Abstract
Cardiac progenitor cells (CPCs) are multipotent cells that may offer tremendous potentials for the regeneration of injured myocardium. To expand the limited number of CPCs for effective clinical regeneration of myocardium, it is important to understand their proliferative potentials. Single-cell based assays were utilized to purify c-kitpos CPCs from human and mouse hearts. MicroRNA profiling identified eight differentially expressed microRNAs in CPCs from neonatal and adult hearts. Notably, the predicted protein targets were predominantly involved in cellular proliferation-related pathways. To directly test this phenotypic prediction, the developmental variance in the proliferation of CPCs was tested. Ki67 protein expression and DNA kinetics were tested in human and mouse in vivo CPCs, and doubling times were tested in primary culture of mouse CPCs. The human embryonic and mouse neonatal CPCs showed a six-fold increase in Ki67 expressing cells, a two-fold increase in the number of cells in S/G2-M phases of cell cycle, and a seven-fold increase in the doubling time in culture when compared to the corresponding adult CPCs. The over-expression of miR-17-92 increased the proliferation in adult CPCs in vivo by two-fold. In addition, the level of retinoblastoma-like 2 (Rbl2/p130) protein was two-fold higher in adult compared to neonatal-mouse CPCs. In conclusion, we demonstrate a differentially regulated cohort of microRNAs that predicts differences in cellular proliferation in CPCs during postnatal development and target microRNAs that are involved in this transition. Our study provides new insights that may enhance the utilization of adult CPCs for regenerative therapy of the injured myocardium.
Keywords: Cardiac progenitor cells, c-kit, microRNA profiling, proliferation
INTRODUCTION
Cardiovascular disease is the leading cause of death in the United States. Since cardiac myocytes have a limited ability to regenerate, their malfunction and/or significant loss during myocardial infarction or cardiomyopathy can lead to life-threatening arrhythmias and/or heart failure. Although considerable progress has been made in the pharmacologic and device management of heart failure in recent decades, the mortality in heart failure patients remains significant [1]. Therefore, there is a compelling need to seek alternative options for the treatment of patients with heart failure.
Recent studies have provided exciting evidence that stem cells may offer an enormous potential for regenerative therapy by replacing cardiac myocytes that are lost during cardiomyopathic processes [2–5]. To date, various types of extra-cardiac stem cells have been used in clinical trials, with each cell type presenting advantages and limitations [6, 7]. In addition, new evidence for the existence of resident cardiac progenitor cells (CPCs) in adult hearts has directly challenged the existing paradigm that myocardium is terminally differentiated and hence, devoid of regenerative capacity [8–11]. These progenitors can be found among differentiated cardiac myocytes and are capable of self-renewal and clonogenicity [9, 10]. Furthermore, CPCs have been shown to differentiate into cardiomyocytes, endothelial and smooth muscle cells in vitro [8, 12]. Thus, CPCs may offer the potential for regeneration and functional recovery of damaged myocardium that may avoid the shortcomings of extra-cardiac stem cells and organ donation [11, 13]. Despite this potential, one of the limiting factors in cardiac regeneration is the limited number and quiescent disposition of CPCs within the normal adult heart. Although the number of CPCs is modestly increased in human cardiomyopathies [14], there are concerns that these progenitors may not be sufficiently effectively in repairing the diseased heart. One recent study demonstrated that transplanted CPCs provided no long-term engraftment or benefit to cardiac function as assessed by multimodality imaging [15]. While other studies have demonstrated that the numbers of CPCs and their myogenic potential may decrease postnatally suggesting that a lack of cell proliferation may be regulated during postnatal development [16–19]. In order to enhance CPCs as an effective clinical target to regenerate myocardium, it is critical to understand the mechanisms responsible for their proliferative as well as cardiomyogenic potentials.
MicroRNAs (miR) are short, non-coding, single stranded regulatory RNA that are 20–23 nucleotides in length that play an important role in the regulation of germline stem cell proliferation in Drosophila [20]. Human embryonic stem cells (ESC) demonstrate a unique set of microRNAs [21] and the expression profile is different in ESC-derived cardiomyocytes [22]. To date, more than 1,000 microRNAs have been identified in the human genome and it is estimated that they may regulate up to 30% of the protein-coding genes in humans [23]. Recent evidence is emerging that cardiac-specific microRNAs are important regulators during development since their deletion leads to defective cardiogenesis [24–26]. Specifically, microRNAs have been shown to regulate critical cardiac regulatory proteins that control the delicate balance between proliferation and differentiation during cardiogenesis [24].
In this study, we hypothesize that the differentially expressed microRNAs between mouse neonatal and adult CPCs can predict phenotypic differences in CPCs from two different developmental stages. Specifically, we identified eight unique microRNAs that are differentially expressed between neonatal and adult mouse CPCs. Using bioinformatic protein target analysis of the differentially expressed microRNAs in mice, we predicted a developmental difference in the cellular proliferation. The difference in the proliferative potential was directly demonstrated between embryonic and adult humans CPCs as well as between neonatal and adult mouse CPCs. Finally, over-expression of miR-17-92 cluster, a member of which is differentially expressed in CPCs during development, increased the proliferation in adult mouse CPCs by two-fold. Taken together, our data supports a functional role for microRNAs in the regulation of the proliferative capacity of CPCs.
MATERIALS AND METHODS
A detailed description of methods is given in the supplemental methods section.
Fluorescent Activated Cell Sorting (FACS) of CPCs
Neonatal and adult mouse hearts were obtained from C57BL/6J mice according to the approved UC Davis Animal Care and Use protocol. To obtain single cell suspensions from mouse neonatal and adult hearts, cardiac tissues were enzymatically digested into single cells and stained with antibodies before FACS.
MicroRNA profiling and target analysis
Expression microarrays identified differentially expressed microRNAs in total RNA isolated from neonatal and adult mouse CPCs. Multiple algorithms (miRecords, miRanda, TargetScan, PicTar and MicroCosm) identified protein targets for the differentially expressed microRNA. The sensitivity and specificity of target prediction was increased by intersecting results from at least three target prediction algorithms.
Flow cytometric analysis of mouse and human CPCs for proliferative capacity
Human atrial appendage specimens from informed consented patients undergoing cardiac bypass surgery were obtained from UC Davis Medical Center in accordance with the approved UC Davis Institutional Review Board (IRB) protocol. Human embryonic cardiac tissue was obtained from informed consented patients in accordance with the Sanford Burnham Medical Research Institute IRB protocol. Single cell suspensions were obtained from human adult and embryonic hearts, as well as from mouse adult [27] and neonatal hearts by enzymatic digestion and stained with antibodies before analyzing by flow cytometry.
Lentiviral over-expression of miR-17 cluster in adult mouse CPCs
Homogenous transmural viral-mediated gene transfer in ten-week-old male C57Bl/6J mice was performed using techniques previously described in large animal models [28]. Right thoracotomy was performed to expose the pericardial sac. Lentiviral solution (100 μl) was delivered into the sac and incubated for 15 min before closing the chest. The solution contained 30 μl of lentiviral constructs expressing miR-17-92 cluster and green fluorescence protein (GFP) or GFP only (control lentivirus), 70 μl of 20% pluronic acid in phosphate buffered saline (PBS), and 1% trypsin. Mice were maintained for 19–20 days when cardiac single cell suspensions were prepared to phenotype the GFPpos CPCs using flow cytometry as described above.
Statistical analysis
All data were expressed as means ±S.E.M. Significant differences were tested with one-way ANOVA for more than two groups, or Student’s unpaired t-test for two groups. A P value <0.05 was considered significant.
RESULTS
Isolation and characterization of c-kitpos CPCs from mouse cardiac explants
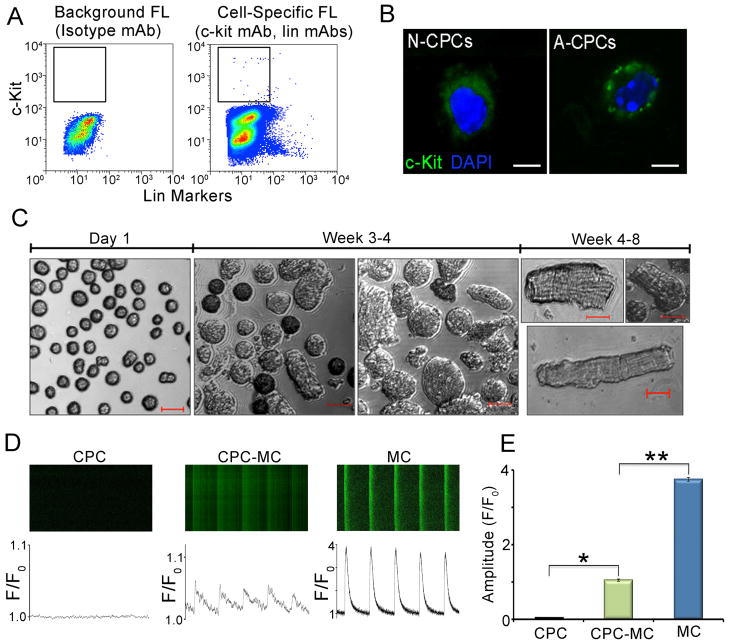
The stem cell antigen, c-kit (CD117), a member of the receptor tyrosine kinase family has been widely used as a marker for CPCs [17, 29]. Lineage-negative c-kit positive CPCs have been shown to be multipotent and capable of differentiating into cells of cardiogenic lineages [8]. We isolated lineage-negative c-kit-positive (linneg c-kitpos) CPCs from adult mouse cardiac tissue using FACS (Figure 1A) with a purity of 99±0.5% (n=3) and determined their myogenic potential in our culture conditions. Figure 1B shows the specificity of the c-kit antibody in labeling the outer cell membrane of CPCs. Sorted adult mouse CPCs were grown in maintenance media for one week and in differentiating media containing growth factors (listed in Supplemental methods) for 4–8 weeks (Figure 1C). CPCs-derived-myocytes (CPC-MC) were detected at a frequency of 1.4% of the sorted CPCs as determined by the rod shape appearance. Functionally, these rod-shaped cells demonstrated a significant increase in the intracellular Ca2+ ([Ca2+]i) transients as measured by line scanning using confocal microscopy (Figure 1D) when compared to CPCs. Figure 1E shows the summary data for the peak amplitude of the ([Ca2+]i) transients (F/Fo ratios, n=6, P<0.001). The amplitude of the [Ca2+]i transients in CPC-MCs remained smaller than that of adult ventricular myocytes (positive control) suggesting that the former were relatively immature when compared to the latter.
Figure 1. Characterization of adult murine c-kitpos CPCs.
(A) Isolation of adult mouse linneg c-kitpos CPCs using FACS. Isotype-matched antibodies were used to control for background fluorescence (FL) and is shown in the left panel labeled as Background FL compared to the Cell-Specific FL shown in the right panel. (B) Immunofluorescence confocal images of neonatal and adult mouse CPCs showing c-kit (green) and DAPI (blue) staining. (C) Photomicrographs of adult mouse CPCs and CPC-derived cardiomyocytes (CPC-MC) at different time points in culture after FACS. (D) Representative examples of confocal line-scan images of intracellular Ca2+ ([Ca2+i]) transient recordings from adult mouse CPCs (CPC), adult mouse CPC-MC, and adult mouse ventricular myocytes (MC). Lower panels show the corresponding [Ca2+i] (measured as F/Fo). Cells were field stimulated at 1 Hz. (E) Summary data of the peak amplitude of the ([Ca2+i] transients (peak F/Fo) recorded from CPC (black), CPC-MC (green), and MC (blue) (n=6). Scale bars are 20 μm. N-CPCs: neonatal CPCs, A-CPCs: adult-CPCs, Lin: lineage, mAb: monoclonal antibody. Error bars represent standard error * and ** P<0.001.
MicroRNA expression in mouse neonatal and adult CPCs
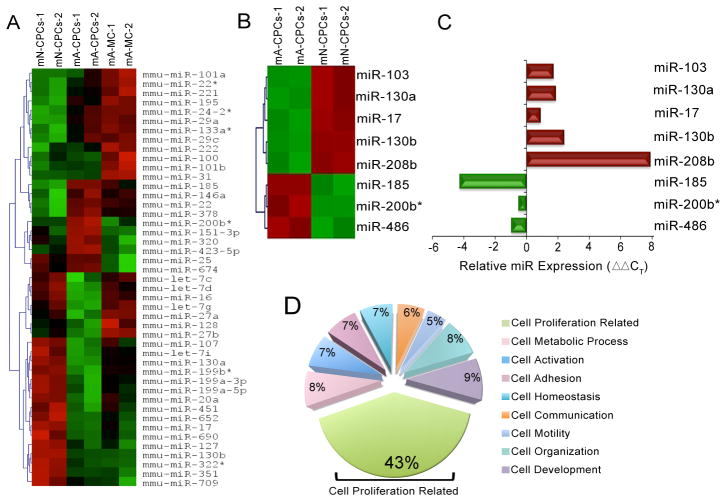
The regulation of mRNA by the interacting microRNAs depends upon the milieu and varies with time [30]. To test for CPC developmental changes in microRNA expression, we studied the microRNA profiles in the CPCs at two developmental time points. Enriched populations of neonatal and adult mouse CPCs were obtained by FACS and microRNA expression profiled using microarray analyses. Freshly harvested CPCs were used to avoid possible artifacts from cell culture. Adult mouse ventricular cardiomyocytes (MC) were also included in this analysis as a separate population to test for differentially expressed microRNAs between cardiomyocytes and CPCs. To ensure the detection of possible differences in microRNA profiles unique to each individual cell type, we utilized the same genetic background for all groups of mice. MicroRNA profiling revealed that 45 microRNAs were differentially expressed (P<0.05) among neonatal and adult CPCs, and adult cardiomyocytes (Heat map, Figure 2A). Of these 45 microRNAs, eight were differentially expressed between the neonatal and adult CPCs (miR-103, -130a, -17, -130b, -208b, -185, -200b* and -486, Heat map Figure 2B) with P<0.05 plus ≥ five-fold difference. The expression pattern of the eight microRNAs was further verified by real-time PCR (Q-PCR), demonstrating the same trend of expression for all eight microRNA as seen in the microarray experiment (Figure 2C).
Figure 2. MicroRNA profiling of neonatal and adult mouse CPCs.
(A) Heat maps showing the differential expression of 45 microRNAs across mouse neonatal, adult CPCs, and adult cardiomyocytes (performed in duplicate, P<0.05). Red to green indicates high to low expression. (B) Heat maps of eight differentially expressed microRNAs in neonatal and adult CPCs (P<0.05 and ≥5-fold difference). For each microarray run, 10 adult and 10–12 neonatal mouse hearts were pooled. (C) Q-PCR validation of the microRNA microarray data of the eight differentially expressed microRNAs represented as ΔΔCT. (D) Summary data of the percentages of proteins relative to the total proteins in each cellular process illustrating the predominant involvement of the predicted target proteins in the cell proliferation-related pathway (green sector, 43%). mN: mouse neonatal, mA: mouse adult, mA-MC: mouse adult myocytes and mmu-miR: mouse microRNA.
To test for cellular processes predicted by the differentially expressed microRNAs across CPC postnatal development, we identified the potential protein targets for these eight microRNAs. Potential protein targets (~1000–2000 for each microRNA) were analyzed using Gene Ontology (GO) classification, Gene Microarray Pathway Profiler, and Map Annotation and Pathway Profiling (MAPP) [31]. Analysis of the cellular process sub-category within the biological process category of GO classification demonstrated that the predicted target proteins of the differentially expressed microRNAs were involved predominantly in cell proliferation-related processes (43% of all proteins). The cell proliferation-related processes include cell cycle, cell division, cell growth, and cell proliferation pathways. This prediction was independent of the direction and magnitude of the changes observed in the eight analyzed microRNAs. The percentages of other proteins predictions were cell organization (8%), cell development (9%), cell metabolic process (8%), cell adhesion (7%), cell homeostasis (7%), cell activation (7%), cell communication (6%), and cell motility (5%) (Figure 2D). The microRNA target proteins analyses showed that 0.07% of total proteins were involved in the sub-process of cardiac muscle differentiation. Taken together, these data suggest that the differentially expressed microRNAs predicted differences in the proliferative phenotype between neonatal and adult CPCs.
Single-cell based assays to phenotype the rare population of CPCs
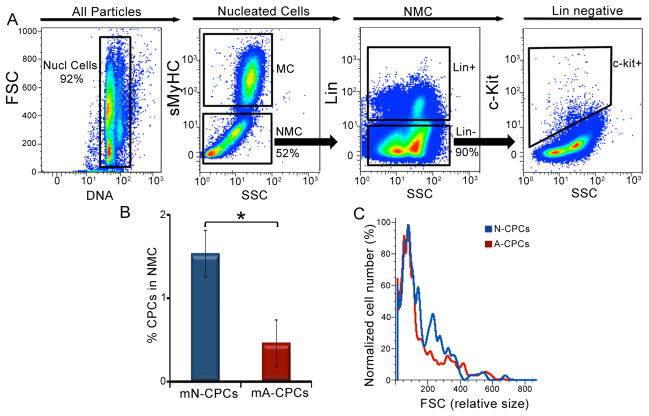
To directly test the prediction from microRNA profiling on the proliferative phenotype of CPCs, a relatively rare population of cells among all other myocardial cells, we used flow cytometric phenotyping as a single-cell based assay to quantify the cell cycle activity (Figure 3A). The nucleated cells from the fresh myocardial preparation (similarly performed for human and mouse hearts) were enumerated based on the incorporation of 7-aminoactinomycin D (7-AAD), a DNA binding fluorescent molecule. On an average, 3.6×105±8×104 mouse nucleated cells were analyzed in each run. A total sarcomeric myosin heavy chain specific antibody or Troponin T antibody were used to label the myocytes (MC) and allowed us to gate and eliminate the MC population which had higher auto fluorescence from the non-myocyte cells (NMC, ~3.0×105±7×104 cells, 51% of mouse nucleated cells) in this analysis [32]. The NMCs are heterogeneous and consist of a mixture of CPCs, fibroblasts, smooth muscle, endothelial, and hematogenous cells. Flow cytometry identifies linneg c-kitpos CPCs (~2.1×103±6×102 cells, 0.6% of mouse nucleated cells) within the NMCs gate (1.53±0.2% neonatal CPCs and 0.43±0.1% adult CPCs, n=8, Figure 3B). Isotype-matched antibodies were used to control for background fluorescence (Figure S1). Using forward scatter (FSC) as an indicator of cell size, we found no appreciable size difference between the CPCs from neonatal and adult mouse hearts (Figure 3C).
Figure 3. Identification of CPCs from mouse cardiac tissue.
Flow cytometric analysis of the heterogeneous population of cells isolated from mouse hearts. (A) The selection of nucleated cells (Nucl cells) from the mixed population based on the incorporation of 7-AAD and the separation of the myocytes (MC) from the non-muscle cells (NMC) using sarcomeric myosin heavy chain (sMyHC) specific antibody. The linneg c-kitpos CPCs are identified from the NMC population using c-kit and lineage (lin)-specific antibodies. X and Y-axes represent arbitrary units. (B) Summary data from A (n=8). (C) A plot of cell size distribution with the percentages of normalized cell number against forward scatter (FSC). Representative results are shown. FSC: forward scatter, SSC: side scatter, mN: mouse neonatal and mA: mouse adult. Error bars represent standard error and *P<0.05.
Proliferative capacity of mouse neonatal and adult CPCs
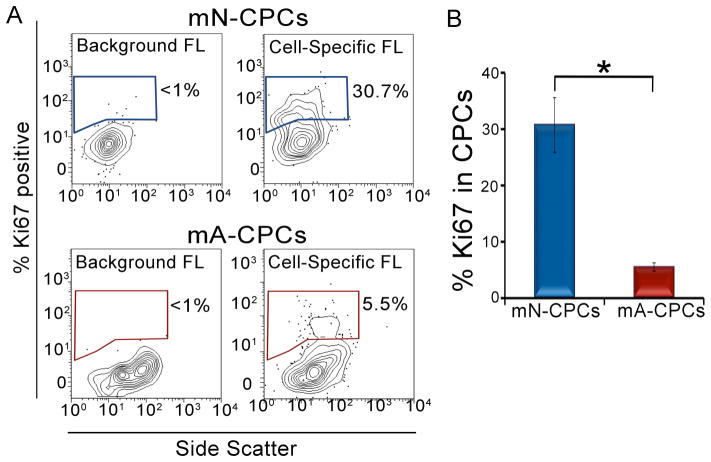
Ki67 is a widely used nuclear antigen that is expressed in actively cycling cells [33]. To directly test the hypothesis that there are significant differences in the proliferative phenotype between neonatal and adult CPCs, we measured the percent of CPCs expressing Ki67. Ki67 is absent in resting G0 phase, increases during the G1/S phase and reaches a maximum during the G2/M phase [34]. We first validated the efficiency of the Ki67 antibody to identify proliferating cells with our flow cytometry protocol by using low density plated, highly proliferating fibroblasts (56% Ki67 positive) and comparing them to high density plated, contact inhibited fibroblasts (11% Ki67 positive, Figure S2). Freshly isolated CPCs from neonatal and adult mice were stained with Ki67-specific antibody and the fraction of proliferating cells was calculated by determining the number of Ki67-positive CPCs over the total number of CPCs (Figure 4A). The adult CPCs (5.5±0.7% Ki67 positive CPCs) showed nearly a six-fold decrease in the Ki67-positive cells compared to the neonatal CPCs (30.7±4.8% Ki67 positive CPCs, n=6, P<0.05), suggesting that the neonatal CPCs have a greater proliferative activity than the adult CPCs (Figure 4B).
Figure 4. Proliferative capacity of neonatal and adult mouse CPCs.
(A) Flow cytometric analysis of neonatal and adult CPCs using proliferative marker, Ki67. The outliers (small dots) in the 10% contour plots represent less than 2% of the cell population. (B) The percentage of Ki67-positive cells in neonatal CPCs is higher than the adult CPCs as represented quantitatively. mN: mouse neonatal, mA: mouse adult and FL: fluorescence. Error bars represent standard error and *P<0.05.
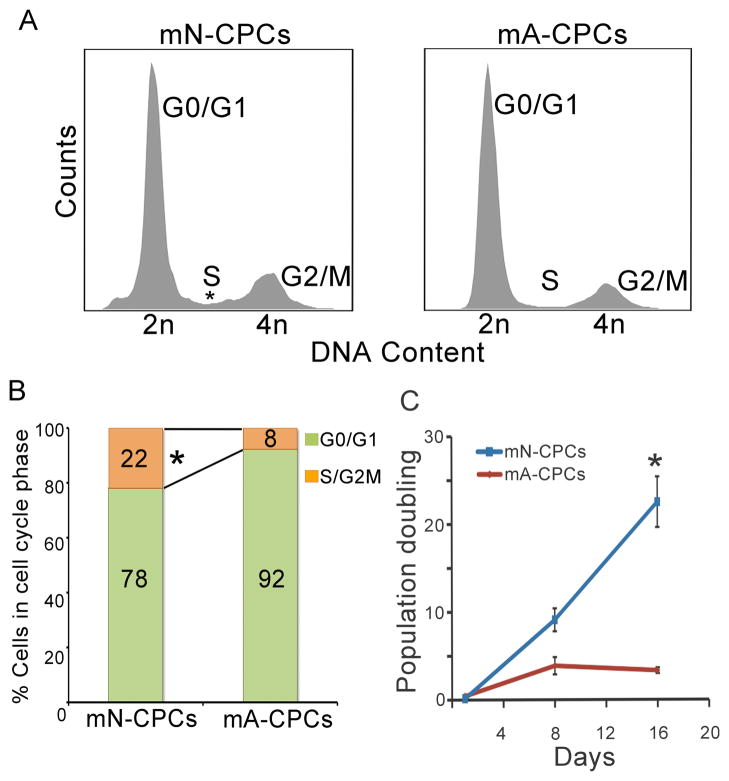
Cell cycle kinetics in mouse neonatal and adult CPCs
To further elucidate the proliferative differences in neonatal and adult CPCs, the relative DNA content and cell cycle phases were measured. Most adult CPCs were found to be in the G0/G1 phase of the cell cycle with 92±1% of them having 2N DNA content. The remainder of the cells (8±0.8%) were in the S/G2M phase with a >2N DNA content (Figure 5A). In contrast, 78±4.5% of the neonatal CPCs were in G0/G1 phase (2N DNA content) while 22±4.5% of the cells were in the S/G2/M phases with >2N DNA content (n=3, P<0.005). This represents a 2.5-fold increase in the percentage of neonatal CPCs in the S/G2/M phases when compared to adult CPCs (Figure 5B).
Figure 5. Cell cycle kinetics in mouse neonatal and adult CPCs.
(A) DNA histograms of neonatal and adult CPCs showing increased cells with a more than 2N DNA in neonatal CPCs. Representative results are shown. (B) Quantitative analysis of data represented in (A) showing G0/G1 (green) and S/G2M (orange) phases. (C) Population doubling of neonatal and adult CPCs, (n=6). mN: mouse neonatal, mA: mouse adult. Error bars represent standard error and *P<0.05.
To determine if the microRNA profile and molecular phenotyping translates into a difference in actual cell division, we examined the doubling time of mouse adult and neonatal CPCs in primary culture. The adult CPCs had 3.8±1 population doublings by 8 days in culture that did not change by 16 days (3.3±0.1 population doublings). In contrast, neonatal CPCs divided at a higher rate on day 8 (9.1±1.3 population doublings) and continued to divide at a higher rate on day 16 (22.5±2.8 population doublings, n=6, P<0.05, Figure 5C). There was a seven-fold increase in cell division of neonatal CPCs at day 16 when compared to the adult CPCs.
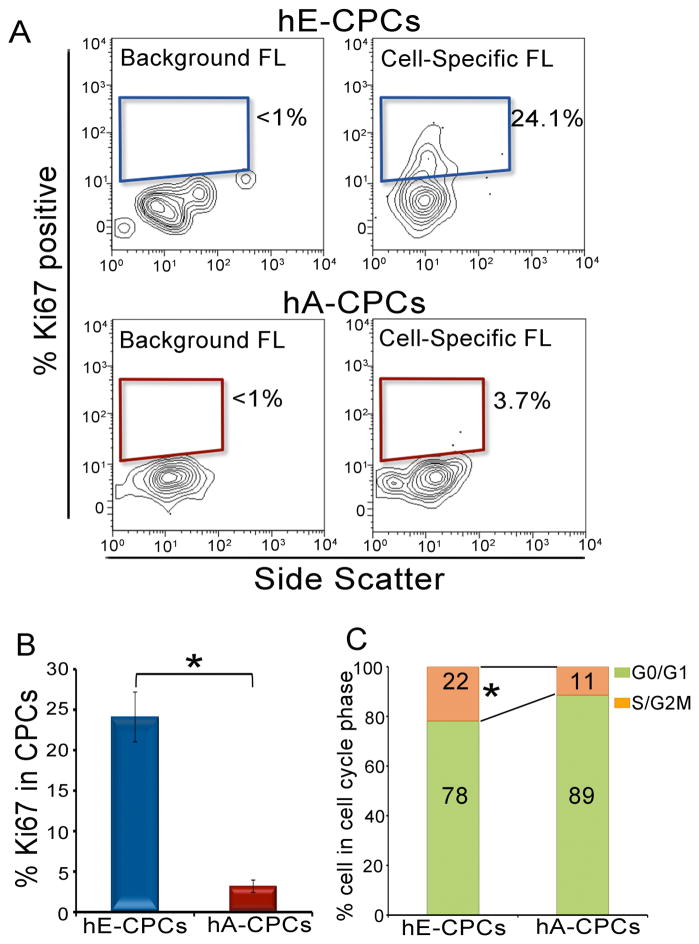
Proliferative capacity of human embryonic and adult CPCs
CPCs were isolated from first trimester (~95 d.p.c.) human embryonic and adult cardiac tissue to evaluate their proliferative phenotype. Nucleated cells were selected based on 7-AAD incorporation and NMCs were separated from the myocytes using troponin T antibody as shown in Figure 3. The proliferating cells were identified by their expression of Ki67 protein in the embryonic and adult human CPCs (Figure 6A). The human adult CPCs (3.7±0.6% Ki67 positive cells) showed a more than 6.5-fold decrease in the Ki67 positive cells when compared to embryonic CPCs (24.1±3% Ki67 positive CPCs; Figure 6B). This six-fold increase in the percentage of Ki67 positive cells is comparable to our finding in the mouse neonatal and adult CPCs supporting the notion that the greater proliferative activity of CPCs in early development occurs across species.
Figure 6. Proliferative capacity of embryonic and adult human CPCs.
(A) Flow cytometric analysis of embryonic and adult human CPCs using Ki67 as presented using 10% contour plots. The percentage of Ki67-positive cells in embryonic CPCs is more than 6.5-fold higher than that of adult CPCs. (B) Summary data of the percentages of Ki67-positive cells in embryonic and adult human CPCs. All the analyses were prepared in triplicates. (C) DNA content analysis of embryonic and adult CPCs demonstrating a two-fold increase in the S/G2-M phases in the embryonic CPCs when compared to the adult CPCs. hE: human embryonic, hA: human adult, FL: fluorescence. Error bars represent standard error and *P<0.05.
To further elucidate differences in the proliferative capacities between human embryonic and adult CPCs, the cell cycle phases were examined. Majority of human adult CPCs (89±0.4%) were in G0/G1 phase and only 11±0.4% of the cells were in S/G2-M phases. In contrast, 78±2% of the human embryonic CPCs were in the G0/G1 phase (2N DNA content) and up to 22±2% of the human embryonic CPCs were in the S/G2-M phases (Figure 6C). The two-fold increase in the percentage of cells in the S/G2-M phases is similar to findings from the mouse neonatal and adult CPCs supporting a similar phenotypic difference between human and mouse CPCs during development.
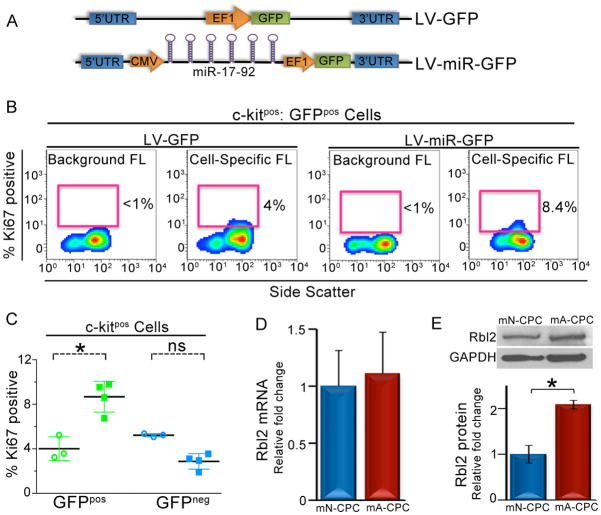
Functional analysis of microRNAs in adult mouse CPCs in vivo
To assess the gain-of-function of the differentially regulated microRNAs on cell proliferation, we tested the proliferative capacity of adult mouse CPCs over-expressing the miR-17-92 cluster under the CMV promoter and GFP under the EF1 promoter (LV-miR-GFP, Figure 7A). A lentiviral construct that expressed only GFP under the EF1 promoter (LV-GFP, Figure 7A) was used as the control. The entire miR-17 cluster (miR-17, -18, -19a, -19b, -20a, -92a) was utilized since the cluster is positioned within a 1kb region on the mouse chromosome 14 and is transcribed as a single product [35]. In addition, our Q-PCR analysis demonstrated a more than two-fold increase in the expression of all members of the miR-17-92 cluster in neonatal CPCs when compared to adult CPCs (Figure S3, P<0.05). Specifically, we hypothesize that the re-expression of miR-17-92 in adult CPCs will increase the proliferative capacity of the GFP-positive (GFPpos) CPCs when compared to the GFP-negative (GFPneg) CPCs from the same hearts.
Figure 7. Functional analysis of microRNAs in adult mouse CPCs in vivo.
A) Schematic representation of the LV-GFP control (top) and LV-miR-GFP (bottom) constructs. B) Flow cytometric analysis of the percentages of Ki67-positive GFPpos CPCs. The percentage of Ki67-positive cells in miR-17-92 over-expressed cells is 2-fold higher than that of control GFPpos CPCs. (C) Summary data of the percentages of Ki67-positive cells in GFPpos (green) and GFPneg (blue) CPCs from control (circles) and miR-17-92 over-expressed (squares) hearts. (D) Summary data of Q-PCR for Rbl2 mRNA from neonatal and adult CPCs (n=3). (E) Representative lanes of western blot assay and summary data (below) for Rbl2 and GAPDH (loading control) performed in neonatal and adult mouse CPCs (n=3). mN: neonatal, mA: adult, FL: fluorescence, ns: not significant, LV: lentivirus, GFP: green fluorescent protein. Error bars represent standard errors and *P<0.05.
Cell type-specific analysis allowed us to identify among the c-kitpos cells, the percentages of Ki67-positivity in GFPpos and GFPneg CPCs in control and miR-17-92 transduced hearts (Figure 7B). The GFPpos CPCs isolated from miR-17-92 over-expressing hearts demonstrated a two-fold increase in the Ki67-positivity compared to the GFPpos CPCs from control hearts (8.7±0.9% vs. 4±0.6% of Ki67 positive CPCs, n=3, P<0.05, Figure 7B and C). In contrast, the GFPneg CPCs isolated from microRNA over-expressing hearts (3.2±0.1% Ki67 positive CPCs) and control hearts (5.2±0.08% Ki67 positive CPCs, n=3, P=NS) exhibited similar level of Ki67 expression to the GFPpos CPCs from control hearts. Taken together, the data suggest that the over-expression of miR-17-92 results in an increased proliferative activity in adult CPCs.
Analysis of the cell cycle protein retinoblastoma-like 2 (Rbl2/p130) in neonatal and adult mouse CPCs
To directly validate the predicted microRNA target proteins involved in the observed differences in the proliferation between neonatal and adult CPCs, the mRNA and protein expression levels of an anti-proliferative cell cycle protein retinoblastoma-like 2 (Rbl2/p130) were analyzed in neonatal and adult mouse CPCs. Rbl2 was chosen as an example of a candidate protein to test because its 3’UTR region predicted an interaction with two microRNAs (miR-17, -20a, Figure S4) which were expressed at a higher levels in the neonatal mouse CPCs (Figure S3). The transcript levels of Rbl2 were found to be similar in both neonatal and adult CPCs (Figure 7D). However, the protein expression levels of Rbl2 in adult CPCs were two fold higher than in neonatal CPCs (Figure 7E) consistent with a possible post-transcriptional regulation of the protein by microRNAs.
DISCUSSION
In the present study, we sought to identify regulated microRNAs in mouse CPCs during development and determined their relationship to phenotypic variations occurring in the postnatal heart. A cohort of 45 microRNAs was differentially regulated between adult myocytes and CPC from both neonatal and adult mouse hearts. Of these, only eight microRNAs were uniquely regulated between neonatal and adult CPCs (Figure 2) using a stringent criteria (≥5-fold change, P<0.05) and validated by Q-PCR. We utilize three bioinformatic strategies, by weighting the number of downstream protein targets regulating different cellular processes, to predict cellular phenotypes that may be regulated in CPCs during development. This prediction was independent of the direction or magnitude of changes in microRNAs once the microRNAs had met the threshold criteria. Using the above strategy, we identify a cohort of eight microRNAs that is regulated during the post-natal development of CPCs that may predict the observed differences in their cellular proliferation. The difference in proliferative potential predicted by the microRNA analysis of mouse neonatal and adult CPCs while in vivo is recapitulated in human embryonic and adult CPC while in vivo, and in mouse neonatal and adult CPCs in culture.
To test the functional relationship between the regulated microRNA cohort and the predicted changes in CPC proliferation, we tested the gain-of-function of the miR-17-92 cluster. These results suggest a role of this cluster in regulating the exiting from cell cycle of CPCs in the adult hearts. Furthermore, the expression pattern of Rbl2, a cell cycle regulatory protein, predicted to be regulated by the differentially expressed microRNA cohort (Figure 7) was analyzed in neonatal and adult CPCs. Together, these results show an experimental strategy that identifies cohorts of differentially expressed miRNAs that predict cellular phenotypes of CPCs and functional regulators of these phenotypes.
This work provides an additional approach to determine the putative regulatory functions of microRNA cohorts in CPCs biology. In this study, we focused our analysis in developmental stages. It is anticipated that similar analysis in the future would lead to the discovery of microRNA cohorts in the injured heart that may regulate CPC proliferation, and their manipulation may translate into a greater cardiac regeneration and/or repair.
A hallmark of microRNAs is their tremendous potential to target a large number of genes, often in common pathways. With an estimated 60% of human mRNA having conserved microRNA targets, microRNAs represent one of the key regulators and informative markers of a set of genes controlling different cellular processes rather than single genes [36, 37]. To comprehensively evaluate the influence of microRNAs on multiple proteins by the integration of multiple downstream genes, the global evaluation of microRNA in the signaling cascade becomes essential [38]. Similarly, we have utilized the overall assessment of the microRNA profile in establishing the possible roles of the differentially expressed microRNAs in the proliferative phenotype of neonatal and adult CPCs.
The miR-17-92 cluster either individually or synergistically has been shown to promote cellular proliferation in multiple tissues [39–41]. However, the functions of the miR-17-92 cluster in the heart are not completely understood. The reduced heart weight and ventricular septal defects in mice deficient in miR-17-92 cluster supports an important role of miR-17-92 cluster in the prenatal developing myocardium [42, 43]. Individual microRNAs from this cluster target a complex network of proliferative cell cycle proteins and modulate proteins that control the transition from G1 to S phase thereby promoting proliferation [35]. Our results suggest a role for this microRNA cluster in cell proliferation of CPCs since its re-expression contributes to a partial re-entry into the cell cycle. Future experiments into the re-expression of this microRNA cluster and others after injury may further elucidate the function of microRNAs in regulating regeneration in the adult myocardium.
A large network of interacting proteins including Rbl2 (an anti-proliferative cell cycle protein) and the associated E2F transcription factors orchestrates the regulation of cell proliferation. Rbl2 belongs to the retinoblastoma pocket protein family and has been previously validated as a target of miR-17 and miR-20a using a variety of different reporter gene assays [35, 41, 44, 45]. Our analysis shows that all the members of the miR-17 cluster are expressed at least 2-fold higher in neonatal CPCs. The transcript levels of Rbl2 remain constant in both neonatal and adult CPCs while the adult CPCs show a two-fold increase in Rbl2 protein when compared to the neonatal CPCs. Taken together, our data suggest that in CPCs, the miR-17 cluster may regulate cell proliferation, at least in part, through its action on one of the known target proteins, Rbl2. Nonetheless, the effect of over-expression of miR-17 cluster on Rbl2 protein level in CPCs was not directly demonstrated in our study due to the limited number of CPCs inherent in our in vivo gain-of-function study design.
Future Directions
Our study demonstrated differential microRNA profiles in murine CPCs that predicted the differences in proliferative capacities in these cells. However, the analyses of human CPCs were limited to the proliferative phenotype of CPCs due to limited amount of tissues. Nonetheless, previous studies have demonstrated that many core cellular processes and microRNA orthologs are conserved between mouse and human [46]. Moreover, the functions of mature microRNAs are phylogenetically conserved between species for example, the muscle-specificity of miR-1 in Drosophila, mouse and humans [26, 47]. Future experiments are required to further evaluate microRNAs regulation in human CPCs. Moreover, previous studies have documented significant differences in the differentiation capacity between neonatal and adult CPCs [18]. The possible roles of microRNAs in these differences are not yet known and will require future studies.
Recent identification of CPCs in adult hearts provides an exciting and promising new paradigm for cardiac regenerative therapy. However, with the loss of about one billion cardiomyocytes in a typical myocardial infarction in patients [48], it is vital to ensure a sufficient number of CPCs for transplantation. It has previously been demonstrated that despite the increase in the CPCs in the failing human hearts of transplant recipients, the repair processes remained limited because patients ultimately required transplantation [49]. The decline in the number and proliferative capacity of CPCs during development from neonate to adult stages [16, 17, 19] may limit the use of CPCs for an efficient cell-based therapy. Therefore, insights into the proliferative capacity may provide new strategies to maintain the cardiac organ homeostasis and repair of the adult heart.
Supplementary Material
Highlights.
MicroRNAs profiles in cardiac progenitor cells (CPCs) are distinct developmentally.
The predicted protein targets are predominantly involved in cellular proliferation.
The proliferative capacity of neonatal CPCs is higher than adult CPCs.
Over-expression of miR-17-92 increases the proliferation in adult CPCs.
A cohort of microRNAs may predict differences in cellular proliferation in CPCs.
Acknowledgments
We are grateful to Dr. Ebenezer Yamoah laboratory members for their suggestions. We thank Carol Oxford for assistance with FACS. The MF20 monoclonal antibody developed by Donald A. Fischman was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA-52242.
FUNDING SOURCES
Supported by NIH/HL85727 and HL85844 and the VA Merit Review Grant (NC), Howard Hughes Medical Institute Med-into-Grad Training Program to UC Davis (PS), American Heart Association Western States Affiliate Predoctoral Fellowship Award (PS), Fellow-to-Faculty Award from Sarnoff Cardiovascular Research Foundation (JEL), American Heart Association Western States Affiliate Beginning Grant-in-Aid (JEL), NIH/HL105194 and California Institute of Regenerative Medicine RB2-01512 (HVC).
Footnotes
DISCLOSURE
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, et al. Heart disease and stroke statistics--2011 update: a report from the American Heart Association. Circulation. 2011 Feb 1;123(4):e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laflamme MA, Zbinden S, Epstein SE, Murry CE. Cell-Based Therapy for Myocardial Ischemia and Infarction: Pathophysiological Mechanisms. Annual Rev of Pathology. 2007;2:307–39. doi: 10.1146/annurev.pathol.2.010506.092038. [DOI] [PubMed] [Google Scholar]
- 3.Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annual Rev of Cell and Dev Biol. 2010;26:667–87. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singla DK, Lyons GE, Kamp TJ. Transplanted embryonic stem cells following mouse myocardial infarction inhibit apoptosis and cardiac remodeling. Amer Jour of Physiol. 2007;293(2):H1308–H214. doi: 10.1152/ajpheart.01277.2006. [DOI] [PubMed] [Google Scholar]
- 5.Layland KS, Strem BM, Jordan MC, Deemedio MT, Hedrick MH, Roos KP, et al. Adipose tissue-derived cells improve cardiac function following myocardial infarction. J Surg Res. 2009;153(2):217–23. doi: 10.1016/j.jss.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wollert KC, Drexler H. Clinical Applications of Stem Cells for the Heart. Circulation research. 2005;96:151–63. doi: 10.1161/01.RES.0000155333.69009.63. [DOI] [PubMed] [Google Scholar]
- 7.Smart N, Riley PR. The Stem cell movement. Circ Res. 2008;102(10):1155–68. doi: 10.1161/CIRCRESAHA.108.175158. [DOI] [PubMed] [Google Scholar]
- 8.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003 Sep 19;114(6):763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 9.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004 Oct 29;95(9):911–21. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 10.Bearzi C. Human cardiac stem cells. PNAS. 2007;104(35):14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajstura J, Urbanek K, Rota M, Bearzi C, Hosoda T, Bolli R, et al. Cardiac stem cells and myocardial disease. J Mol Cell Cardiol. 2008 Oct;45(4):505–13. doi: 10.1016/j.yjmcc.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Davis DR, Zhang Y, Smith RR, Cheng K, Terrovitis J, Malliaras K, et al. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 2009;4(9):e7195. doi: 10.1371/journal.pone.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamuleau SA, Vrijsen KR, Rokosh DG, Tang XL, Piek JJ, Bolli R. Cell therapy for ischaemic heart disease: focus on the role of resident cardiac stem cells. Neth Heart J. 2009 May;17(5):199–207. doi: 10.1007/BF03086247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urbanek K, Torella D, Sheikh F, De Angelis A, Nurzynska D, Silvestri F, et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci U S A. 2005 Jun 14;102(24):8692–7. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Lee A, Huang M, Chun H, Chung J, Chu P, et al. Imaging survival and function of transplanted cardiac resident stem cells. J Am Coll Cardiol. 2009 Apr 7;53(14):1229–40. doi: 10.1016/j.jacc.2008.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir G, Ma X, Reddy VM, Hanley FL, Reinhartz O, Ramamoorthy C, et al. Dynamics of human myocardial progenitor cell populations in the neonatal period. Ann Thorac Surg. 2008 Oct;86(4):1311–9. doi: 10.1016/j.athoracsur.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 17.Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci U S A. 2009 Feb 10;106(6):1808–13. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic Potential of C-Kit–Expressing Cells Derived From Neonatal and Adult Mouse Hearts. Circ Research. 2010;121:1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra R, Vijayan K, Colletti EJ, Harrington DA, Matthiesen TS, Simpson D, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011 Feb 1;123(4):364–73. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Cartew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435(16):974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 21.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004 Jun 15;270(2):488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, Abilez OJ, et al. Dynamic MicroRNA Expression Programs During Cardiac Differentiation of Human Embryonic Stem Cells. Circulation. 2010;3:426–35. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bostjancic E, Zidar N, Stajer D, Glavac D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010;115(3):163–9. doi: 10.1159/000268088. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 25.Chen JF, Mandel EM, Thomson M, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Ransom JF, Li A, Vedantham V, Drehle MV, Muth AN, et al. Dysregulation of Cardiogenesis, cardiac conduction and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Timofeyev V, Tuteja D, Xu D, Lu L, Zhang Q, et al. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. The Journal of Physiology. 2009;587(5):1087–100. doi: 10.1113/jphysiol.2008.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikuchi K, McDonald AD, Sasano T, Donahue JK. Targeted modification of atrial electrophysiology by homogeneous transmural atrial gene transfer. Circulation. 2005 Jan 25;111(3):264–70. doi: 10.1161/01.CIR.0000153338.47507.83. [DOI] [PubMed] [Google Scholar]
- 29.Cottage CT, Bailey B, Fischer KM, Avitable D, Collins B, Tuck S, et al. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ Res. 2010 Mar 19;106(5):891–901. doi: 10.1161/CIRCRESAHA.109.208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rooij E. The art of microRNA Research. Circ Res. 2011 Jan 21;108(2):219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 31.Dahlquist KD. Using GenMAPP and MAPPFinder to view microarray data on biological pathways and identify global trends in the data. Curr Protoc Bioinformatics. 2004 May;Chapter 7(Unit 7):5. doi: 10.1002/0471250953.bi0705s05. [DOI] [PubMed] [Google Scholar]
- 32.Lopez JE, Myagmar BE, Swigart PM, Montgomery MD, Haynam S, Bigos M, et al. {beta}-Myosin Heavy Chain Is Induced by Pressure Overload in a Minor Subpopulation of Smaller Mouse Cardiac Myocytes. Circ Res. 2011 Sep 2;109(6):629–38. doi: 10.1161/CIRCRESAHA.111.243410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epting CL, Lopez JE, Pedersen A, Brown C, Spitz P, Ursell PC, et al. Stem cell antigen-1 regulates the tempo of muscle repair through effects on proliferation of alpha7 integrin-expressing myoblasts. Exp Cell Res. 2008 Mar 10;314(5):1125–35. doi: 10.1016/j.yexcr.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landberg G, Tan EM, Roos G. Flow cytometric multiparameter analysis of proliferating cell nuclear antigen/cyclin and Ki-67 antigen: a new view of the cell cycle. Exp Cell Res. 1990 Mar;187(1):111–8. doi: 10.1016/0014-4827(90)90124-s. [DOI] [PubMed] [Google Scholar]
- 35.Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, et al. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9(8):R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009 Jan;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006 Oct 9;25(46):6220–7. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 38.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010 Apr;11(4):252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 39.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005 Nov 1;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 40.Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, et al. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9(8):R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, et al. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 2011;6(1):e16138. doi: 10.1371/journal.pone.0016138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009 Aug;11(8):1031–8. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 43.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008 Mar 7;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007 Oct 15;310(2):442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2889–94. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie W, Rajasekhar M, Flamant S, Rasko JE. Conserved expression patterns predict microRNA targets. PLoS Comput Biol. 2009 Sep;5(9):e1000513. doi: 10.1371/journal.pcbi.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005 Dec 27;102(52):18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinecke H, Minami E, Zhu WZ, Laflamme MA. Cardiogenic differentiation and transdifferentiation of progenitor cells. Circ Res. 2008 Nov 7;103(10):1058–71. doi: 10.1161/CIRCRESAHA.108.180588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo H, Jaleel N, Kumarapeli A, Berretta RM, Bratinov G, Shan X, et al. Increased Cardiac Myocyte Progenitors in Failing Human Hearts. Circulation. 2008;118:649–57. doi: 10.1161/CIRCULATIONAHA.107.761031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.