4a14, and Cyp1a1.
Fig. 6.
Two-way hierarchical clustering of expression profiles for Cyp1 to Cyp4 genes in livers of male (left) and female (right) mice (Ward's minimal variance; JMP 8.0). Trees describe the relationships between different gene expression profiles (left trees) and different ages (top trees). Distances between genes reflect the significance of associations. Shaded squares, average expression data for five animals per gender at each age. Red, relatively high expression; blue, relatively low expression.
Female P450s also fall into four distinct groups. Group 1, the members of which were all highly expressed at neonatal ages (from 0 to 5 days of age), consists of Cyp1a1, Cyp2j11, Cyp4a14, Cyp4f39, Cyp2d34, Cyp2t4, Cyp2j13, Cyp2j8, Cyp2j7, and Cyp2w1. Group 2, which exhibited highest mRNA expression levels at 10 to 20 days of age, consists of Cyp1b1, Cyp2a22, Cyp2g1, Cyp2c66, Cyp2b19, Cyp2b9, Cyp3a44, Cyp4f15, Cyp2d26, Cyp2e1, Cyp2b23, Cyp4a29, Cyp4f16, Cyp4f18, Cyp4x1, Cyp4f17, Cyp2j12, Cyp4a30b, Cyp4f40, and Cyp3a16. Group 3, which exhibited highest levels of mRNA expression in adults (45 days of age), consists of Cyp1a2, Cyp4v3, Cyp2a5, Cyp2c29, Cyp2a4, Cyp2f2, Cyp4f14, Cyp2u1, Cyp2b10, Cyp2c54, Cyp2c40, Cyp2c67, Cyp2c69, Cyp2c38, Cyp2b13, Cyp2c39, Cyp3a41b, Cyp2d40, Cyp2d10, Cyp2j5, Cyp2c44, Cyp3a41a, Cyp2d9, Cyp4a10, Cyp4a32, Cyp4a31, and Cyp4a12a/b. Genes in group 4 achieved high levels of expression after 20 days of age, and levels remained relatively high thereafter. The genes in group 4 include Cyp2c37, Cyp2c50, Cyp3a25, Cyp3a59, Cyp2c55, Cyp2c70, Cyp3a13, Cyp2c68, Cyp2d12, Cyp2d11, Cyp4f13, Cyp2j9, Cyp2d22, Cyp3a11, Cyp2j6, and Cyp4b1.
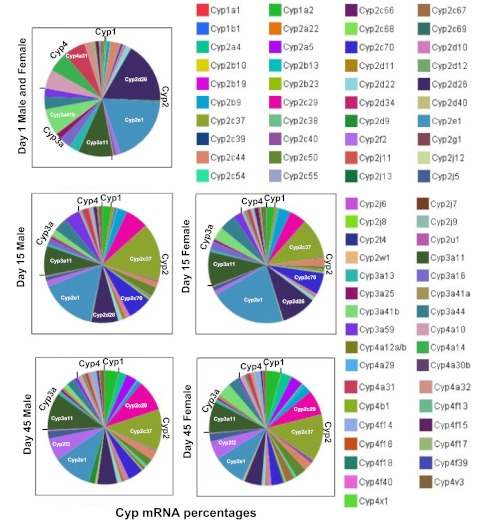
The relative proportions of the P450 mRNAs at selected ages, with day 1 representing a neonatal age, day 15 representing an adolescent age, and day 45 representing an adult age, are shown in Fig. 7. It appears that Cyp2e1 and Cyp3a11 are the two most enriched P450 isoforms at all ages and in both genders. Cyp3a41b, Cyp2d26, and Cyp4a31 are most highly expressed in newborn livers, Cyp2c37, Cyp2c70, and Cyp2d26 in adolescent livers, and Cyp2c29, Cyp2c37, and 2f2 in adult livers.
Fig. 7.
Relative proportions of P450 mRNAs at selected ages (days 1, 15, and 45, which represent neonatal, adolescent, and adult ages, respectively). Genders were not separated for day 1. The P450 mRNA levels were normalized to Gapdh mRNA levels, and the normalized mean values were used for calculation of the proportions at each age. The images were generated with JMP 8.0. The five P450s with the highest expression levels at each age are labeled.
Discussion
Although the ontogeny of a few P450s was characterized previously by different research groups (Lacroix et al., 1997; Treluyer et al., 1997; Sonnier and Cresteil, 1998; de Wildt et al., 1999; Koukouritaki et al., 2004; Blake et al., 2005; Stevens et al., 2008; Williams and Flavell, 2008), the present study provides large amounts of novel information by systematically characterizing 75 isozymes from P450 families Cyp1 to Cyp4 and identifies four distinct ontogenic patterns. The majority of P450s form a total of eight genomic clusters, and certain segments of the P450s within the same cluster on a chromosome tended to show similar ontogenic expression patterns, which indicates that neighboring P450s may be regulated by common pathways during liver maturation.
The similarity in the ontogenic expression patterns for mouse and human P450 homologs was reported previously for CYP1A1, CYP1A2, and CYP2E1 during fetal and adult stages (Choudhary et al., 2005). In the ontogeny of CYP2C19 in human livers, the CYP2C19 protein and catalytic activity levels were low before birth, did not change at birth, increased linearly over the first postnatal months, and varied 21-fold from 5 months to 10 years of age, when adult levels were observed (Koukouritaki et al., 2004). In mice, there are three homologs of human CYP2C19, namely, Cyp2c37, Cyp2c50, and Cyp2c54. The present study showed that all three Cyp2c genes in mice were expressed at low levels before birth and in the neonatal period, but levels increased markedly after 10 days of age in mouse liver. The mRNA levels for Cyp2c37 and Cyp2c50 reached peaks at 20 days of age, whereas Cyp2c54 mRNA levels reached a peak at 45 days of age. Therefore, human CYP2C19 and the mouse Cyp2c homologs share certain similarities in their ontogenic expression patterns. The human CYP2D6 levels were higher in the third-trimester samples and in neonates than in the first- and second-trimester samples. Human CYP2D6 expression in postnatal samples (>7 days of age) was substantially higher than in any earlier age category (Stevens et al., 2008). In mice, the present study showed that levels of the mRNA for the mouse homolog of human CYP2D6, namely, Cyp2d22 mRNA, were low before birth, increased markedly after birth, and reached adult levels at 45 days of age. Therefore, human CYP2D6 and mouse Cyp2d22 appeared to have similar ontogenic expression patterns, although the time scale in mice was condensed. Other P450s, such as Cyp3a11 and Cyp4a10, are also somewhat similar to the human homologs (Hines, 2007).
Although mice and humans share certain similarities in the ontogeny of some P450s during liver development, strain differences have been observed for other P450s. Many mouse P450s (such as Cyp2c67–Cyp2c70) have no human homlogs. Human CYP3A7 was highly expressed in the prenatal and neonatal stages and then was minimally expressed or silenced after 1 to 2 years of age (Lacroix et al., 1997; de Wildt et al., 1999), whereas mRNA levels for the mouse ortholog, Cyp3a16, remained high from neonatal to adolescent ages (0–20 days of age in mice). No mouse homolog has been identified for human CYP3A5, and CYP3A5 is expressed at highly variable levels during development but independent of age (Wrighton et al., 1990). Caution needs to be used in interpretation of the P450 ontogeny data for mice and prediction of the ontogeny of human P450s in liver.
Another interesting finding in the postnatal enrichment of all P450s in families 1 to 4 is their gender-divergent gene expression. Female-predominant expression was observed for many genes in the Cyp2b and Cyp2c subfamilies (such as Cyp2b9, Cyp2b10, Cyp2b13, Cyp2c40, Cyp2c67, Cyp2c68, and Cyp2c69). The present findings are generally consistent with the gender differences reported for these genes in the literature (Kawamoto et al., 2000; Löfgren et al., 2009; Waxman and Holloway, 2009). The expression of CAR protein also shows a female-predominant pattern, and it has been found to be responsible for the female-predominant expression of Cyp2b10 in mouse liver (Kawamoto et al., 2000). Findings indicate that CAR also regulates the basal expression of Cyp2c29 and Cyp2b13 (Hernandez et al., 2009). Nuclear receptors may contribute to the age- and gender-specific gene expression of P450 isoforms during development, given that many nuclear receptors, which are well known to regulate P450 gene expression in adults, begin to be expressed in livers after birth in both rats and mice (Balasubramaniyan et al., 2005; Cui et al., 2010). Therefore, it is possible that the postnatal enrichment and female-predominant expression of CAR contribute to the female-specific expression of other novel P450 isoforms in the Cyp2b and Cyp2c subfamilies, including Cyp2b9, Cyp2b13, Cyp2c39, Cyp2c40, Cyp2c67, Cyp2c68, and Cyp2c69 (Figs. 2 and 3). Löfgren et al. (2009) examined the roles of sex and growth hormones in P450 gene expression and demonstrated that the androgen-dependent, pituitary growth hormone secretory pattern is the primary regulator of male-specific expression in the liver, which suggests that there is a higher order of gene regulation mediated by pituitary hormones, beyond the local regulation of P450s by nuclear receptors. The postnatal enrichment of P450s may be an adaptive mechanism for biotransformation of increased amounts of xenobiotics from the environment, compared with the prenatal period, when the fetus is largely protected from these chemicals by the placenta and fetal membranes (Aleksunes et al., 2008).
It is interesting to note that some P450s within the same genomic cluster may exhibit similar expression patterns. The spatial organization of the genome is thought to play an important part in the coordination of gene regulation, and the coordinate expression of these P450s in each segment indicates that common spatial regulatory mechanisms may exist. Conversely, on the boundary regions between segments in the same P450 cluster, there may be distinct regulators that prevent the spread of transcription from one segment to another at certain ages. It is well known that the two human CYP3A members, namely, CYP3A4 and CYP3A7, exhibit a developmental switch in gene expression during liver maturation (Schuetz et al., 1994). In mice, by using chromatin immunoprecipitation tiling-array technology assays for three epigenetic markers for gene regulation, namely, DNA and histone H3 lysine-27 trimethylation for gene suppression and histone H3 lysine-4 dimethylation for gene activation, it was determined that the developmental switch between the perinatal Cyp3a isoform and adult Cyp3a genes is likely attributable to the dynamic changes in histone H3 lysine-4 dimethylation and histone H3 lysine-27 trimethylation during postnatal liver maturation but is likely not attributable to DNA methylation (Li et al., 2009). It is possible that distinct chromatin epigenetic markers modify other P450 gene loci with segment-specific signatures within a certain cluster, resulting in common expression patterns within the same segment.
The protein ontogeny of many P450s in human and mouse livers has been determined. For example, in humans, the enzyme contents of six key drug-metabolizing P450s were quantified in 240 human liver samples during development, and both quantitative Western blotting and enzyme activity assays with probe substrates were performed when possible (Hines, 2007). For the CYP2C subfamily, the ontogenic patterns of human liver CYP2C9 and CYP2C19 were quantified with Western blotting and with probe substrates, and the data showed postnatal increase patterns in the protein expression and activities of the two CYP2Cs (Koukouritaki et al., 2004). For the CYP3A family, the enzyme activities of the adult-specific CYP3A4 and the fetal CYP3A7 were determined (Lacroix et al., 1997; de Wildt et al., 1999). In mice, corresponding to the mRNA data, the protein levels for mouse Cyp3a11 were very low in fetal liver but increased markedly after birth (80% of adult levels on postnatal day 4) and reached maximal levels in adult liver (Li et al., 2008). The protein expression and/or enzyme activity levels for CYP1A1/2, CYP2B1/2, CYP2E1, CYP3A1/2, and CYP4A1 were also determined in rat liver during development (Li et al., 2008). Several approaches, including immunoquantification of individual enzymes and profiling of substrate activities through immunoinhibition using highly specific polyclonal and monoclonal antibodies, have been used to investigate the ontogeny of P450s in the livers of rodents and humans. Evidence suggests that P450 enzymes in humans are regulated in a manner similar to that in other animals (Rich and Boobis, 1997). In summary, the protein expression and/or enzyme activities of many major P450s during liver development have been well characterized. Gene expression at the protein level was not determined for novel P450s included in the present study, because of lack of specific antibodies and substrates for most of these novel mouse P450 proteins. The protein ontogeny of these P450s will be determined in future studies.
Although much progress has been made in characterizing P450 gene isoforms, many issues are still unresolved. For example, are there any novel splicing isoforms of P450 mRNAs during liver development or in disease situations? What transcription factors contribute to the temporal and spatial regulation of various P450 isoforms? Fifty years ago, we thought there were only one or two P450 isoforms. The completion of the Human Genome Project has broadened our perspective with a relatively complete assembly of P450s. Next-generation sequencing has allowed fast characterization of the transcriptome and transcription factor-DNA binding (Werner, 2010). The characterization of age-dependent P450 mRNA splicing isoforms and the regulatory factors for transcription will be determined in future studies.
Supplementary Material
Acknowledgments
We thank all of the members of Dr. Curtis Klaassen's laboratory group for technical support and reading of the manuscript.
This work was supported by the National Institutes of Health National Institute of Environmental Sciences [Grants ES-019487, ES-009716, ES-009649] (to C.D.K.); and the National Institutes of Health National Center for Research Resources [Grant RR-021940] (to C.D.K.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- P450
- cytochrome P450
- CAR
- constitutive androstane receptor
- Gapdh
- glyceraldehyde-3-phosphate dehydrogenase
- RT
- reverse transcription
- PCR
- polymerase chain reaction.
Authorship Contributions
Participated in research design: Cui, Renaud, and Klaassen.
Conducted experiments: Cui and Renaud.
Performed data analysis: Cui and Renaud.
Wrote or contributed to the writing of the manuscript: Cui, Renaud, and Klaassen.
References
- Aleksunes LM, Cui Y, Klaassen CD. (2008) Prominent expression of xenobiotic efflux transporters in mouse extraembryonic fetal membranes compared with placenta. Drug Metab Dispos 36:1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93:242–255 [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101:51–64 [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Petrick JS, Klaassen CD. (2006) Tissue distribution and ontogeny of organic cation transporters in mice. Drug Metab Dispos 34:477–482 [DOI] [PubMed] [Google Scholar]
- Balasubramaniyan N, Shahid M, Suchy FJ, Ananthanarayanan M. (2005) Multiple mechanisms of ontogenic regulation of nuclear receptors during rat liver development. Am J Physiol Gastrointest Liver Physiol 288:G251–G260 [DOI] [PubMed] [Google Scholar]
- Blake MJ, Castro L, Leeder JS, Kearns GL. (2005) Ontogeny of drug metabolizing enzymes in the neonate. Semin Fetal Neonatal Med 10:123–138 [DOI] [PubMed] [Google Scholar]
- Caldwell J. (2004) Pharmacogenetics and individual variation in the range of amino acid adequacy: the biological aspects. J Nutr 134:1600S–1604S [DOI] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. (2009) Tissue distribution, ontogeny, and hormonal regulation of xenobiotic transporters in mouse kidneys. Drug Metab Dispos 37:2178–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Chen C, Klaassen CD. (2005) Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps). Drug Metab Dispos 33:1062–1073 [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Schenkman JB, Sarfarazi M, Stoilov I. (2003) Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Arch Biochem Biophys 414:91–100 [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. (2005) Expression patterns of mouse and human CYP orthologs (families 1–4) during development and in different adult tissues. Arch Biochem Biophys 436:50–61 [DOI] [PubMed] [Google Scholar]
- Cui JY, Choudhuri S, Knight TR, Klaassen CD. (2010) Genetic and epigenetic regulation and expression signatures of glutathione S-transferases in developing mouse liver. Toxicol Sci 116:32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. (2009) Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab Dispos 37:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. (1999) Cytochrome P450 3A: ontogeny and drug disposition. Clin Pharmacokinet 37:485–505 [DOI] [PubMed] [Google Scholar]
- Estabrook RW. (2003) A passion for P450s (remembrances of the early history of research on cytochrome P450). Drug Metab Dispos 31:1461–1473 [DOI] [PubMed] [Google Scholar]
- Fouts JR, Adamson RH. (1959) Drug metabolism in the newborn rabbit. Science 129:897–898 [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Baker DW, Totah RA, Gaedigk R, Pearce RE, Vyhlidal CA, Zeldin DC, Leeder JS. (2006) Variability of CYP2J2 expression in human fetal tissues. J Pharmacol Exp Ther 319:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD, Zhong XB. (2009) Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab Dispos 37:116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Huang W, Moore DD, Baldwin WS. (2009) Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR). Toxicology 256:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines RN. (2007) Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol 21:169–175 [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. (2007) Gene expression profiling reveals a regulatory role for ROR α and ROR γ in phase I and phase II metabolism. Physiol Genomics 31:281–294 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Kakizaki S, Yoshinari K, Negishi M. (2000) Estrogen activation of the nuclear orphan receptor CAR (constitutive active receptor) in induction of the mouse Cyp2b10 gene. Mol Endocrinol 14:1897–1905 [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN. (2004) Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308:965–974 [DOI] [PubMed] [Google Scholar]
- Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T. (1997) Expression of CYP3A in the human liver–evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 247:625–634 [DOI] [PubMed] [Google Scholar]
- Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, Pearce RE. (2005) Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther 314:626–635 [DOI] [PubMed] [Google Scholar]
- Li X, Schuler MA, Berenbaum MR. (2007) Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol 52:231–253 [DOI] [PubMed] [Google Scholar]
- Li XY, Zhang C, Wang H, Ji YL, Wang SF, Zhao L, Chen X, Xu DX. (2008) Tumor necrosis factor α partially contributes to lipopolysaccharide-induced downregulation of CYP3A in fetal liver: its repression by a low dose LPS pretreatment. Toxicol Lett 179:71–77 [DOI] [PubMed] [Google Scholar]
- Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB. (2009) Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol Pharmacol 75:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfgren S, Baldwin RM, Carlerös M, Terelius Y, Fransson-Steen R, Mwinyi J, Waxman DJ, Ingelman-Sundberg M. (2009) Regulation of human CYP2C18 and CYP2C19 in transgenic mice: influence of castration, testosterone, and growth hormone. Drug Metab Dispos 37:1505–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Cherrington NJ, Cheng X, Klaassen CD. (2005) Tissue distribution and hepatic and renal ontogeny of the multidrug resistance-associated protein (Mrp) family in mice. Drug Metab Dispos 33:947–955 [DOI] [PubMed] [Google Scholar]
- Nebert DW, Russell DW. (2002) Clinical importance of the cytochromes P450. Lancet 360:1155–1162 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14:1–18 [DOI] [PubMed] [Google Scholar]
- Petrick JS, Klaassen CD. (2007) Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab Dispos 35:1806–1815 [DOI] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M, Klaassen CD. (2011) Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci 124:261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich KJ, Boobis AR. (1997) Expression and inducibility of P450 enzymes during liver ontogeny. Microsc Res Tech 39:424–435 [DOI] [PubMed] [Google Scholar]
- Rowell M, Zlotkin S. (1997) The ethical boundaries of drug research in pediatrics. Pediatr Clin North Am 44:27–40 [DOI] [PubMed] [Google Scholar]
- Schuetz JD, Beach DL, Guzelian PS. (1994) Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver. Pharmacogenetics 4:11–20 [DOI] [PubMed] [Google Scholar]
- Sonnier M, Cresteil T. (1998) Delayed ontogenesis of CYP1A2 in the human liver. Eur J Biochem 251:893–898 [DOI] [PubMed] [Google Scholar]
- Stevens JC, Marsh SA, Zaya MJ, Regina KJ, Divakaran K, Le M, Hines RN. (2008) Developmental changes in human liver CYP2D6 expression. Drug Metab Dispos 36:1587–1593 [DOI] [PubMed] [Google Scholar]
- Treluyer JM, Gueret G, Cheron G, Sonnier M, Cresteil T. (1997) Developmental expression of CYP2C and CYP2C-dependent activities in the human liver: in-vivo/in-vitro correlation and inducibility. Pharmacogenetics 7:441–452 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Holloway MG. (2009) Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. (2000) The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407:920–923 [DOI] [PubMed] [Google Scholar]
- Werner T. (2010) Next generation sequencing in functional genomics. Brief Bioinform 11:499–511 [DOI] [PubMed] [Google Scholar]
- Williams A, Flavell RA. (2008) The role of CTCF in regulating nuclear organization. J Exp Med 205:747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton SA, Brian WR, Sari MA, Iwasaki M, Guengerich FP, Raucy JL, Molowa DT, Vandenbranden M. (1990) Studies on the expression and metabolic capabilities of human liver cytochrome P450IIIA5 (HLp3). Mol Pharmacol 38:207–213 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.